Abstract
In the past 10 years, transcriptome and proteome analyses have provided valuable data on global gene expression and cell functional networks. However, when integrated, these analyses revealed partial correlations between mRNA expression levels and protein abundance thus suggesting that post-transcriptional regulations may be in part responsible for this discrepancy. In the present work, we report the development of a functional, integrated, and quantitative method to measure post-transcriptional regulations that we named FunREG. This method enables (i) quantitative measure of post-transcriptional regulations mediated by selected 3′-untranslated regions and exogenous small interfering-RNA or micro-RNAs and (ii) comparison of these regulatory processes in physiologically relevant systems (e.g. cancer versus primary untransformed cells). We applied FunREG to the study of liver cancer, and we demonstrate for the first time the differential regulatory mechanisms controlling gene expression at a post-transcriptional level in normal and tumoral hepatic cells. As an example, translation efficiency mediated by heparin-binding epidermal growth factor 3′-untranslated region was increased 3-fold in liver cancer cells compared with normal hepatocytes, whereas stability of an mRNA containing a portion of Cyclin D1 3′-untranslated region was increased more than 2-fold in HepG2 cells compared with normal hepatocytes. Consequently we believe that the method presented herein may become an important tool in fundamental and medical research. This approach is convenient and easy to perform, accessible to any investigator, and should be adaptable to a large number of cell type, functional and chemical screens, as well as genome scale analyses. Finally FunREG may represent a helpful tool to reconcile transcriptome and proteome data.
One of the current challenges in modern biology aims at understanding how cells work at a genomic scale and how particular cellular contexts (environment, differentiation, transformation, etc.) influence global gene expression, reorganize protein networks, and consequently condition cell fate. To this end, mRNA and protein expression levels have been widely studied using large scale transcriptomics and proteomics approaches. Besides the amount of valuable information generated by such analyses, data from combined transcriptomics and proteomics analyses revealed discrepancies in the correlation between mRNA expression levels and protein abundance (1–5). For example, a comparative proteomics and transcriptomics profiling of hepatocellular carcinoma (HCC),1 a primitive liver cancer, showed that the abundance of some proteins, whose expression varies between HCC and the adjacent non-tumoral tissue, was poorly correlated to mRNA expression changes (5). Elsewhere correlation between transcriptomics and proteomics comparisons in developing embryonic stem cells showed that significant changes in protein amounts were observed in the absence of any variation of mRNA abundance (2). These findings therefore limit the usefulness of mRNA expression values as an index of genomic expression as proteins, the causative effectors, are more likely to play this role. The reasons for the observed discrepancies between mRNA and protein expression levels can be multiple (4). One reason could be the selective degradation of proteins by the proteasome. On this point, Yen et al. (6) developed an approach for proteome scale protein turnover analysis using a lentiviral and fluorescent reporter strategy. They remarkably demonstrated that more than 80% of the cellular proteins are degraded through a proteasome-dependent pathway. On the other hand, the role of regulatory events occurring at the post-transcriptional level may certainly be accountable for transient and adaptable expression of mRNA translation products (7–9).
Post-transcriptional regulations are key mechanisms in the control of gene expression. Messenger RNA turnover and translation are of particular importance as these features directly govern the amount of protein being produced by the cell (10, 11). Messenger RNAs are post-transcriptionally informative molecules that contain many cis-acting RNA sequences (cisARSs) located throughout the transcripts. Specialized trans-regulatory factors (transRFs) associate with cisARSs to produce specific biological effects (11, 12). The nature of these associations determines the fate of mRNAs (transport, degradation, storage, etc.) as well as their translation efficiency (10–14). The number of transRFs, previously thought to be exclusively of proteinaceous origin, was recently revised with the discovery that natural ribonucleic factors (i.e. micro-RNAs (miRNAs)) could control gene expression at a post-transcriptional level (9, 15).
Most cisARSs have been described as located in the 3′-untranslated region (UTR) of transcripts (11, 16). Among those, AU-rich elements (AREs) are present in short lived mRNAs (11, 17, 18), and miRNA-binding sites (9, 15) currently represent the center of interest for a significant part of the scientific and medical communities (9, 15, 19, 20). Both AREs and miRNA-binding sites control the expression of many genes involved in transient and adaptable cellular processes (7, 9, 11, 15, 17, 18, 21–23). With little exceptions, they act as apparent repressors of gene expression as they induce mRNA degradation, translational repression, or both. The functional capacities of these cisARSs depend on specific transRFs, namely the ARE-binding proteins (ARE-BPs) (11, 17) and the miRNAs (in association with Argonaute) (9, 15, 24), respectively. Interestingly ARE-BPs and miRNAs can collaborate to control mRNA decay and translation suggesting complex interrelations between these two post-transcriptional mechanisms (9, 11, 15). Alternatively a number of ARE-BPs, exemplified by ELAVL1/HuR (embryonic lethal abnormal vision-like protein 1/Hu antigen R), inhibit ARE- or miRNA-mediated regulations by competing with other ARE-BPs or miRNAs for binding to cisARS (9, 15, 17, 19).
Most of the strategies developed to study post-transcriptional mechanisms in mammalian cells are based on the expression of reporter transgenes, either luciferase or enhanced green fluorescent protein (eGFP), transiently transfected into target cells (23, 25). Even if stable clones or clone populations can be established, this requires several weeks of antibiotic-based selection and clone amplification. Moreover the transgene expression is insertion-dependent and therefore clone-specific. Finally the above mentioned approaches do not allow comparative post-transcriptional studies using normal and pathological cells because primary cells in culture are hardly transfectable using methods with non-virus-based vector. Finally the choice of specific reporter genes may sometimes induce experimental biases as illustrated by the popular firefly luciferase reporter gene that contains a cryptic promoter sequence in its coding region (26).
In this study we developed lentivirus-based delivery of an eGFP reporter system combined with real time quantitative PCR to study post-transcriptional mechanisms. This allowed us to (i) compare the function of selected cisARSs in normal and tumoral cells and (ii) build a novel experimental pipeline (named “functional, integrated, and quantitative method to measure post-transcriptional regulations” (FunREG)) that circumvents the problems raised when using transfection-based approaches. We applied FunREG to the study of post-transcriptional events taking place in the course of liver carcinogenesis and used primary human hepatocytes as well as human primary liver cancer-derived HuH7 and HepG2 cells. This experimental system represents a model of choice not only because primary liver cancer represents one of the deadliest cancer in the world but also because significant discrepancies between transcriptomes and proteomes have been reported using hepatic cells from different sources or contexts (i.e. cancerous versus non-cancerous hepatic tissue) (5, 27–29). This analysis revealed the differential post-transcriptional regulatory mechanisms controlling the expression of heparin-binding epidermal growth factor (HBEGF) and Cyclin D1 (CCND1) in cancer versus primary cells. The FunREG approach described in this study may consequently become a universal tool (i) to study post-transcriptional regulations in a broad spectrum of cellular models, (ii) to compare post-transcriptional mechanisms in normal and pathological contexts, and (iii) to provide some elements of the answer to address the existing discrepancy between mRNA and protein expression levels reported in the literature (1–5, 27–29).
EXPERIMENTAL PROCEDURES
Lentiviral Plasmid Constructs
The pTRIPdeltaU3-EF1α-GFP (pTRIP-eGFP) lentiviral plasmid was a gift from Pierre Charneau (Institut Pasteur, Paris, France) (30). The eGFP expression is driven by the constitutive EF1α promoter. The pTRIP-eGFP and pTRE-eGFP plasmids with the different 3′-UTRs were constructed as described in the supplemental data.
Cell Lines, Primary Hepatocyte Cultures, and siRNA Transfection
The adenocarcinoma HeLa, osteosarcoma MG63, HCC-derived HuH7, and hepatoblastoma HepG2 cell lines were grown in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% FCS and penicillin/streptomycin antibiotics. Fresh primary human hepatocytes (Biopredic SA, Rennes, France) were grown following the provider's instructions. The miRNAs were from Perbio Sciences. The small interfering-RNAs (siRNAs) anti-eGFP and anti-luciferase were from Applied Biosystems and Eurogentec, respectively. Small RNAs were transferred in the target cells using Lipofectamine RNAiMAX (Invitrogen).
Lentiviral Production, Titration, and Cell Transduction
Production and titration of infectious lentiviral particles were as detailed previously (31) and in the supplemental data. Lentiviral particles were added to the target cells and incubated for 24 h. Then the cells were washed twice in PBS and grown in the presence of medium for 6 days before experimental use. Biosafety considerations when using and handling lentivirus as well as safety procedures and policies can be found in the literature (32–35).
Flow Cytometry Analyses
One week after transduction, cells were washed in PBS, detached with trypsin/EDTA, collected, and analyzed by flow cytometry (FCM) using an EPICS XL flow cytometer (Coulter) and the Expo 32 analysis software. Ten thousand cellular events were gated using forward and side scatter settings. Cells transduced with the non-fluorescent reporter expressing plasmid pTRIP-0 were used as negative control to gate the eGFP-positive cell populations (% eGFP+ (CT), arbitrarily fixed at 2%) and to measure the basal mean fluorescence intensity (MFI CT) of the whole cell population. The eGFP expression (called “eGFP protein”) was determined in each condition by measuring the MFI of the whole transduced cell population (MFI test) by comparison with the control cell population as follows: ((MFI test − MFI CT)/MFI CT). The percentage of the eGFP-positive cells (% eGFP+ cells) obtained from data plots was obtained as follows: (% eGFP+ (Test) − % eGFP+ (CT)).
Real Time Quantitative PCR and RT-PCR
Total DNA and RNA were extracted from cells using the Nucleospin Tissue and Nucleospin II RNA kits (Macherey-Nagel), respectively. Real time quantitative PCR amplifications were performed in 25-μl multiplex PCRs containing 1× iQTM SYBR® Green Supermix (Bio-Rad), two primers (see supplemental Table 2 for details), and either 50 ng of total DNA or cDNAs from 100 ng of reverse transcribed total RNA as described in the supplemental data. The albumin gene, RPLP0 mRNA, or α-tubulin mRNA served as internal control for normalization. Subsequent data analyses were performed using the Mx4000 Multiplex Quantitative PCR System equipped with Version 4.2 software (Stratagene).
Statistical Analyses
Data are represented as mean ± S.D. from three independent experiments. The non-parametric Mann-Whitney test was used for the comparison of two values within small samples. One-way analysis of variance was used for the comparison of multiple means with an α level of 0.05 and was followed by the Dunnett multiple comparisons post-test if a significant F ratio was obtained. p < 0.05 was considered statistically significant. All analyses were done using GraphPad Prism 5.0.
RESULTS
In an attempt to address the problems mentioned in the Introduction, we developed a method allowing for (i) measuring post-transcriptional regulations in cellulo and (ii) comparing those events in normal and disease-derived human cells. To this end, lentivirus particles were used to deliver transgenes of interest in target cells. Indeed retroviruses have been shown to efficiently transduce dividing or non-dividing human cells from different tissues including the liver. Moreover integration of the transgene into the genome of the target cell leads to stable expression (35–38). To detect the expression of the transgene of interest, the eGFP reporter gene was used because its expression level can be analyzed in live cells by FCM (23).
FunREG
FunREG is schematically summarized in Fig. 1. FunREG integrates three major steps as follows.
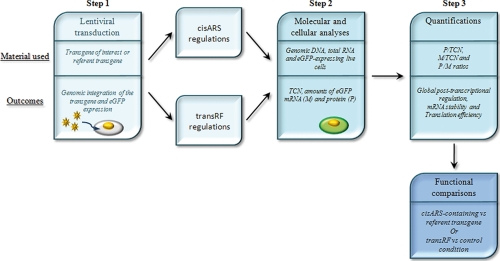
Fig. 1.
FunREG. A schematic representation of the FunREG experimental pipeline is shown. The three major steps are depicted by boxes as indicated. Boxes are recognized by a title (top panel), and some, corresponding to the three major steps of the method, contain the material used (middle panel) and the biological or experimental outcomes (bottom panel), as indicated. Step 1, target cells are transduced with lentiviral particles (as illustrated) containing either the transgene of interest or the reference. Following transduction, the transgenes are integrated into the host genome, and eGFP is expressed. At this stage, the objective is either to study the functioning of known or putative cisARSs as post-transcriptional elements in selected cells, evaluate the involvement of transRFs in a cisARS-mediated post-transcriptional regulation, or perform both analyses in parallel (cisARS and transRF regulations boxes as indicated). Step 2, molecular and cellular analyses are done by measuring the “TCN” and the amounts of “M” by qPCR and qRT-PCR using genomic DNA and total RNA extracted from the whole transduced cell population, respectively. The amount of “P” is measured by flow cytometry using eGFP-expressing live cells (as illustrated). Step 3, the three ratios, P/TCN, M/TCN, and P/M, are calculated. These ratios are indicative of the global post-transcriptional regulation, the relative mRNA stability, and the relative translation efficiency, respectively. Finally the values obtained with every transgene or in every condition are compared.
Step 1: Lentiviral Transduction
Lentiviral particles containing either the transgene of interest or the reference transgene are used to transduce cells in culture. The transgenes are integrated into the host genome and followed by eGFP (selected as reporter) expression. Transgene selection can be based on the study of (i) functional ability of known or putative cisARSs in selected cells, (ii) transRF involvement in cisARS-mediated post-transcriptional regulation, or (iii) both configurations (Fig. 1, “cisARS and transRF regulations” boxes).
Step 2: Molecular and Cellular Analyses
Transgene copy number (TCN) and eGFP mRNA expression levels are measured by qPCR and qRT-PCR using, respectively, genomic DNA and reverse transcribed total RNA extracted from transduced cells. eGFP protein expression is then measured by FCM using eGFP-expressing live cells (Fig. 1, green cell).
Step 3: Quantification
The ratios, respectively indicating the global post-transcriptional regulation, the mRNA stability, and the translation efficiency, are calculated. At last, the values obtained with each transgene or in each condition are compared, allowing for functional interpretation of the biological processes (mRNA stability, translation efficiency, or both) controlling the post-transcriptional regulation.
FunREG Quality Control
To ensure FunREG accuracy and specificity, several points had to be ascertained. At first, we made sure that the expression of a given transgene should be directly proportional to the number of copies of this transgene in the selected cell types. Provided that this is the case, any difference in the expression of this transgene consecutive to the insertion of a specific regulatory sequence (i.e. a 5′- or 3′-UTR) upstream or downstream the eGFP ORF is expected to originate from a post-transcriptional event and not from a transcriptional or post-translational regulation. Indeed it is unlikely that such an insertion would influence the transcription efficiency and eGFP protein turnover from the transgene. Second, we ensured that transgene expression should be stable as a function of time. The eGFP-GLO transgene, which contains the rabbit β-globin 3′-UTR inserted downstream of the eGFP coding region (Fig. 3A, top panel), was used to prove these points because this 3′-UTR is generally used as a control in post-transcriptional studies (23, 39–41). Finally we assessed by cell sorting that the level of eGFP proteins in cells had no influence on the regulation of its level.
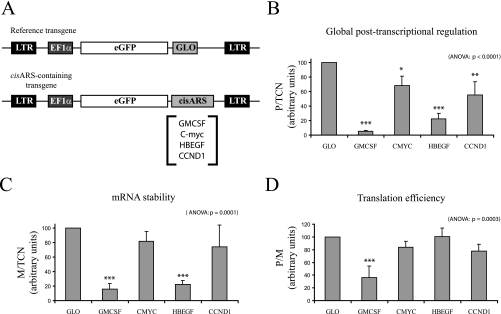
Fig. 3.
Measurement of 3′-UTR-mediated post-transcriptional regulations by FunREG. A, schematic representations of the different transgenes used in this study. The referent eGFP-GLO transgene is shown in the top panel. The transgenes containing selected cisARSs are shown in the bottom panel. LTR, long terminal repeat. B–D, HuH7 cells were transduced once with lentiviral particles expressing the indicated transgene. After 1 week, the transgene copy number and eGFP mRNA amount were determined by qPCR and qRT-PCR using genomic DNA or reverse transcribed total RNA extracted from each transduced cell population, respectively. The eGFP protein amount was determined by flow cytometry on each transduced cell population. B, global post-transcriptional regulation. C, mRNA stability. D, translation efficiency. In this and the following figures, the different ratios (in arbitrary units) are shown on the y axes, error bars represent S.D., and analysis of variance (ANOVA) p values are indicated at the top right of the figure (n = 3). Significant variations using Dunnett post-test are represented by asterisks above the corresponding bar when comparing the referent (or control) condition and the indicated condition or above the line when comparing the two indicated conditions: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
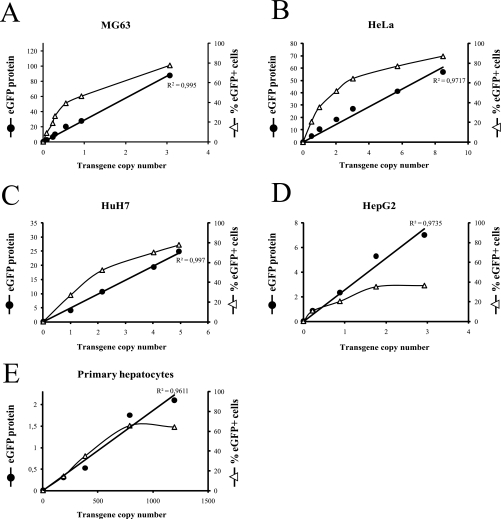
Five cells types (MG63, HeLa, HuH7, HepG2, and fresh primary hepatocytes) were transduced with increasing multiplicities of infection (m.o.i.) of eGFP-GLO-expressing lentiviral particles. m.o.i. ranging from 0 to 5 were used to transduce the four cell lines. To transduce the primary hepatocytes and obtain a percentage of eGFP+ cells similar to that obtained with the four cell lines, m.o.i. ranging from 0 to 75 were used. One week after transduction, cells expressing eGFP-GLO were analyzed by FCM in each case (see corresponding histogram in supplemental Fig. S1). The MFI, representing the eGFP protein expression (eGFP protein), was measured on the whole cell population, and the percentage of eGFP-positive cells (supplemental Fig. S1) was obtained by comparison with the eGFP-negative cell population (transduced with pTRIP-0 lentiviral particles). Following FCM analysis, genomic DNA was extracted from the different transduced cell populations. Then the average number of lentiviral transgene copies per cell (TCN) was measured by qPCR in each condition using the two copies of the albumin gene as reference. As shown in Fig. 2, A–E (closed circles), eGFP protein expression fully correlated with the TCN in the five cell types tested, including hepatocytes (linear regression curves; R2 > 0.96). However, eGFP expression was not identical in the different cell types tested (Fig. 2, A–E, compare eGFP protein expression on the left y axes) supporting the idea that the global eGFP protein expression is cell-specific. This “eGFP protein expression:TCN” correlation was also obtained in independent experiments using HuH7 cells expressing eGFP transgenes under the control of a selected 3′-UTR (see supplemental Fig. S2). On the other hand, the percentage of the eGFP+ cells (Fig. 2, A–E, open triangles) did show some correlation with the number of integrated copies when using low m.o.i. However, these correlations were rapidly lost when m.o.i. increased (Fig. 2, A–E, open triangles). These results support the idea that some cells of the population integrated more than one copy of eGFP-GLO transgene, whereas others remained non-transduced. From these experiments, we concluded that in a given cell type eGFP protein expression was directly proportional to the number of lentiviral transgene copies. Therefore, when comparing the expression of cisARS-containing transgenes with that of a reference transgene (i.e. globin), any variation in eGFP expression can be expected to originate exclusively from cisARS-dependent post-transcriptional regulations using data normalized to the number of transgene copies. As in a given cell type post-transcriptional mechanisms are influenced by the abundance of a given mRNA (which itself depends on its intrinsic stability) and its capacity to undergo translation, the average quantity of eGFP mRNA was also measured by qRT-PCR using total RNA extracted from each transduced cell population.
Fig. 2.
EGFP protein expression correlates with the transgene copy number in the different cell types. MG63 (A), HeLa (B), HuH7 (C), and HepG2 (D) cells and fresh primary hepatocytes (E) were transduced once with increasing amounts of eGFP-GLO-expressing lentiviral particles. After 1 week, eGFP protein expression (MFI) and percentage of eGFP+ cells were determined in each condition by flow cytometry. TCN was determined by qPCR using genomic DNA and normalizing to albumin gene. The curves represent the “percentage of eGFP+ cells:transgene copy number” correlation (▵ curve) or the “eGFP protein:transgene copy number” correlation (● curve). The R2 corresponding to eGFP protein:transgene copy number correlation curve is as indicated.
In a second set of experiments, we monitored the expression of the eGFP-GLO transgene and that of four 3′-UTR-containing transgenes (Fig. 3A, bottom panel) in HuH7 cells cultured for 6 weeks. In all cases, the TCN, eGFP mRNA expression, and percentage of eGFP+ cells remained stable (see data in supplemental Fig. S3, A, B, and C, respectively). We then concluded that none of the transgene copies were lost during this period. It should be noted, however, that eGFP protein expression from the different transgenes concomitantly fluctuated (supplemental Fig. S3D). These variations suggested that cell culture conditions and handling did influence transgene expression independently of the 3′-UTR located downstream of the eGFP coding region. Therefore, eGFP protein expression from the different transgenes of interest was normalized to that of referent eGFP-GLO transgene. In this case (supplemental Fig. S3E), eGFP protein expression became as stable as the other parameters (shown in supplemental Figs. S3, A–C). In conclusion, these results highlighted the necessity to systematically and concomitantly use a reference transgene (here eGFP-GLO) to correct nonspecific variations of eGFP expression due to cellular environment or context. As a consequence, the next series of experiments were systematically performed using eGFP-GLO as the reference transgene.
Finally we assessed that, in our conditions, the gross amount of eGFP protein present in a cell did not influence the regulation of its levels. For that, HuH7 cells were transduced with eGFP-c-MYC-expressing lentiviral particles at an m.o.i. giving more than 90% eGFP+ cells (m.o.i. = 6). One week later, cell subpopulations expressing low, medium, or high levels of eGFP proteins per cell were selected by cell sorting (supplemental Fig. S4) and grown for 1 additional week. Then the eGFP protein expression and the eGFP mRNA amount were determined in each eGFP-expressing HuH7 cell subpopulation by FCM and qRT-PCR, respectively, as described above. As shown in supplemental Table 3, the amount of eGFP protein per mRNA was similar in the three subpopulations. Therefore we concluded that, in our conditions, the expression level of ectopic eGFP protein and mRNA in individual cells did not affect the regulatory mechanisms measured by our method.
Based on these values, we calculated three ratios. The “eGFP protein/transgene copy number” ratio (P/TCN) represents the quantity of protein produced per transgene. The “eGFP protein/eGFP mRNA” ratio (P/M) represents the quantity of protein produced per mRNA. Finally the “eGFP mRNA/transgene copy number” ratio (M/TCN) represents the quantity of mRNA produced per transgene (or steady-state level of mRNA). When comparing the expression from two transgenes (one being the reference), P/TCN and P/M were indicative of the global post-transcriptional regulation and the relative translation efficiency, respectively. As the eGFP transgene expression is considered equivalent in terms of transcription and protein turnover when performed in a given cell type and condition (see above), M/TCN is an index of relative mRNA stability. Such an assumption was demonstrated in complementary experiments (see below and supplemental Fig. S6).
To directly test the capacity of FunREG to monitor post-transcriptional regulation, we repressed eGFP expression using siRNAs targeting eGFP ORF. siRNA-mediated repression very efficiently decreased eGFP protein expression by 92% when compared with the control (supplemental Fig. S5A). FunREG allowed demonstrating that this strong repression was due to a 65% decrease in mRNA amount (supplemental Fig. S5B) and a 75% reduction of translation efficiency (supplemental Fig. S5C).
Validation of FunREG to Measure Post-transcriptional Regulations Mediated by Either Selected 3′-UTRs or Exogenous transRFs
We then tested FunREG to measure post-transcriptional regulations mediated by specific cisARSs (AREs or miRNA-binding sites) located in the 3′-UTR of selected mRNAs. To this end, specific regions present in c-myc, HBEGF, or CCND1 mRNA 3′-UTRs were inserted downstream of eGFP ORF (Fig. 3A, bottom panel). These 3′-UTRs were selected because the corresponding genes were found to be up-regulated in liver cancer tissues and to contain known or putative post-transcriptional elements (42–45). The granulocyte macrophage colony-stimulating factor (GM-CSF) ARE (Fig. 3A) was used as a positive control because it mediates rapid mRNA decay and translation repression (17, 23). HuH7 cells were transduced once with viral particles expressing each transgene at m.o.i. favoring unique lentiviral integration (Fig. 2). This was carried out to minimize functional biases due to either eGFP overexpression or multiple lentiviral integrations. Then eGFP protein expression, eGFP mRNA expression, and the transgene copy number were measured in each cell population, and ratios were calculated as described above. Comparisons with the reference eGFP-GLO transgene (Fig. 3A, top panel) revealed that post-transcriptional regulation is conditioned by the selected 3′-UTR. Indeed eGFP expression was significantly inhibited (94, 77, 43, and 30% reductions) when GM-CSF ARE and HBEGF, Cyclin D1, and c-myc 3′-UTRs, respectively, were inserted downstream of eGFP ORF (Fig. 3B). As expected, the presence of GMCSF ARE induced both robust mRNA destabilization (85% decrease; Fig. 3C) and repression of translation (64% reduction; Fig. 3D). c-myc and Cyclin D1 3′-UTRs displayed significant post-transcriptional regulations (Fig. 3B). However, we were unable to statistically determine whether this effect was due to change in mRNA stability, in translation efficiency, or both (Fig. 3, C and D). Finally the strong negative post-transcriptional regulation mediated by the HBEGF 3′-UTR mainly originated from a significant change in mRNA stability (78% reduction; Fig. 3C), whereas translational efficiency remained unchanged (Fig. 3D).
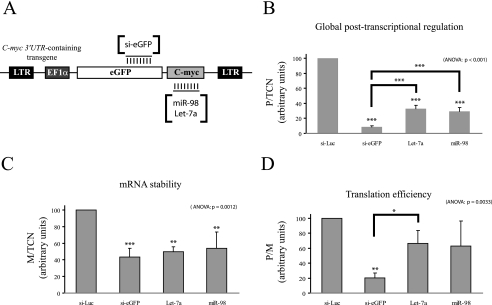
We then tested the ability of miRNAs to mediate post-transcriptional regulations using the FunREG pipeline (Fig. 1) and compared the result with that obtained previously with siRNA against eGFP mRNA (supplemental Fig. S5). let-7a and miR-98 were selected because they were reported to regulate c-myc expression in a post-transcriptional manner through its 3′-UTR (46). HuH7 cells expressing standardized copies of either eGFP-GLO or eGFP-c-MYC transgenes were transfected with various siRNAs and miRNAs (Fig. 4A). Three days later, eGFP mRNA and protein expression levels were measured by qRT-PCR and FCM, respectively. Then ratios were calculated (Fig. 4, B–D). Compared with control siRNA (si-Luc), the siRNA directed against eGFP mRNA (si-eGFP) significantly down-regulated eGFP-c-MYC expression (92% reduction) by decreasing mRNA stability (57% reduction) and repressing translation (80% reduction) (Fig. 4, B–D). These results were in agreement with those previously obtained with eGFP-GLO transgene (supplemental Fig. S5). As expected (46), let-7a and miR-98 significantly down-regulated eGFP-c-MYC expression (67 and 71% reductions, respectively; Fig. 4B) by inducing mRNA destabilization (50 and 46% reductions, respectively; Fig. 4C) and did not display any effect on eGFP-GLO expression (not shown). Finally both miRNAs had no significant effect on eGFP-c-MYC translation besides a tendency to repress it (Fig. 4D). Part of these results was in agreement with the regulatory roles previously described for these two types of small regulatory RNAs (9, 15, 22, 47, 48), thus demonstrating that effects of transRFs on mRNA stability and translation ability could be efficiently assessed by FunREG.
Fig. 4.
Measurement of siRNA- and miRNA-mediated post-transcriptional regulations by FunREG. A, schematic representation of eGFP-c-MYC transgene. Parts of the eGFP-c-MYC mRNA targeted by the si-eGFP and miRNAs (miR-98 and let-7a) are as shown. B–D, the eGFP-c-MYC-expressing HuH7 cells (from Fig. 3; transgene copy number known) were transfected with the indicated siRNA or miRNA. Three days later, the eGFP protein and eGFP mRNA amounts were determined as described in Fig. 3 except that α-tubulin mRNA was used as internal control. B, global post-transcriptional regulation. C, mRNA stability. D, translation efficiency. ANOVA, analysis of variance.
Application of FunREG to Study Post-transcriptional Deregulations during Liver Carcinogenesis
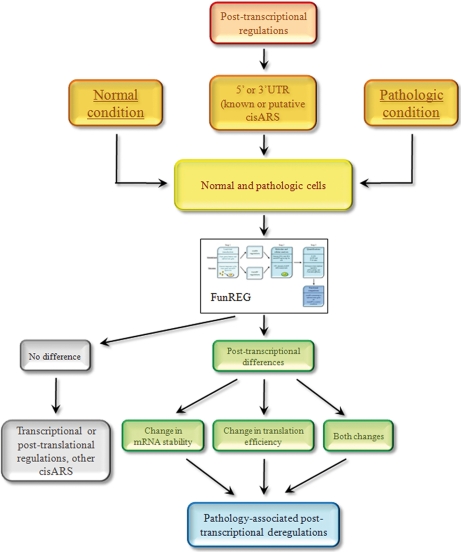
One of the major advantages of FunREG is that functional comparisons can be carried out in normal versus diseased cells, thus allowing for detection of potential post-transcriptional regulations specific of one case or the other (Fig. 5). The function of a cisARS of interest (from either the 5′- or 3′-UTR), deriving from a gene abnormally expressed in disease, can be evaluated through the FunREG pipeline in relevant cellular models (Fig. 5). The expression levels of the reporter can then be monitored in the different cell types and then compared. The absence of a post-transcriptional difference observed in a disease-relevant cellular model may suggest that differential gene expression observed in situ could reflect transcriptional, post-translational, or both kinds of regulations together. In addition, we cannot exclude the possibility of the presence of a cisARS not located in the 5′- or 3′-UTR but rather within the gene ORF itself (Fig. 5). On the contrary, the measurement of a post-transcriptionally occurring difference supports the idea that the selected UTR contains a cisARS whose functionality is altered in diseased cells and provides information on the molecular mechanism (mRNA stability, translation efficiency, or both) at the origin of this expression change (Fig. 5).
Fig. 5.
Functional comparisons of selected 3′-UTRs in primary human hepatocytes and liver cancer-derived human cell lines using FunREG. Among genes differentially expressed in normal and pathologic conditions (left and right orange boxes), those susceptible to being post-transcriptionally regulated (red box) by known or putative cisARSs located into their 5′- or 3′-UTR (middle orange box) are selected. The corresponding transgenes (bearing either the 5′- or 3′-UTR of interest) are transferred into normal and pathologic cells used as models by lentiviral transduction (yellow box). Then both types of transduced cells are “injected” in the FunREG pipeline. Two outcomes can be achieved. First there is no functional difference between normal and pathologic cells (top gray box). Therefore the transgene of interest does not contain the cisARS responsible for the differential gene expression, or the deregulation is due to a transcriptional or post-translational mechanism (bottom gray box). On the other hand, there is a functional difference between normal and pathologic cells (top green box) indicative of a post-transcriptional deregulation associated with the pathology (blue box). Following FunREG, the origin of the molecular mechanism (mRNA stability, translation efficiency, or both) responsible for the post-transcriptional deregulation is determined (bottom green boxes).
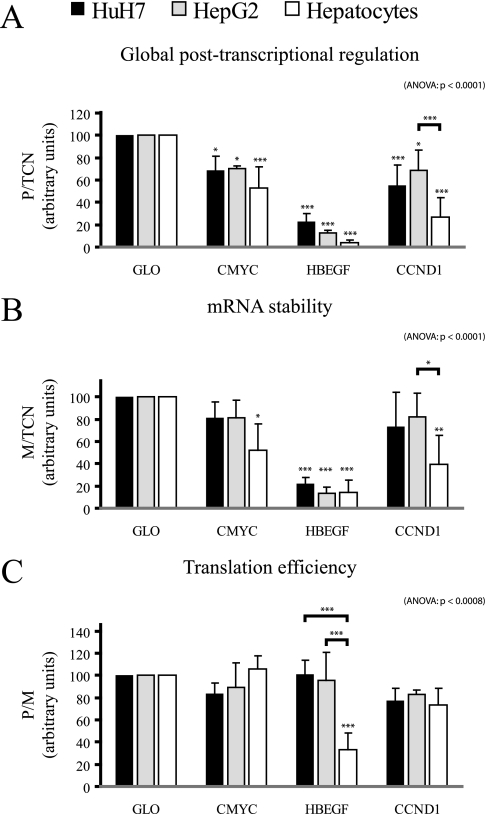
To test the capacity of FunREG to measure potential post-transcriptional differences existing between primary human hepatocytes and liver cancer-derived human cell lines, primary hepatocytes, HepG2, and HuH7 cells were transduced with lentiviral particles containing each transgene described above (Fig. 3A) with the exception of the GM-CSF ARE that was used as positive control in previous experiments. Interestingly some post-transcriptional regulations were cell-specific as illustrated with eGFP-CCND1, which displayed a significant functional variability between HepG2 and hepatocytes (Fig. 6A). Whereas the presence of CCND1 3′-UTR in the transgene reduced eGFP expression by 28% in HepG2, this decrease was dramatically enhanced in primary hepatocytes (72% decrease). Our data revealed that this effect was due to a significant decrease in mRNA stability (Fig. 6B) because the translation efficiency of the transgene product was not affected (Fig. 6C). Another interesting observation was made when using the HBEGF 3′-UTR. Indeed the insertion of this 3′-UTR downstream of the eGFP ORF greatly decreased eGFP expression in every cell type. The expression of eGFP was not significantly different in liver cancer-derived cells and in hepatocytes possibly because of the dominant effect of this 3′-UTR (Fig. 6A). However, translation efficiency was significantly decreased in hepatocytes when compared with the two liver cancer-derived cell types (Fig. 6C). In conclusion, our system allowed a direct comparison of post-transcriptional regulations mediated by 3′-UTRs in normal and disease-derived cells revealing significant functional differences.
Fig. 6.
Comparative analyses of post-transcriptional regulations in normal and tumoral hepatic cells. HuH7 cells, HepG2 cells, and hepatocytes (as indicated) were transduced once with lentiviral particles expressing the indicated transgene. After 1 week, eGFP protein and mRNA amounts as well as transgene copy number were determined as described in Fig. 3. A, global post-transcriptional regulation. B, mRNA stability. C, translation efficiency. ANOVA, analysis of variance.
DISCUSSION
In the present study, we developed a method to (i) accurately measure post-transcriptional regulations mediated by cisARS from selected messenger RNAs in mammalian cells, (ii) assess the role of exogenous transRFs in these regulations, (iii) perform post-transcriptional and functional comparisons in normal versus pathological cells, and finally (iv) measure the contribution of mRNA stability and translation ability to the observed post-transcriptional variations.
This method, named FunREG (Fig. 1), presents many advantages over those currently used (23, 25, 49). Among those, the first advantage of FunREG is represented by the use of lentivirus-mediated transgene delivery. Indeed added to ease of use, delivery efficiency, versatility, very low cellular toxicity, and straightforward stable cell line establishment, laboratory-produced lentiviruses are totally safe as they cannot replicate or produce viral RNA and proteins in cellulo (32, 34, 50). Their use to transduce different cell types does not require individual technical adaptations because infectious particles enter easily into the target cells, even into non-dividing cells (36, 37). It should be noted that, in agreement with other studies (36, 51), (i) 100-fold more infectious particles were needed to efficiently transduce primary hepatocytes when compared with the four cell lines used in this study, and (ii) a low percentage of lentiviral transgene copies was likely integrated into the transduced hepatocytes (Fig. 2E). Nevertheless besides the fact that qPCR analysis did not discriminate between the integrated and non-integrated forms of the lentiviral transgene, the results presented in Fig. 2E showed that eGFP-GLO expression fully correlated with the transgene copy number in hepatocytes. Finally based on the lentivirus-mediated delivery of eGFP-expressing transgenes, FunREG has the major technical advantage, over methods currently used, to be adapted to functional and chemical screens using an automated flow cytometer.
Besides the use of lentivirus-based transgene delivery, FunREG also presents other significant advantages over existing methodologies (23, 41, 49). Indeed this method allows the unbiased analysis of a cell population statistically containing one randomly integrated transgene, therefore bypassing biases resulting from eGFP overexpression, multiple lentiviral integrations, or artifacts due to virus integration. In this case, global transgene expression is similar from one cell population to another, thus allowing for experimental and functional comparisons. Moreover FunREG allows (i) the integrated functional evaluation of post-transcriptional regulations mediated by either cisARSs or exogenous transRFs and (ii) the direct and concomitant comparisons of mRNA half-life and translation efficiency in a “one-time point experiment” using different regulatory sequences, host cells, or experimental conditions. In addition to the study of cisARSs localized in the 3′-UTR, FunREG can also be adapted to study those localized in the 5′-UTR and evaluate the effects of their associated transRFs (9, 15, 52). However, investigations on cisARSs localized in the coding region (e.g. c-fos coding region determinant of instability (13)) are not applicable to FunREG as described in Fig. 1. Indeed insertion of coding sequences may affect the turnover of the chimeric eGFP protein excluding functional comparisons with the referent eGFP-GLO transgene. A recent method by Yen et al. (6) is more adequate to such protein turnover studies. Investigators may still use FunREG in a partial FunREG version for such studies by comparing the mRNA stability (M/TCN) of the eGFP mRNA with a coding region-derived cisARS with that of the eGFP-GLO referent.
Valuable applications could also be envisaged by developing global methods based on FunREG. Large scale post-transcriptional analyses may be performed by combining FunREG with either a 5′- or 3′-UTR lentiviral library from human when available, giving birth to a new “omics” field, named either post-transcriptomics or UTRomics as described for Caenorhabditis elegans (53). As shown herein, FunREG accurately measured translational efficiency from mRNA bearing selected 3′-UTRs in different cells or contexts. An analysis combining FunREG with a UTR-derived library may help to shed light on the weak correlations between mRNA expression and protein abundance reported in combined transcriptomics and proteomics analyses (1–5, 27–29).
At a physiopathological level, FunREG allowed comparative post-transcriptional studies in normal and pathological cells. By using three 3′-UTRs deriving from genes overexpressed in liver cancer and likely to be involved in liver carcinogenesis (42, 43, 45) as models, we showed that the ability of two of these 3′-UTRs to control eGFP transgene expression was reduced in primary liver cancer-derived cells when compared with normal hepatocytes (Fig. 6). This is consistent with the overexpression of the respective transcripts in primary liver cancers (42, 43, 45). Therefore, these post-transcriptional deregulations are in direct agreement with the role and the higher expression of these genes in cancerous hepatic cells (42–45, 54). Such tumor-specific post-transcriptional alterations may be explained by the implication of cell-specific transRFs (protein or miRNAs). This is indeed the case with either ARE-BPs or miRNAs as their deregulated expression has been linked to carcinogenesis (19, 20, 55). On the other hand, disease-associated mutations/deletions in the 3′-UTR of particular transcripts have been shown to abrogate ARE- and miRNA-mediated regulations leading to overexpression of the corresponding genes (for a review, see Ref. 19).2 By generalizing the use of FunREG in physiopathological studies, the reported post-transcriptional deregulations may constitute new markers for the prognosis or the classification of specific pathologies such as cancers. In addition, the function of a specific post-transcriptional mechanism may be evaluated in one particular disease using different cell lines with various genetic backgrounds.
With the help of FunREG, profound functional insight about the different post-transcriptional controls taking place in normal and pathological cells could be gained. As post-transcriptional dysfunctions are strongly suspected in human pathologies such as cancer or chronic inflammatory diseases (19, 56), FunREG may represent a valuable tool to reveal disease-associated deregulations or evaluate the responsibility of particular transRFs (mainly ARE-BPs and miRNAs (19, 20)) opening the road to new areas of investigation in human pathologies. Besides its usefulness in the understanding of physiopathology, FunREG may also provide an efficient way to reconcile results from combined transcriptome and proteome as well as opening the way to a new omics field, therefore participating in the understanding of the molecular networks governing cell fate and properties.
Acknowledgments
We thank M. Gorospe, P. Malik, and P. Charneau for providing reagents. E. Chevet, S. Jalvy, and J. Rosenbaum are warmly acknowledged for critical reading of the manuscript. We thank the cytometry core facility of Institut Fédératif de Recherche 66 and V. Pitard for technical assistance with cell analysis and sorting.
Footnotes
* This work was supported by grants from INSERM, the French Ministry of Research, the Ligue Régionale contre le Cancer-Comités Aquitaine, the Association pour la Recherche contre le Cancer, and the Agence Nationale pour la Recherche-Programme Jeunes Chercheurs (Grant JC07_184264 to C. G.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
2 D. Simon, B. Laloo, C. Grosset, and B. Arveiler, unpublished data.
1 The abbreviations used are:
- HCC
- hepatocellular carcinoma
- ARE-BP
- ARE-binding protein
- ARE
- AU-rich element
- cisARS
- cis-acting RNA sequence
- CCND1
- Cyclin D1
- CT
- control
- GFP
- enhanced green fluorescent protein
- FCM
- flow cytometry
- GM-CSF
- granulocyte macrophage colony-stimulating factor
- GLO
- globin
- HBEGF
- heparin-binding epidermal growth factor
- M
- eGFP mRNA
- MFI
- mean fluorescence intensity
- miRNA
- micro-RNA
- m.o.i.
- multiplicity(ies) of infection
- TCN
- transgene copy number
- P
- eGFP protein
- siRNA
- small interfering-RNA
- transRF
- trans-regulatory factor
- UTR
- untranslated region
- FunREG
- functional, integrated, and quantitative method to measure post-transcriptional regulations
- qPCR
- quantitative PCR
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1.Gallardo K., Firnhaber C., Zuber H., Héricher D., Belghazi M., Henry C., Küster H., Thompson R. ( 2007) A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol. Cell. Proteomics 6, 2165– 2179 [DOI] [PubMed] [Google Scholar]
- 2.Williamson A. J., Smith D. L., Blinco D., Unwin R. D., Pearson S., Wilson C., Miller C., Lancashire L., Lacaud G., Kouskoff V., Whetton A. D. ( 2008) Quantitative proteomics analysis demonstrates post-transcriptional regulation of embryonic stem cell differentiation to hematopoiesis. Mol. Cell. Proteomics 7, 459– 472 [DOI] [PubMed] [Google Scholar]
- 3.Beyer A., Hollunder J., Nasheuer H. P., Wilhelm T. ( 2004) Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics 3, 1083– 1092 [DOI] [PubMed] [Google Scholar]
- 4.Greenbaum D., Colangelo C., Williams K., Gerstein M. ( 2003) Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minagawa H., Honda M., Miyazaki K., Tabuse Y., Teramoto R., Yamashita T., Nishino R., Takatori H., Ueda T., Kamijo K., Kaneko S. ( 2008) Comparative proteomic and transcriptomic profiling of the human hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 366, 186– 192 [DOI] [PubMed] [Google Scholar]
- 6.Yen H. C., Xu Q., Chou D. M., Zhao Z., Elledge S. J. ( 2008) Global protein stability profiling in mammalian cells. Science 322, 918– 923 [DOI] [PubMed] [Google Scholar]
- 7.Bakheet T., Frevel M., Williams B. R., Greer W., Khabar K. S. ( 2001) ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 29, 246– 254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakheet T., Williams B. R., Khabar K. S. ( 2006) ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 34, D111– 114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz W., Bhattacharyya S. N., Sonenberg N. ( 2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102– 114 [DOI] [PubMed] [Google Scholar]
- 10.Eulalio A., Behm-Ansmant I., Izaurralde E. ( 2007) P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8, 9– 22 [DOI] [PubMed] [Google Scholar]
- 11.Garneau N. L., Wilusz J., Wilusz C. J. ( 2007) The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113– 126 [DOI] [PubMed] [Google Scholar]
- 12.Glisovic T., Bachorik J. L., Yong J., Dreyfuss G. ( 2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977– 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosset C., Chen C. Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A. B. ( 2000) A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103, 29– 40 [DOI] [PubMed] [Google Scholar]
- 14.Khaleghpour K., Kahvejian A., De Crescenzo G., Roy G., Svitkin Y. V., Imataka H., O'Connor-McCourt M., Sonenberg N. ( 2001) Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 21, 5200– 5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L., Belasco J. G. ( 2008) Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell 29, 1– 7 [DOI] [PubMed] [Google Scholar]
- 16.Guhaniyogi J., Brewer G. ( 2001) Regulation of mRNA stability in mammalian cells. Gene 265, 11– 23 [DOI] [PubMed] [Google Scholar]
- 17.Barreau C., Paillard L., Osborne H. B. ( 2005) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33, 7138– 7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espel E. ( 2005) The role of the AU-rich elements of mRNAs in controlling translation. Semin. Cell Dev. Biol. 16, 59– 67 [DOI] [PubMed] [Google Scholar]
- 19.Lopez de Silanes I., Quesada M. P., Esteller M. ( 2007) Aberrant regulation of messenger RNA 3′-untranslated region in human cancer. Cell. Oncol. 29, 1– 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong A. W., Nemunaitis J. ( 2008) Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 15, 341– 355 [DOI] [PubMed] [Google Scholar]
- 21.Capurro M. I., Xiang Y. Y., Lobe C., Filmus J. ( 2005) Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 65, 6245– 6254 [DOI] [PubMed] [Google Scholar]
- 22.Brodersen P., Sakvarelidze-Achard L., Bruun-Rasmussen M., Dunoyer P., Yamamoto Y. Y., Sieburth L., Voinnet O. ( 2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185– 1190 [DOI] [PubMed] [Google Scholar]
- 23.Grosset C., Boniface R., Duchez P., Solanilla A., Cosson B., Ripoche J. ( 2004) In vivo studies of translational repression mediated by the granulocyte-macrophage colony-stimulating factor AU-rich element. J. Biol. Chem. 279, 13354– 13362 [DOI] [PubMed] [Google Scholar]
- 24.Hutvagner G., Simard M. J. ( 2008) Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22– 32 [DOI] [PubMed] [Google Scholar]
- 25.Benjamin D., Colombi M., Stoecklin G., Moroni C. ( 2006) A GFP-based assay for monitoring post-transcriptional regulation of ARE-mRNA turnover. Mol. Biosyst. 2, 561– 567 [DOI] [PubMed] [Google Scholar]
- 26.Vopálenský V., Masek T., Horváth O., Vicenová B., Mokrejs M., Pospísek M. ( 2008) Firefly luciferase gene contains a cryptic promoter. RNA 14, 1720– 1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurbatov L. K., Cheglakov I. B., Iarygin K. N., Sukhikh G. T., Vartanian G. V., Toropygin I. Iu., Archakov A. I. ( 2008) [Comparative transcriptomic and proteomic analysis of the fetal and adult human liver]. Biomed. Khim. 54, 140– 153 [PubMed] [Google Scholar]
- 28.Mijalski T., Harder A., Halder T., Kersten M., Horsch M., Strom T. M., Liebscher H. V., Lottspeich F., de Angelis M. H., Beckers J. ( 2005) Identification of coexpressed gene clusters in a comparative analysis of transcriptome and proteome in mouse tissues. Proc. Natl. Acad. Sci. U.S.A. 102, 8621– 8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastorelli R., Carpi D., Campagna R., Airoldi L., Pohjanvirta R., Viluksela M., Hakansson H., Boutros P. C., Moffat I. D., Okey A. B., Fanelli R. ( 2006) Differential expression profiling of the hepatic proteome in a rat model of dioxin resistance: correlation with genomic and transcriptomic analyses. Mol. Cell. Proteomics 5, 882– 894 [DOI] [PubMed] [Google Scholar]
- 30.Sirven A., Ravet E., Charneau P., Zennou V., Coulombel L., Guétard D., Pflumio F., Dubart-Kupperschmitt A. ( 2001) Enhanced transgene expression in cord blood CD34(+)-derived hematopoietic cells, including developing T cells and NOD/SCID mouse repopulating cells, following transduction with modified trip lentiviral vectors. Mol. Ther. 3, 438– 448 [DOI] [PubMed] [Google Scholar]
- 31.Robert-Richard E., Richard E., Malik P., Ged C., de Verneuil H., Moreau-Gaudry F. ( 2007) Murine retroviral but not human cellular promoters induce in vivo erythroid-specific deregulation that can be partially prevented by insulators. Mol. Ther. 15, 173– 182 [DOI] [PubMed] [Google Scholar]
- 32.Bauer G., Dao M. A., Case S. S., Meyerrose T., Wirthlin L., Zhou P., Wang X., Herrbrich P., Arevalo J., Csik S., Skelton D. C., Walker J., Pepper K., Kohn D. B., Nolta J. A. ( 2008) In vivo biosafety model to assess the risk of adverse events from retroviral and lentiviral vectors. Mol. Ther. 16, 1308– 1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Debyser Z. ( 2003) Biosafety of lentiviral vectors. Curr. Gene Ther. 3, 517– 525 [DOI] [PubMed] [Google Scholar]
- 34.Sinn P. L., Sauter S. L., McCray P. B., Jr. ( 2005) Gene therapy progress and prospects: development of improved lentiviral and retroviral vectors–design, biosafety, and production. Gene Ther. 12, 1089– 1098 [DOI] [PubMed] [Google Scholar]
- 35.Trono D. ( 2000) Lentiviral vectors: turning a deadly foe into a therapeutic agent. Gene Ther. 7, 20– 23 [DOI] [PubMed] [Google Scholar]
- 36.Nguyen T. H., Oberholzer J., Birraux J., Majno P., Morel P., Trono D. ( 2002) Highly efficient lentiviral vector-mediated transduction of nondividing, fully reimplantable primary hepatocytes. Mol. Ther 6, 199– 209 [DOI] [PubMed] [Google Scholar]
- 37.Géronimi F., Richard E., Lamrissi-Garcia I., Lalanne M., Ged C., Redonnet-Vernhet I., Moreau-Gaudry F., de Verneuil H. ( 2003) Lentivirus-mediated gene transfer of uroporphyrinogen III synthase fully corrects the porphyric phenotype in human cells. J. Mol. Med. 81, 310– 320 [DOI] [PubMed] [Google Scholar]
- 38.Salmon P., Trono D. ( 2006) Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. Suppl. 37, Unit 4.21 [DOI] [PubMed] [Google Scholar]
- 39.Chen C. Y., Shyu A. B. ( 1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20, 465– 470 [DOI] [PubMed] [Google Scholar]
- 40.Chen C. Y., Xu N., Shyu A. B. ( 1995) mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15, 5777– 5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu N., Loflin P., Chen C. Y., Shyu A. B. ( 1998) A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26, 558– 565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inui Y., Higashiyama S., Kawata S., Tamura S., Miyagawa J., Taniguchi N., Matsuzawa Y. ( 1994) Expression of heparin-binding epidermal growth factor in human hepatocellular carcinoma. Gastroenterology 107, 1799– 1804 [DOI] [PubMed] [Google Scholar]
- 43.Joo M., Kang Y. K., Kim M. R., Lee H. K., Jang J. J. ( 2001) Cyclin D1 overexpression in hepatocellular carcinoma. Liver 21, 89– 95 [DOI] [PubMed] [Google Scholar]
- 44.Sorensen B. S., Ornskov D., Nexo E. ( 2006) The chemotherapeutic agent VP16 increases the stability of HB-EGF mRNA by a mechanism involving the 3′-UTR. Exp. Cell Res. 312, 3651– 3658 [DOI] [PubMed] [Google Scholar]
- 45.Xu X. R., Huang J., Xu Z. G., Qian B. Z., Zhu Z. D., Yan Q., Cai T., Zhang X., Xiao H. S., Qu J., Liu F., Huang Q. H., Cheng Z. H., Li N. G., Du J. J., Hu W., Shen K. T., Lu G., Fu G., Zhong M., Xu S. H., Gu W. Y., Huang W., Zhao X. T., Hu G. X., Gu J. R., Chen Z., Han Z. G. ( 2001) Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc. Natl. Acad. Sci. U.S.A. 98, 15089– 15094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sampson V. B., Rong N. H., Han J., Yang Q., Aris V., Soteropoulos P., Petrelli N. J., Dunn S. P., Krueger L. J. ( 2007) MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 67, 9762– 9770 [DOI] [PubMed] [Google Scholar]
- 47.Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. ( 2008) The impact of microRNAs on protein output. Nature 455, 64– 71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nilsen T. W. ( 2008) Endo-siRNAs: yet another layer of complexity in RNA silencing. Nat. Struct. Mol. Biol. 15, 546– 548 [DOI] [PubMed] [Google Scholar]
- 49.Sureban S. M., Murmu N., Rodriguez P., May R., Maheshwari R., Dieckgraefe B. K., Houchen C. W., Anant S. ( 2007) Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology 132, 1055– 1065 [DOI] [PubMed] [Google Scholar]
- 50.Amado R. G., Chen I. S. ( 1999) Lentiviral vectors–the promise of gene therapy within reach? Science 285, 674– 676 [DOI] [PubMed] [Google Scholar]
- 51.Wu Y. ( 2004) HIV-1 gene expression: lessons from provirus and non-integrated DNA. Retrovirology 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilusz C. J., Wormington M., Peltz S. W. ( 2001) The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2, 237– 246 [DOI] [PubMed] [Google Scholar]
- 53.Mangone M., Macmenamin P., Zegar C., Piano F., Gunsalus K. C. ( 2008) UTRome.org: a platform for 3′UTR biology in C. elegans. Nucleic Acids Res. 36, D57– 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozaki I., Zhang H., Mizuta T., Ide Y., Eguchi Y., Yasutake T., Sakamaki T., Pestell R. G., Yamamoto K. ( 2007) Menatetrenone, a vitamin K2 analogue, inhibits hepatocellular carcinoma cell growth by suppressing cyclin D1 expression through inhibition of nuclear factor kappaB activation. Clin. Cancer Res. 13, 2236– 2245 [DOI] [PubMed] [Google Scholar]
- 55.Fawal M., Armstrong F., Ollier S., Dupont H., Touriol C., Monsarrat B., Delsol G., Payrastre B., Morello D. ( 2006) A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood 108, 2780– 2788 [DOI] [PubMed] [Google Scholar]
- 56.Stoecklin G., Anderson P. ( 2006) Posttranscriptional mechanisms regulating the inflammatory response. Adv. Immunol. 89, 1– 37 [DOI] [PubMed] [Google Scholar]