Abstract
Signal transduction in metazoans regulates almost all aspects of biological function, and aberrant signaling is involved in many diseases. Perturbations in phosphorylation-based signaling networks are typically studied in a hypothesis-driven approach, using phospho-specific antibodies. Here we apply quantitative, high-resolution mass spectrometry to determine the systems response to the depletion of one signaling component. Drosophila cells were metabolically labeled using stable isotope labeling by amino acids in cell culture (SILAC) and the phosphatase Ptp61F, the ortholog of mammalian PTB1B, a drug target for diabetes, was knocked down by RNAi. In total we detected more than 10,000 phosphorylation sites in the phosphoproteome of Drosophila Schneider cells and trained a phosphorylation site predictor with this data. SILAC-based quantitation after phosphatase knock-down showed that apart from the phosphatase, the proteome was minimally affected whereas 288 of 6,478 high-confidence phosphorylation sites changed significantly. Responses at the phosphotyrosine level included the already described Ptp61F substrates Stat92E and Abi. Our analysis highlights a connection of Ptp61F to cytoskeletal regulation through GTPase regulating proteins and focal adhesion components.
Information processing in biological systems relies heavily on activation and inactivation of proteins by phosphorylation. This key post-translational modification is involved in the regulation of most cellular processes and mediates many rapid responses as well as long-term gene expression changes in response to stimuli. Protein kinases and protein phosphatases coordinately regulate this highly dynamic and reversible modification. Phosphorylation is usually studied in a candidate-based approach by in vitro kinase assays or by immune techniques employing phospho-specific antibodies. Despite the success of this reductionist approach, it does not afford a systems-wide observation of the effects upon perturbations of signaling networks.
Recent advances in MS-based1 proteomics now allow the identification of thousands of phosphorylation sites from complex protein mixtures (1–3). Most large-scale phosphoproteomics studies have been qualitative rather than quantitative; however, isotope-based methods enable precise quantitation of phosphorylation sites between two or more cellular states (4–6). Our group has applied the metabolic labeling technology termed stable isotope labeling by amino acids in cell culture (SILAC) (7) for the quantitative comparison of phosphoproteomes. For example, we quantified phosphorylation dynamics in response to epidermal growth factor stimulation. Out of a measured phosphoproteome of several thousand sites only a minority (about 10%) was regulated by the signal, highlighting the importance of quantitation in pinpointing specific systems responses (8).
Drosophila is a well established model system to study key players in cell signaling and development. Genetic studies have been performed for decades whereas more recently also RNA interference (RNAi) has been employed for gene function studies using a highly efficient silencing protocol (9). A further advantage of Drosophila as a model system is the lower degree of functional redundancy compared with higher vertebrates while maintaining a high level of conservation of human genes linked to disease (10).
Two large-scale, non-quantitative Drosophila phosphoproteome studies were carried out in embryonic Kc167 cells (11) and embryos (12). Both studies identified more than 10,000 sites of the Drosophila phosphoproteome.
We have recently adapted the SILAC methodology for quantitative proteomics to Drosophila. Schneider line 2 (SL2) cells were treated with either mock dsRNA or dsRNA against ISWI, a component of chromatin remodeling complexes. The combination of RNAi and SILAC allows the unbiased “phenotypization” of the gene knock-down directly at the proteome level (13).
Here we determined a high-quality basal phosphoproteome in SL2 cells and characterized its structural and evolutionary properties. We compared kinase substrate motives between Drosophila and human and trained a Drosophila phosphorylation site predictor.
To explore the potential of quantitative phosphoproteomics in a systems-wide manner, we focused on the Drosophila non-transmembrane tyrosine phosphatase Ptp61F. This phosphatase is the ortholog of mammalian PTB1B, which is thought to be involved in type 2 diabetes, obesity, and cancer (14), and which is the target of several ongoing drug development projects (15). Ptp61F is a negative regulator of JAK/STAT signaling (16, 17) and, together with the Ableson kinase (Abl), involved in the regulation of the Abl interacting protein (Abi) and lamella formation (18). Both PTP1B and Ptp61F are among the best studied protein tyrosine phosphatases in their respective organisms; however the characterization of their substrates is still far from complete. Two recent mass spectrometric studies employed substrate trapping to identify direct substrates of PTP1B and Ptp61F (19, 20). The PTP1B study was combined with phosphotyrosine peptide enrichment, which led to site-specific detection of potential PTP1B targets. PTP1B function was additionally investigated by quantitative phosphotyrosine proteomics comparing wild type and PTP1B-deficient fibroblasts. In contrast, the Ptp61F study identified potential substrates without site-specific information. One of these was PVR, the Drosophila homolog of VEGFR and PDGFR, suggesting that Ptp61F - like its mammalian counterpart - counteracts receptor tyrosine kinase signaling. Apart from Abi, further components of the SCAR/WAVE complex as well as its regulatory kinase Abl were identified as potential Ptp61F substrates. This supports an involvement of Ptp61F in the regulation of actin reorganization and remodeling.
To study the role of Ptp61F in a global and unbiased approach we combined global quantitative phosphoproteome analysis with RNA interference. We profiled tyrosine, serine and threonine phosphorylation changes upon ablation of Ptp61F by RNAi. In parallel, we quantified changes in the proteome, which allowed us to normalize changes in phosphorylation sites to corresponding changes at the protein level. Interestingly, we observed increased tyrosine phosphorylation of the protein tyrosine kinase Abl which suggests an enhanced Abl activity upon Ptp61F RNAi. We additionally detected up-regulated phosphotyrosine sites on GTPase regulating proteins (like RhoGAP15B and Vav) and constituents of focal adhesions (like Paxillin and Lasp) which expand the proposed involvement of Ptp61F in the regulation of cytoskeleton organization. Our work represents proof-of-principle that the combination of large-scale phosphoproteomics and a loss-of-function approach can contribute significantly to elucidating the role of key players in phosphorylation-dependent signaling. Importantly, this systems-wide approach measures the net effect of the perturbation on the entire signaling network, without the need to define specific substrate-kinase or substrate -phosphatase relationships or other direct functional mechanisms.
EXPERIMENTAL PROCEDURES
Cell Culture
Drosophila Schneider SL2 cells were a kind gift from Peter Becker's laboratory. Cells were grown in custom-made Schneider media at 26 °C (13). Schneider media were supplemented by yeast extract and fetal bovine serum dialyzed overnight using a molecular weight cutoff of 1.2 kDa to remove free amino acids. “Heavy” and “light” SILAC media were prepared by adding 0.4 g/liter 13C615N4 l-arginine (Arg-10) and 1.65 g/liter 13C615N2 l-lysine (Lys-8) (Sigma Isotec) or the corresponding non-labeled amino acids, 12C614N4 l-arginine (Arg-0) and 12C614N2 l-lysine (Lys-0), respectively. Media were always prepared freshly before the experiments. To obtain two labeled cell populations the cells were grown for 8–9 days in heavy or light SILAC media.
Phosphatase Inhibitor Treatment of SILAC-labeled SL2 Cells
SILAC-labeled SL2 cells were treated with an inhibitor mixture for 15 min. The mixture contained 0.3 mm Pervanadate (prepared from 100 mm sodium orthovanadate (Sigma-Aldrich) mixed with 100 mm H2O2), 33 nm Calyculin A (Millipore), and 3 μm deltamethrin (Merck).
RNAi of SILAC-labeled SL2 Cells
7.7 × 106 SL2 cells were resuspended in 7 ml of heavy or light serum-free SILAC media and seeded in a 75 cm2 tissue culture flask. 210-μg Ptp61F dsRNA was added to the heavy or 210-μg control GST dsRNA to the light condition, respectively (less than 200 nm). The cells were incubated for one hour at 26 °C and subsequently 14 ml of heavy or light serum-containing SILAC media were added. After 4 days two-third of the SILAC media were refreshed, and after 8 days the cells were harvested. The Ptp61F dsRNAs (forward: 5′-TTAATACGACTCACTATAGGGAGAGGATGTGAATCCGTACGATCAT-3′; reverse: 5′-TTAATACGACTCACTATAGGGAGAGAACTGCATCACTTCACGACTCT-3′) and GST dsRNAs (forward: 5′-TTAATACGACTCACTATAGGGAGAATGTCCCCTATACTAGGTTA-3′; reverse: 5′-TTAATACGACTCACTATAGGGAGAACGCATCCAGGCACATTG-3′) were synthesized according to Worby et al. (9) using MEGAshortscript™ kit (Ambion).
Preparation of Cell Extract
Heavy and light SL2 cells were counted, and equal cell numbers were combined. The cell mixture was pelleted by centrifugation at 1000 rpm for 5 min, washed with cold phosphate-buffered saline, and lysed under rotation for 60 min at 4 °C using radioimmune precipitation assay buffer (containing 1% Triton, 0.1% SDS, 0.1% sodium deoxycholate, 140 mm NaCl, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, 10 mm Tris, pH 8) supplemented with protease inhibitors (complete tablets; Roche Diagnostics) and phosphatase inhibitors (1 mm sodium orthovanadate, 5 mm NaF, 5 mm beta-glycerophosphate). The lysate was cleared by centrifugation, and the resulting supernatant was subjected to digestion.
Protein Digestion
For in-gel digestion the whole cell lysate was separated by one-dimensional SDS-PAGE (4–12% Novex mini-gel; Invitrogen) and visualized by colloidal Coomassie staining. For the proteome analysis 100-μg whole cell lysate was separated in two adjacent lanes and identically cut into 15 slices whereas for the phosphoproteome analysis 300 μg of cell lysate was separated in one lane and cut into 8 slices. All gel slices were subjected to in-gel digestion with trypsin (21, 22). Resulting tryptic peptides were extracted with 30% ACN in 3% TFA. For phosphoproteome analysis extracted peptides were subjected to TiO2 chromatography. For proteome analysis samples were concentrated until full evaporation of organic solvent and reconstituted with 1% TFA and 5% ACN. Subsequently peptides were concentrated and desalted on reversed phase C18 StageTips (23, 24). Peptides were eluted by 30-μl buffer B (80% ACN in 0.5% acetic acid) solution into a sample 96-well plate (Abgene), concentrated in a SpeedVac until removal of the organic solvent and reconstituted with a one-to-one mix of buffer A (0.5% acetic acid) and buffer A* (2% ACN in 1% TFA).
For in-solution digestion whole cell extracts were mixed with four volumes of ice-cold acetone to precipitate proteins. Precipitated proteins were collected by centrifugation and dissolved in 6 m urea, 2 m thiourea, 10 mm HEPES or 10 mm Tris and 1% n-octyl glucoside. 2-mg (phosphoproteome with phosphatase inhibitor treatment) or 20-mg proteins (phosphoproteome of Ptp61F RNAi) were reduced with 1 mm dithiothreitol for 45 min at RT and then alkylated with 5.5 mm iodoacetamide for 30 min. Reduced and alkylated proteins were digested with 200-μg endopeptidase Lys-C (Waco) for 3 h. Subsequently the peptide mixture was diluted with four volumes deionized water and digested with 200-μg sequencing grade modified trypsin (Promega) overnight at RT. Trypsin was inactivated by acidification with TFA to pH < 3, and the peptide mixture was subjected to phosphopeptide enrichment.
Phosphopeptide Fractionation and Enrichment
Phosphopeptides were fractionated and enriched from in-solution digested peptides by the combination of strong cation exchange chromatography (SCX) and TiO2 chromatography or by multiple rounds of TiO2 chromatography in batch mode. Alternatively phosphopeptides were enriched by TiO2 chromatography from in-gel digested peptide fractions.
SCX was carried out as described (25). The resulting 15 fractions were pooled according to the 230-nm absorbance intensity. For the phosphatase inhibitor phosphoproteome experiment 10 fractions and the flow-through were further enriched for phosphopeptides by TiO2 chromatography (26). For the Ptp61F RNAi experiment 15 fractions and the flow-through (loading 20 mg at once) and 13 fractions and the flow-through (performing two separate SCX runs with 10 mg each for better separation) were subjected to TiO2 chromatography (25). For the phosphatase inhibitor phosphoproteome experiment both the TiO2 beads (kindly provided by GL Sciences) and the samples were pre-incubated with 5 mg/ml 2,5-dihydroxybenzoic acid (DHB) in 80% ACN before the sample, and ∼5 mg of the TiO2 beads were combined and incubated batchwise with end-over-end rotation for 1–3 h. For the Ptp61F phosphoproteome the beads were pre-incubated with 30 mg/ml DHB and added directly to the SCX fraction to reduce the amount of DHB in the samples.
After incubation, the beads were washed twice with 10% ACN in 0.1% TFA and once with 80% ACN in 0.1% TFA solution. Bound peptides were eluted from TiO2 beads once with 1% ammonium solution, pH 10.5, in 20% ACN and twice with 1% ammonium solution, pH 10.5, in 40% ACN into 20 μl of 2% ACN in 1% TFA solution. Eluates were subsequently dried to ∼5 μl in a SpeedVac and reconstituted with 2% ACN in 1% TFA solution for LC-MS/MS analysis.
LC-MS/MS Analysis
The samples were analyzed by a nanoflow HPLC (Agilent 1100 or 1200; Agilent Technologies) coupled on-line via a nanoelectrospray ion source (Proxeon Biosystems) to a LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptide mixtures were loaded onto a C18-reversed phase column (15 cm long, 75-μm inner diameter, packed in-house with ReproSil-Pur C18-AQ 3-μm resin) in buffer B with a flow rate of 500 nl/min for 20 min and eluted with a linear gradient from 2% to 40% buffer B (80% ACN and 0.5% acetic acid solution) at a flow rate of 250 nl/min over two hours. After each sample the column was washed with 90% buffer B and re-equilibrated with buffer A.
Mass spectra were acquired in the positive ion mode applying a data-dependent automatic switch between survey scan and tandem mass spectra (MS/MS) acquisition. Proteome samples were analyzed with a top5 method, acquiring one Orbitrap survey scan in the mass range of m/z 300–2000 followed by MS/MS of the five most intense ions in the LTQ. The target value in the LTQ-Orbitrap was 1,000,000 for survey scan at a resolution of 60,000 at m/z 400 using lock masses for recalibration to improve the mass accuracy of precursor ions (27). Fragmentation in the LTQ was performed by collision-induced dissociation with a target value of 5,000 ions. Ion selection threshold was 500 counts. Selected sequenced ions were dynamically excluded for 90 s.
For the phosphoproteome acquisition a total scan cycle consisted of three mass ranges, 350–1050, 850–1750, and 350–1750 with MS/MS of the five most intense ions for the first two ranges and MS/MS of the seven most intense ions for the last range. Multi-stage activation was enabled upon detection of a neutral loss of phosphoric acid (97.97, 48.99, or 32.66 amu) (28) for further ion fragmentation. Selected sequenced ions were dynamically excluded for 300 s after sequencing. Otherwise settings were the same as described before for the proteome acquisition.
Data Analysis
Mass spectrometric data were analyzed with the in-house developed software MaxQuant (version 1.0.11.5 for the total basal SL2 phosphoproteome and version 1.0.12.33 for the spectra extraction as well as for the proteome and phosphoproteome of Ptp61F RNAi) (29, 30). MS/MS spectra were searched by Mascot (version 2.2.04, Matrix Science) against the FlyBase (version 5.4) (containing 20,822 entries) combined with 175 common contaminants and concatenated with the reversed versions of all sequences. The following parameters were set for the Mascot searches. Trypsin allowing for cleavage N-terminal to proline and cleavage between aspartic acid and proline was chosen as enzyme specificity. Cysteine carbamidomethylation was selected as a fixed modification, whereas protein N-terminal acetylation, methionine oxidation and serine, threonine and tyrosine phosphorylation were selected as variable modifications. Depending on a priori knowledge about the number of Arg and Lys in the precursor ion determined by MaxQuant before the search, Arg-10 and Lys-8 were used as additional fixed or variable modifications. Maximally three (version 1.0.11.5) or two missed cleavages (version 1.0.12.33) and three labeled amino acids were allowed. Initial mass deviation of precursor ion and fragment ions were up to 7 ppm and 0.5 Da, respectively. MaxQuant automatically quantified SILAC peptides and proteins. SILAC protein ratios were calculated as the median of all peptide ratios assigned to the protein. A false discovery rate of 0.01 was required for proteins and peptides with a minimum length of 6 amino acids. In addition a posterior error probability for each MS/MS spectrum below or equal to 0.1 was required. In case the identified peptides of two proteins were the same or the identified peptides of one protein included all peptides of another protein, these proteins (e.g. isoforms and homologs) were combined by MaxQuant and reported as one protein group. Phosphorylation sites were made non-redundant with regards to their surrounding peptide sequence. Nevertheless all alternative proteins that matched a particular phosphosite were reported as one group. The PTM score was used for assignment of the phosphorylation site(s) as described (8). Class I phosphorylation sites are defined by a localization probability of 0.75, and probability localization score difference higher or equal to 5 (8).
Quantitative RT-PCR
Total RNA was extracted from 1.5-ml cell suspension 8 days after dsRNA treatment using PrepEaseTM RNA spin kit (USB). The protocol included a DNA digestion step with DNase I on the spin column. Reverse transcription was carried out from 2-μg total RNA using first strand cDNA synthesis kit (Fermentas) according to manufacturer's protocol with oligo(dT)18 primers. Quantitative real-time PCR was performed using iQ™ SYBR® Green Supermix (BIO-RAD) in the MyiQ Real-Time PCR System (BIO-RAD). Each reaction mix (25 μl) contained 10 ng of cDNA, 12.5 μl of SYBER Green Supermix, and 12.5 pmol of forward and reverse primers. Two different primer pairs were used to analyze Ptp61F mRNA levels (Ptp61F 1 forward: 5′-AGCTGCACGATCCCACCTT-3′, Ptp61F 1 reverse: 5′-GTGGCAGATTAAGCGATTGGA-3′; Ptp61F 2 forward: 5′-CAGCACGACATGATCCACGA-3′, Ptp61F 2 reverse: 5′-TGTCGTCCTCGTCATCGTCAT-3′). GAPDH levels were measured for normalization via 2−ΔΔCT method (31) (GAPDH forward: 5′-ATGAAGGTGGTCTCCAACGC-3′; reverse: 5′-TCATCAGACCCTCGACGATCT-3′). Three technical replicates of each sample (Ptp61F RNAi) and control (GST RNAi) were performed.
Motif Extraction
Motif-X was used to derive potentially new phosphorylation motifs in silico and to confirm the observations of the χ2-Test. A probability value of 0.0001 was considered significant. In addition, a minimum occurrence of 20 was required to derive a significant consensus sequence. The approach was performed residue-specific on all Class I sites using the entire fly proteome as a background model.
Phosphorylation Site Prediction
A support vector machine was trained separately for each amino acid on high-confidence phosphorylation sites as described in Gnad et al. (32). The essential feature of each instance that was used for this machine learning approach was the raw sequence: the phosphorylated residue along with its surrounding sequence (±6 residues). This yields 260 dimensions in the feature space reflecting the surrounding amino acids. Each feature dimension presents a certain residue on a certain position within the given 13 amino acid sequence window. To generate a negative set of the same size, we randomly chose sites from fly proteins that were not, to date, detected to be phosphorylated. The positive and negative datasets were split into a training set (90%) and a test set (10%). The parameters C and σ were optimized by varying them from 2−10 to 210 in multiplicative steps of two on the basis of a 5-fold cross validation on the training set. The optimal C and σ parameters for predicting phosphoserines were 32 and 0.125, respectively. In the case of phosphothreonines the best parameters were 4 and 0.125, respectively. We trained the optimal model for each set of each phospho amino acid separately using the radial basis function.
Data Availability
Identified phosphorylated peptides and phosphorylated sites along with further information ranging from evolutionary conservation to structural constraints and Swiss-Prot annotations were uploaded to Phosida, the phosphorylation site database (32). Besides the fly phosphoproteome, Phosida also contains phosphorylation sites of mouse, human, and various prokaryotes and now provides the described Drosophila-specific phosphorylation site predictor. Furthermore, the genome annotation section of Phosida links directly to the EnsEMBL database, which displays our proteomic data on the basis of the DAS source management. The setup of Phosida DAS layers for a given gene of interest in EnsEMBL is described in the “Background” section of Phosida and in EnsEMBL.
Moreover, the proteomic data (identified peptides) were uploaded to MAPU - Max-Planck Unified Proteome Database. The detected peptides, which have been assigned to gene entries as described in this study, are also available via the MAPU DAS source in the EnsEMBL database.
RESULTS AND DISCUSSION
High-confidence Determination of the in Vivo SL2 Phosphoproteome
Most of the phosphoproteomics experiments are performed in a qualitative manner. Here we employed the SILAC method instead to measure quantitative changes in the levels of in vivo phosphorylation sites. In a first SILAC experiment, we compared basal phosphorylation to phosphatase inhibitor mixture-induced phosphorylation. This involves application of cell permeable inhibitors of serine/threonine and tyrosine protein phosphatases (see below) and has an analytical rather than a functional goal; it is advantageous because we trigger the identification of very low abundant endogenous phosphorylation sites that are up-regulated in response to the inhibitor treatment. Furthermore, the presence and mass spacing of the SILAC pair distinguish peptides from noise and allow counting the number of labeled amino acids, thereby decreasing the search space in database identification (30). The second SILAC experiment in which we quantified changes in the phosphoproteome upon RNAi-mediated knock-down of a phosphatase (see below) has both an analytical and a functional goal: it increases the measured endogenous phosphoproteome, which we subsequently analyzed bioinformatically, and it supplies information on the cellular roles of the phosphatase. Analytically, these two experiments lead to the detection of different sets of the Drosophila phosphoproteome (Fig. 1 and supplemental Tables 8 and 9). All phosphorylation sites in either SILAC experiments that are present in the control state (manifest as SILAC pairs) thus reflect the endogenous phosphorylation status of the Drosophila SL2 cells here termed “basal phosphoproteome”.
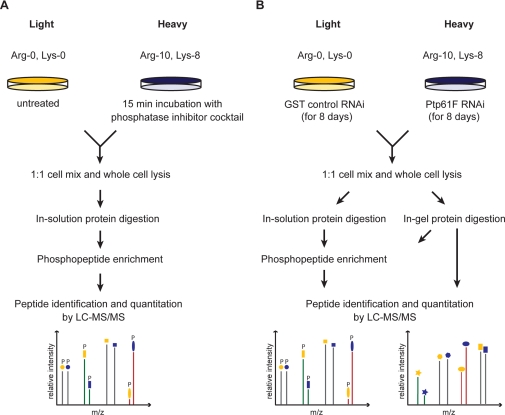
Fig. 1.
Basal phosphoproteome of Drosophila SL2 cells. A, experimental procedure for quantitative phosphoproteomics with phosphatase inhibitor mixture treatment of the heavy labeled SL2 cell population. Phosphopeptides were enriched by applying two different strategies (supplemental Fig. 1) and analyzed by high-resolution mass spectrometry. B, experimental procedure for quantitative proteomics and phosphoproteomics in response to Ptp61F deletion by RNA interference. For proteome analysis extracted proteins were digested in gel after one-dimensional SDS-gelelectrophoresis. For phosphoproteome analysis extracted proteins were both in-gel and in-solution digested and enriched for phosphopeptides (supplemental Fig. 3).
First we SILAC-labeled SL2 cells and treated the heavy cell population with an inhibitor mixture against serine/threonine and tyrosine protein phosphatases for 15 min. The two SILAC populations were combined in equal amounts, and their total protein extract was in-solution digested. To enrich phosphopeptides, we employed both multiple rounds of TiO2 chromatography in batch mode and the combination of SCX followed by TiO2 chromatography (supplemental Fig. 1). Phosphopeptide fractions were separated by reverse-phase chromatography and on-line detected in a high-resolution linear ion trap-orbitrap instrument (LTQ-Orbitrap) (LC-MS/MS) (Fig. 1A). A phosphorylation site not responding to inhibitor treatment is detected as a phosphopeptide peak pair with the same peak intensities (ratio H/L = 1). An up-regulated phosphorylation site has a ratio H/L > 1 and a down-regulated one H/L < 1. In total, we identified 6,752 high-confidence phosphorylation sites (Class I sites: 99% peptide identification confidence and >75% amino acid localization confidence (8)). Of the phosphotyrosine sites, 88% were regulated upon phosphatase inhibitor treatment whereas only 44% of the phosphoserine/phosphothreonine sites changed by more than 1.5-fold (supplemental Fig. 2). This likely reflects the high efficiency of pervanadate in inhibiting tyrosine phosphatases (33, 34).
Next, we applied quantitative phosphoproteomics to RNAi of the phosphatase Ptp61F by down-regulating its expression in the heavy cell population. Cells were SILAC-labeled and dsRNA treated from the same day because both processes take about the same number of cell doublings and are completely independent. Both in-gel digestion coupled with TiO2 chromatography and in-solution digestion in combination with SCX and TiO2 chromatography were performed (supplemental Fig. 3). Since dsRNA treatment for 8 days can also alter the proteome, we additionally analyzed global protein expression changes (Fig. 1B). This allows normalization of the phosphorylation changes with respect to protein levels.
All quantitative MS data were analyzed together as described (29, 30, 33, 35). In total, we detected 10,043 high-confidence phosphorylation sites on 2,379 phosphoproteins of which 9,749 were quantifiable and represented our basal phosphoproteome (supplemental Table 9). This number is similar to other recent large-scale Drosophila phosphoproteome datasets (11, 12). There is a good overlap of phosphorylated proteins (65.9% to Zhai et al. 12 and 75.2% to Bodenmiller et al. 11) (supplemental Fig. 4A), especially because the other investigations used a different cell line (Kc167) under various conditions or homogenized embryos. However, while 1,274 phosphorylated proteins were identified in all three studies, 334 phosphoproteins were exclusively found in our study indicating that none of the Drosophila phosphoproteomes is exhaustive yet. This observation is even more striking at the level of phosphorylation sites. We detected 4,691 novel phosphorylation sites, while 5,051 sites (51.8%) were already covered by the other two studies (supplemental Fig. 4B).
Bioinformatics Analysis of the SL2 Phosphoproteome
Insects are at considerable evolutional distance to mammals, in which most phosphoproteomic studies have been done. Drosophila, in particular, serves as an important model organism, but so far it is not known how closely the phosphoproteome resembles that of mammals. We therefore investigated the overall properties of our high-confidence SL2 phosphoproteome using Gene Ontology (GO), kinase motif analysis, structural constraints, and evolutionary conservation.
Overall Properties of the Drosophila Phosphoproteome Are Similar to the Human Phosphoproteome
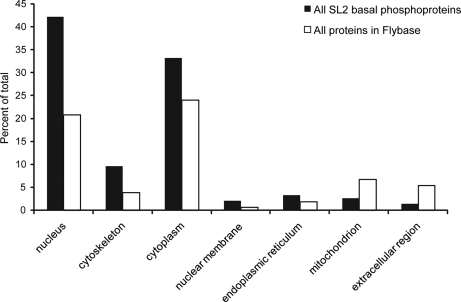
GO cellular compartment analysis, using Cytoscape (36) and BiNGO (37), revealed mitochondria and extracellular compartment to be underrepresented while nucleus, cytoplasma, and cytoskeleton were overrepresented (all with p < 0.001) (Fig. 2 and supplemental Table 1). Regulatory protein classes such as transcription factors, protein kinases, and GTPase regulating proteins were highly overrepresented in GO molecular function analysis (p < 10−15; supplemental Table 1). Furthermore, GO analysis after combining our dataset with that of Bodenmiller et al. (11) and Zhai et al. (12) (which yielded 4,871 phosphoproteins), showed similar trends, suggesting they are valid for the entire Drosophila phosphoproteome (supplemental Table 2). They also mirror recent analyses of a human and mouse cell line (8, 33).
Fig. 2.
Over- and under-represented cellular compartments in the basal phosphoproteome resulting from Gene Ontology analysis.
Phosphorylation Substrates in Drosophila Can Largely be Predicted by Human Kinase Motives
To ask whether SL2 phosphorylation sites match significantly with known human kinase motifs, we employed the χ2-Test as described in Pan et al. (33). The χ2-Test assesses the significance of the variance between observed and expected frequencies. This showed that fly phosphorylation sites matched significantly to most of the known kinase motifs, derived from human cells (supplemental Table 3). As an example, the CDK1 motif p[ST]-P-X-[KR] matches with the surrounding amino acids of identified phosphorylation sites six times more frequently than expected by chance. In the case of human the fold enrichment was twenty.
As described in Schwartz et al. (38), Motif-X is an iterative approach to derive significantly overrepresented motifs from large-scale datasets. A motif is built via recursive extraction of significant amino acid combinations in a given foreground set (surrounding sequence of phosphorylation sites) compared with a given background set. The application of Motif-X confirmed the findings of the χ2-Test (supplemental Table 4). For example, the second most significant consensus sequence was the CDK1 motif. We next applied Motif-X to phosphorylation sites detected in human HeLa cells (8). Overall, 27 significant sequence motifs matched exactly with those derived from the Drosophila set (supplemental Fig. 5) indicating a high degree of conservation of kinases and their signaling pathways ranging from CDK to ERK. This is in concordance with a previous comparison of various eukaryotic kinomes to the human kinome by Manning et al. (39). For example, fly and human share several kinase families involved in immunity, neurobiology as well as basic cellular functions such as the cell cycle (40).
Phosphorylation Sites Are Largely Confined to Loops and Hinges on the Protein Surface
Next we assigned secondary structure and accessibility constraints to phosphorylation sites employing the prediction method SABLE 2.0 (41). SABLE assigns the predicted secondary structure (coil, helix, or β-sheet) to each residue of a given input sequence along with calculated accessibility values ranging from 0 (fully buried) to 9 (fully exposed). As in our previous analysis of the human phosphoproteome (32), phosphorylation sites are almost exclusively located in coil regions of the surfaces of proteins (supplemental Fig. 6A). In total, 91% of phosphorylated serines were found to be located in loop regions in comparison to 77% of the serines that were not identified to be phosphorylated on the same proteins. The resulting degrees of accessibilities calculated by SABLE 2.0 were also significantly higher for phosphorylated residues compared with their non-phosphorylated counterpart (supplemental Fig. 6B). This is important for kinase and phosphatase substrate accessibility, as well as for subsequent functional effects on the substrate protein.
Drosophila melanogaster Phosphorylation Site Predictor
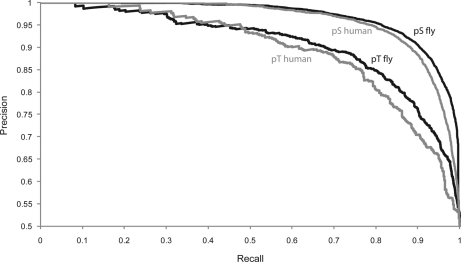
Because none of the Drosophila phosphoproteome datasets reaches completion, we used a machine learning approach to predict phosphorylation events in silico (32). We trained a support vector machine on the full dataset of 7,756 Ser(P), 1,427 Thr(P), and 325 Tyr(P) along with their surrounding sequences. Serines, threonines, and tyrosines that have not been proven to be phosphorylated were randomly chosen from the FlyBase database and defined as negative set. We used 90% of the given instances for training and 10% for testing the accuracy of prediction (5-fold cross validation). We found that 89.8% in the serine set and 81.1% in the threonine set were predicted correctly (Fig. 3). The accuracy of the prediction of phosphotyrosines was 63.0%, due to their smaller number in the training set. Note that this accuracy assumes equal numbers of phosphorylated and non-phosphorylated sites; therefore the accuracy for a given residue in the proteome will be lower. The predictor may prove useful to classify potential phosphorylation sites on proteins of interest, for example, for subsequent mutational studies.
Fig. 3.
Precision-Recall curves reflecting the accuracy of phosphoserine and phosphothreonine prediction using the fly-specific predictor. The recall reflects the proportion of true positives to the sum of true positives and false negatives, whereas the precision describes the number of true positives out of all predicted positives.
Application of the Drosophila-specific predictor to the human phospho dataset (8) showed only somewhat inferior performance compared with the predictor trained on human data (Fig. 3). This further supports the conclusion that human and insect phosphoproteome are generally similar overall. The predictor is available online via the Phosida database. Phosida also lists matching kinase motifs for any given predicted and experimentally found phosphorylation site, thus suggesting potential responsible kinases.
Evolutionary Conservation of the Drosophila Phosphoproteome
We used FlyBase (42) to assign our proteomic data to the genome and EnsEMBL (43) for chromosomal localization. The SL2 phosphoproteome (quantified and non-quantified sites) encompassed 2,454 gene transcripts, corresponding to 2,411 distinct genes. For annotation of the Drosophila genome, we employed the DAS source system using the Proserver technology (44, 45). This combination allows easy checking of individual genes and proteins starting from the genome as well as visualizing overall distribution patterns in the entire genome. As expected, our dataset did not show a specific pattern in the genome (supplemental Fig. 7).
The EnsEMBL Compara database contains phylogenetic relationships between fly and around 40 eukaryotes ranging from yeast to human. Besides the phylogenetic classifications between homologous proteins, it also stores global cDNA alignments. With assigned gene transcripts in hands, we compared the conservation of detected phosphorylated proteins with the one of all proteins from FlyBase that are defined as “known” due to former experimental evidence. We found that phosphorylated proteins show more orthologs throughout all eukaryotes than all other annotated proteins (supplemental Table 5 and supplemental Fig. 8). For example, around 70% of proteins that are phosphorylated in fly have orthologous counterparts in mosquito; in comparison, around 60% of the other known proteins are homologous to mosquito proteins. The same tendency was found for the homology to more distantly related species such as human or even yeast. The results of the analysis on the inter-species level are in concordance with ones of the intra-species level. We found that more than half (55%) of all fly proteins show paralogs, whereas a mere 27% of the phosphorylated subset shows homology within the same species. The higher proportion of non-phosphorylated proteins showing paralogy reflects faster evolution.
In addition, we assessed the preservation of the protein sequence for each homologous protein pair using the rates of synonymous and non-synonymous base pair substitutions (dN/dS ratio) (46). The median dN/dS ratio of orthologous phosphoproteins was found to be 0.25 compared with 0.33 in the case of all homologous fly proteins, indicating that there is negative selection for amino acid changes within the sequence of phosphorylated proteins.
In conclusion, evolutionary analysis indicates strong conservation of phosphoproteins, presumably because of their key regulatory functions; for example, in cell signaling. Phylogenetic relationships and global alignments between homologous proteins are displayed in the Phosida database.
Proteome-wide Analysis of Phosphatase Ptp61F RNAi
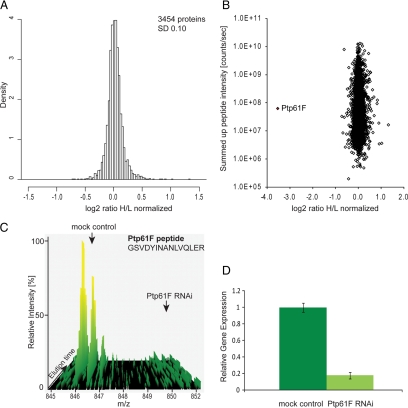
To in-depth characterize proteome- and phosphoproteome-wide responses of the cell to the depletion of the phosphatase Ptp61F, we combined SILAC-based proteomics with RNA interference. The quantitative MS data of the proteome and phosphoproteome analysis of Ptp61F RNAi (Fig. 1B) were analyzed together as described (29, 30, 33, 35). In total 4,132 proteins were identified with a false discovery rate of 1% and at least two unique peptides; out of these 3,454 proteins were quantified with a minimum of three quantitation events (three SILAC pairs) (supplemental Table 10). The histogram of the normalized H/L SILAC ratios is shown in Fig. 4A and is somewhat narrower than a normal distribution, indicating that no gross proteome changes occurred. In Fig. 4B the same ratios are plotted as a function of added peptide signal. The highest significant outlier is the phosphatase, which is down-regulated more than 90% (Fig. 4, B and C). This agrees with 80% decrease of its mRNA in real-time PCR measurements (Fig. 4D).
Fig. 4.
RNAi of the tyrosine phosphatase Ptp61F and the effect on the proteome. A, histogram of log2-transformed normalized protein ratios for all quantified proteins in the Ptp61F RNAi experiment. B, plot of the normalized ratios of all quantified proteins plotted against their summed heavy and light peptide intensities. C, MS- based analysis of RNAi depletion: a peptide-pair for Ptp61F from the SILAC-based Ptp61F RNAi proteome experiment, where the phosphatase was depleted in the heavy samples. The Ptp61F peptide (GSVDYINANLVQLER) shows a heavy to light ratio (H/L) of 0.073 corresponding to a more than 10-fold reduction of the protein level. D, quantitative RT-PCR of Ptp61F transcript derived from mock control and Ptp61F RNAi cells. Ptp61F mRNA level is reduced to less than 20% by RNA interference. Error bars represent the standard deviation of three different technical replicates. GAPDH transcript levels were used for normalization.
The tight distribution around the one-to-one ratio indicates the high accuracy of protein quantitation. We defined significantly regulated proteins as those that changed at least 1.5-fold and the fold change of which had an intensity-dependent statistical significance of at least 0.002 (termed “significance B”). Applying this level of stringency the detected proteome changes are restricted to sixteen up-regulated proteins and six down-regulated proteins (supplemental Table 6). This suggests - at least at the level of the proteome that was quantified in our experiments - that the response to the depletion of the phosphatase has limited effect at the proteome level, and that our RNAi had few if any off-target effects. The proteins found up-regulated are mainly proteins involved in metabolic pathways like the glycolytic enzyme phosphofructokinase or the glutamine synthase 2.
An important purpose of the proteome analysis was the possibility to correct changes at the phosphorylation level by changes at the protein expression level. In our dataset 77% of the phosphorylation sites could be normalized to their corresponding protein expression levels (supplemental Table 10). Interestingly, the remaining phosphorylation sites were not significantly associated with low intensity phosphopeptides. Thus they probably reflect the different selectivity of phosphopeptide enrichment compared with the whole proteome measurement.
Global Phosphoproteome Response to Ptp61F RNAi
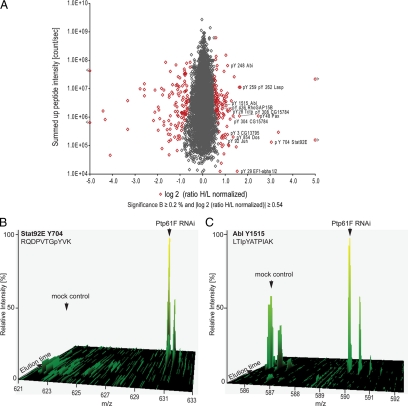
In total 6,478 phosphorylation sites on 1,904 proteins were detected upon Ptp61F RNAi (supplemental Table 10). Appling the same criteria as for the proteome, 288 phosphorylation sites changed significantly, comprising 217 serines, 45 threonines, and 26 tyrosines (Fig. 5A). Thus, in total 4.4% of all detected phosphorylation sites were affected upon depletion of a single tyrosine phosphatase. Direct targets of a tyrosine phosphatase should show up-regulated tyrosine phosphorylation upon phosphatase RNAi. (Note that we use “up-regulated phosphorylation” to denote increase in phosphorylation after accounting for protein expression changes.) We detected 15 proteins with 19 up-regulated tyrosine phosphorylation sites (supplemental Table 7 and Fig. 6). The sole Drosophila Stat (STAT92E) serves as positive control because Ptp61F is a known suppressor of STAT92E-dependent transcription (16, 17). The phosphorylation site Tyr-704 of this transcription factor determines its dimerization and accumulation in the nucleus (47, 48). It was found 10-fold up-regulated upon Ptp61F RNAi (Fig. 5B), which represents the most extreme tyrosine responder in the entire dataset.
Fig. 5.
Quantitative phosphoproteome upon Ptp61F RNAi. A, plot of normalized ratios of all quantified phosphorylation sites against their summed heavy and light peptide intensities (n = 6537). B, SILAC peptide pair of STAT92E, the most up-regulated phosphotyrosine peptide upon Ptp61F RNAi. C, SILAC peptide- pair of Abl, 1.9-fold up-regulation on a C-terminal autophosphoylation site.
Fig. 6.
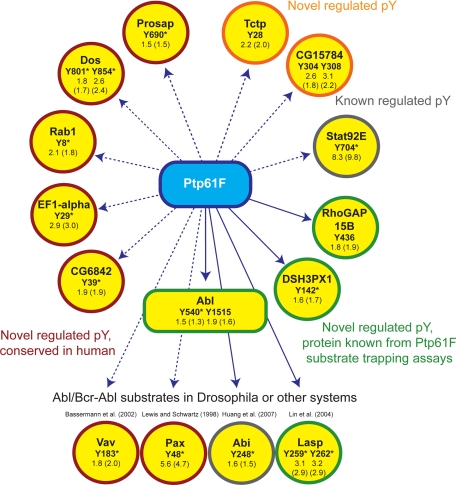
Direct and indirect targets of Ptp61F. The H/L phosphopeptide ratios upon Ptp61F RNAi and in brackets their corresponding values normalized to the protein expression are indicated. Here only phosphotyrosines with normalization and a ratio belonging to a singly phosphorylated peptide are shown. Tyrosines with asterisk are conserved in human (supplemental Fig. 9).
One category of proteins with up-regulated tyrosine phosphorylation sites are likely direct substrates of Ptp61F because they are also found in a recent substrate trapping assay (20). These are the non-receptor protein tyrosine kinase Abl, its interacting protein Abi (also found by Huang et al. (18)), the focal adhesion protein Lasp, the GTPase activating protein RhoGAP15B, and the sorting nexin DSH3PX1. However, the substrate trapping experiment did not identify specific sites, and our experiment now provides this important information (Fig. 6). The kinase Ack was also identified by substrate trapping. The high-confidence tyrosine phosphorylation site Tyr-987 of Ack was up-regulated 1.3-fold but was not statistically significant by our definition (Fig. 5A). This was also the case for the tyrosine phosphorylation sites of SCAR (1.3-fold) and Pvr (1.2-fold), suggesting that our significant thresholds are conservative.
The focal adhesion component Paxillin and the guanine-nucleotide exchange factor Vav are potential novel targets of Ptp61F with a functional link to cytoskeleton organization. In support, mammalian Paxillin was identified in a PTP1B substrate assay experiment (19). Furthermore, the same study found increased tyrosine phosphorylation of mammalian Vav3 comparing wild type and in PTP1B-deficient mouse fibroblasts by quantitative phosphotyrosine proteomics. The tyrosines of the identified Paxillin and Vav phosphorylation site are conserved but not yet described as PTP1B target sites. The translation elongation factor EF1-alpha, the vacuolar protein sorting 4 homolog CG6842, the adaptor protein Dos, the GTPase Rab1, and Prosap are potential novel Ptp61F substrates with conserved tyrosine sites in human (supplemental Fig. 10). The positive regulator of Stat92E-dependent signaling CG15784 and the guanine-nucleotide exchange factor Tctp are novel potential substrates where either the protein or the detected regulated phosphorylation site is not conserved in human.
Our approach is designed to measure systems-wide effects of pathway perturbation in an unbiased way, rather than to discriminate direct and indirect actions of the phosphatase. In addition to up-regulation of direct targets, changes in the tyrosine phosphorylation levels can also be due to activation of substrate tyrosine kinases. Illustrating this point, we found the direct target Abl with increased phosphorylation on Tyr-540 within the activation loop and on Tyr-1515 within a potential autophosphorylation sequence at the C terminus (Fig. 5C). The activation loop phosphorylation of Abl has been detected in oncogenic Abl kinases in vivo (49), and this is the first direct observation of endogenous activation at this site upon Ptp61F depletion. Enhanced tyrosine phosphorylation of Abl in general and activation loop phosphorylation in particular increases its kinase activity (50, 51). Activated Abl propagates the effects of the Ptp61F RNAi to a second layer of effector molecules. Several proteins with up-regulated tyrosine phosphorylation in our experiment are known to be phosphorylated by Bcr-Abl or c-Abl in human cell lines. These include Paxillin (52, 53) and Vav1 (54).
Abi is an interesting example because its phosphorylation is regulated by both Abl and Ptp61F (18). Thus its increased phosphorylation upon Ptp61F RNAi is the combined effect of increased Abl activity and depleted phosphatase. Our in vivo screen shows that the net effect is a 1.5-fold up-regulation of Tyr-248. Similarly, the up-regulation of Paxillin and Vav phosphorylation sites, in our experiment could reflect the separate or combined action of Abl and Ptp61F. Since mammalian Paxillin was also identified in a PTP1B substrate assay experiment (19) a dual action of Abl and Ptp61F on Paxillin is likely.
With regards to the up-regulation of several metabolic enzymes at the protein level, it is interesting that Bcr-Abl signaling has been linked to up-regulation of phosphofructokinase expression level (55). Furthermore, Bcr-Abl signaling activates c-Jun NH2-terminal kinase (JNK) (56), possibly providing the mechanism for increased threonine phosphorylation on the fly homolog dJun within the transactivation domain (57).
Altogether, we detected 101 up- and 115 down-regulated phosphorylation sites upon Ptp61F RNAi (supplemental Table 7), emphasizing the broad effects of the phosphatase on the phosphoproteome. There are a number of serine/threonine kinases whose phosphorylation level is altered such as Fray (1.6-fold up) and MEKK-1 (1.3-fold up), which although not all passing our threshold for significance could be the mechanistic link to these broad effects. Note that phosphorylation of a kinase can either enhance or inhibit its activity.
Conclusion
Here we have, for the first time, combined quantitative proteomics and phosphoproteomics to study the effects of perturbing phosphorylation-dependent signaling network by ablating one of its components. This approach differs from “chemical proteomics” involving ATP-based enrichment or classical in vitro approaches in that it measures the net effect of the perturbation on the signaling network and the expressed proteome in a completely generic and hypothesis-free approach. Effects on the phosphorylation pattern are captured at the quantitative phosphoproteomics level, whereas resulting gene expression changes are measured by quantitative proteomics. These effects are both upstream and downstream of the intermediate output measured by mRNA-based technologies.
We also introduced the concept of parallel acquisition of a proteome and a phosphoproteome for the system-wide analysis of a perturbed system. This is generally necessary in perturbations that last longer than the time required for protein expression changes (generally 20–30 min) because it enables assigning changes to the stoichiometry of the phosphorylation site (phosphorylation change relative to protein amount) and the absolute phosphorylation changes resulting from a protein expression change. Biologically, the total amount of phosphorylation, the relative fraction of phosphorylation, or both, may be important.
We chose to ablate the Drosophila homolog of a disease relevant phosphotyrosine phosphatase, Ptp61F by RNAi. The detected potential substrates were enriched for the categories cytoskeleton regulation and adhesion signaling and thus our data further support and expand a role of Ptp61F in these processes. Our results show a limited effect of the Ptp61F RNAi on the proteome in contrast to a pronounced effect on the phosphoproteome. This indicates that the phosphatase predominantly affects phosphorylation-mediated signaling rather than downstream gene expression. Therefore a microarray experiment may not have been efficient at defining the most pertinent effects of this phosphatase knock-down.
Our proof of principle study clearly demonstrates the power of combined quantitative proteomics and phosphoproteomics to study system-wide effects of signaling network perturbation. With increasing throughput and automation of MS, signaling networks could routinely be probed in this way. Although we used RNAi, other obvious perturbations include gene knock-outs of kinases and phosphatases as well as small molecule inhibitors.
Acknowledgments
Juergen Cox and Nadin Neuhauser helped in the analysis of large-scale datasets, and Jesper Olsen advised on the phosphoproteome acquisition. Shubin Ren contributed to the construction of the Drosophila phosphorylation site predictor. We thank Reinhard Faessler and Ralf Boettcher of the Department for Molecular Medicine for critical reading and suggestions. We thank Ewan Birney, Andrew Jenkinson, and Albert Bertan of the European Bioinformatics Institute for help with establishing the DAS server and the use of the European Bioinformatics Institute Compara database.
Footnotes
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- MS
- mass spectrometry
- Abl
- Ableson kinase
- Abi
- Abl interacting protein
- DHB
- 2,5-dihydroxybenzoic acid
- GO
- Gene Ontology
- LC-MS
- liquid chromatography-mass spectrometry
- MS/MS
- tandem mass spectra
- RNAi
- RNA interference
- SCX
- strong cation exchange chromatography
- SILAC
- stable isotope labeling by amino acids in cell culture
- TFA
- trifluoroacetic acid
- RT
- reverse transcriptase
- SL2
- Schneider line 2
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GST
- glutathione S-transferase
- dsRNA
- double-stranded RNA
- DAS
- distributed annotation system.
REFERENCES
- 1.Aebersold R., Mann M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198– 207 [DOI] [PubMed] [Google Scholar]
- 2.Schmelzle K., White F. M. ( 2006) Phosphoproteomic approaches to elucidate cellular signaling networks. Curr. Opin. Biotechnol. 17, 406– 414 [DOI] [PubMed] [Google Scholar]
- 3.Witze E. S., Old W. M., Resing K. A., Ahn N. G. ( 2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798– 806 [DOI] [PubMed] [Google Scholar]
- 4.Ong S. E., Mann M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252– 262 [DOI] [PubMed] [Google Scholar]
- 5.Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. ( 2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301– 305 [DOI] [PubMed] [Google Scholar]
- 6.White F. M. ( 2008) Quantitative phosphoproteomic analysis of signaling network dynamics. Curr. Opin. Biotechnol. 19, 404– 409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. ( 2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376– 386 [DOI] [PubMed] [Google Scholar]
- 8.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. ( 2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635– 648 [DOI] [PubMed] [Google Scholar]
- 9.Worby C. A., Simonson-Leff N., Dixon J. E. ( 2001) RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE 2001, PL1. [DOI] [PubMed] [Google Scholar]
- 10.Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., Fortini M. E., Li P. W., Apweiler R., Fleischmann W., Cherry J. M., Henikoff S., Skupski M. P., Misra S., Ashburner M., Birney E., Boguski M. S., Brody T., Brokstein P., Celniker S. E., Chervitz S. A., Coates D., Cravchik A., Gabrielian A., Galle R. F., Gelbart W. M., George R. A., Goldstein L. S., Gong F., Guan P., Harris N. L., Hay B. A., Hoskins R. A., Li J., Li Z., Hynes R. O., Jones S. J., Kuehl P. M., Lemaitre B., Littleton J. T., Morrison D. K., Mungall C., O'Farrell P. H., Pickeral O. K., Shue C., Vosshall L. B., Zhang J., Zhao Q., Zheng X. H., Lewis S. ( 2000) Comparative genomics of the eukaryotes. Science 287, 2204– 2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodenmiller B., Malmstrom J., Gerrits B., Campbell D., Lam H., Schmidt A., Rinner O., Mueller L. N., Shannon P. T., Pedrioli P. G., Panse C., Lee H. K., Schlapbach R., Aebersold R. ( 2007) PhosphoPep–a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 3, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhai B., Villén J., Beausoleil S. A., Mintseris J., Gygi S. P. ( 2008) Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7, 1675– 1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonaldi T., Straub T., Cox J., Kumar C., Becker P. B., Mann M. ( 2008) Combined use of RNAi and quantitative proteomics to study gene function in Drosophila. Mol. Cell 31, 762– 772 [DOI] [PubMed] [Google Scholar]
- 14.Dubé N., Tremblay M. L. ( 2005) Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: from diabetes, obesity to cell cycle, and cancer. Biochim. Biophys. Acta 1754, 108– 117 [DOI] [PubMed] [Google Scholar]
- 15.Johnson T. O., Ermolieff J., Jirousek M. R. ( 2002) Protein tyrosine phosphatase 1B inhibitors for diabetes. Nat. Rev. Drug Discov. 1, 696– 709 [DOI] [PubMed] [Google Scholar]
- 16.Baeg G. H., Zhou R., Perrimon N. ( 2005) Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 19, 1861– 1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Müller P., Kuttenkeuler D., Gesellchen V., Zeidler M. P., Boutros M. ( 2005) Identification of JAK/STAT signaling components by genome-wide RNA interference. Nature 436, 871– 875 [DOI] [PubMed] [Google Scholar]
- 18.Huang C. H., Lin T. Y., Pan R. L., Juang J. L. ( 2007) The involvement of Abl and PTP61F in the regulation of Abi protein localization and stability and lamella formation in Drosophila S2 cells. J. Biol. Chem. 282, 32442– 32452 [DOI] [PubMed] [Google Scholar]
- 19.Mertins P., Eberl H. C., Renkawitz J., Olsen J. V., Tremblay M. L., Mann M., Ullrich A., Daub H. ( 2008) Investigation of protein tyrosine phosphatase 1B function by quantitative proteomics. Mol. Cell. Proteomics 7, 1763– 1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y. C., Lin S. Y., Liang S. Y., Pan K. T., Chou C. C., Chen C. H., Liao C. L., Khoo K. H., Meng T. C. ( 2008) Tyrosine phosphoproteomics and identification of substrates of protein tyrosine phosphatase dPTP61F in Drosophila S2 cells by mass spectrometry-based substrate trapping strategy. J. Proteome Res. 7, 1055– 1066 [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko A., Wilm M., Vorm O., Mann M. ( 1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850– 858 [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M. ( 2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856– 2860 [DOI] [PubMed] [Google Scholar]
- 23.Rappsilber J., Mann M., Ishihama Y. ( 2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896– 1906 [DOI] [PubMed] [Google Scholar]
- 24.Rappsilber J., Ishihama Y., Mann M. ( 2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663– 670 [DOI] [PubMed] [Google Scholar]
- 25.Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. ( 2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697– 707 [DOI] [PubMed] [Google Scholar]
- 26.Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. ( 2005) Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol. Cell. Proteomics 4, 873– 886 [DOI] [PubMed] [Google Scholar]
- 27.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. ( 2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010– 2021 [DOI] [PubMed] [Google Scholar]
- 28.Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. ( 2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590– 3598 [DOI] [PubMed] [Google Scholar]
- 29.Cox J., Mann M. ( 2007) Is proteomics the new genomics? Cell 130, 395– 398 [DOI] [PubMed] [Google Scholar]
- 30.Cox J., Mann M. ( 2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367– 1372 [DOI] [PubMed] [Google Scholar]
- 31.Livak K. J., Schmittgen T. D. ( 2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402– 408 [DOI] [PubMed] [Google Scholar]
- 32.Gnad F., Ren S., Cox J., Olsen J. V., Macek B., Oroshi M., Mann M. ( 2007) PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 8, R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan C., Gnad F., Olsen J. V., Mann M. ( 2008) Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics 8, 4534– 4546 [DOI] [PubMed] [Google Scholar]
- 34.Deleted in proof
- 35.Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. ( 2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672– 683 [DOI] [PubMed] [Google Scholar]
- 36.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. ( 2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498– 2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maere S., Heymans K., Kuiper M. ( 2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448– 3449 [DOI] [PubMed] [Google Scholar]
- 38.Schwartz D., Gygi S. P. ( 2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391– 1398 [DOI] [PubMed] [Google Scholar]
- 39.Manning G., Plowman G. D., Hunter T., Sudarsanam S. ( 2002) Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514– 520 [DOI] [PubMed] [Google Scholar]
- 40.Bettencourt-Dias M., Giet R., Sinka R., Mazumdar A., Lock W. G., Balloux F., Zafiropoulos P. J., Yamaguchi S., Winter S., Carthew R. W., Cooper M., Jones D., Frenz L., Glover D. M. ( 2004) Genome-wide survey of protein kinases required for cell cycle progression. Nature 432, 980– 987 [DOI] [PubMed] [Google Scholar]
- 41.Adamczak R., Porollo A., Meller J. ( 2005) Combining prediction of secondary structure and solvent accessibility in proteins. Proteins 59, 467– 475 [DOI] [PubMed] [Google Scholar]
- 42.Grumbling G., Strelets V. ( 2006) FlyBase: anatomical data, images and queries. Nucleic Acids Res. 34, D484– D488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flicek P., Aken B. L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Down T., Dyer S. C., Eyre T., Fitzgerald S., Fernandez-Banet J., Gräf S., Haider S., Hammond M., Holland R., Howe K. L., Howe K., Johnson N., Jenkinson A., Kähäri A., Keefe D., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Prlic A., Rice S., Rios D., Schuster M., Sealy I., Slater G., Smedley D., Spudich G., Trevanion S., Vilella A. J., Vogel J., White S., Wood M., Birney E., Cox T., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Hubbard T. J., Kasprzyk A., Proctor G., Smith J., Ureta-Vidal A., Searle S. ( 2008) Ensembl 2008. Nucleic Acids Res. 36, D707– D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowell R. D., Jokerst R. M., Day A., Eddy S. R., Stein L. ( 2001) The distributed annotation system. BMC Bioinformatics 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn R. D., Stalker J. W., Jackson D. K., Kulesha E., Clements J., Pettett R. ( 2007) ProServer: a simple, extensible Perl DAS server. Bioinformatics 23, 1568– 1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z., Li J., Zhao X. Q., Wang J., Wong G. K., Yu J. ( 2006) KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomic Proteomics Bioinformatics 4, 259– 263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan R., Small S., Desplan C., Dearolf C. R., Darnell J. E., Jr. ( 1996) Identification of a Stat gene that functions in Drosophila development. Cell 84, 421– 430 [DOI] [PubMed] [Google Scholar]
- 48.Arbouzova N. I., Zeidler M. P. ( 2006) JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development 133, 2605– 2616 [DOI] [PubMed] [Google Scholar]
- 49.Woodring P. J., Hunter T., Wang J. Y. ( 2003) Regulation of F-actin-dependent processes by the Abl family of tyrosine kinases. J. Cell Sci. 116, 2613– 2626 [DOI] [PubMed] [Google Scholar]
- 50.Henkemeyer M. J., Bennett R. L., Gertler F. B., Hoffmann F. M. ( 1988) DNA sequence, structure, and tyrosine kinase activity of the Drosophila melanogaster Abelson proto-oncogene homolog. Mol. Cell. Biol. 8, 843– 853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanis K. Q., Veach D., Duewel H. S., Bornmann W. G., Koleske A. J. ( 2003) Two distinct phosphorylation pathways have additive effects on Abl family kinase activation. Mol. Cell. Biol. 23, 3884– 3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis J. M., Schwartz M. A. ( 1998) Integrins regulate the association and phosphorylation of paxillin by c-Abl. J. Biol. Chem. 273, 14225– 14230 [DOI] [PubMed] [Google Scholar]
- 53.Salgia R., Brunkhorst B., Pisick E., Li J. L., Lo S. H., Chen L. B., Griffin J. D. ( 1995) Increased tyrosine phosphorylation of focal adhesion proteins in myeloid cell lines expressing p210BCR/ABL. Oncogene 11, 1149– 1155 [PubMed] [Google Scholar]
- 54.Bassermann F., Jahn T., Miething C., Seipel P., Bai R. Y., Coutinho S., Tybulewicz V. L., Peschel C., Duyster J. ( 2002) Association of Bcr-Abl with the proto-oncogene Vav is implicated in activation of the Rac-1 pathway. J. Biol. Chem. 277, 12437– 12445 [DOI] [PubMed] [Google Scholar]
- 55.Hickey F. B., Cotter T. G. ( 2006) Identification of transcriptional targets associated with the expression of p210 Bcr-Abl. Eur. J. Haematol. 76, 369– 383 [DOI] [PubMed] [Google Scholar]
- 56.Raitano A. B., Halpern J. R., Hambuch T. M., Sawyers C. L. ( 1995) The Bcr-Abl leukemia oncogene activates Jun kinase and requires Jun for transformation. Proc. Natl. Acad. Sci. U. S A. 92, 11746– 11750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franklin C. C., Sanchez V., Wagner F., Woodgett J. R., Kraft A. S. ( 1992) Phorbol ester-induced amino-terminal phosphorylation of human JUN but not JUNB regulates transcriptional activation. Proc. Natl. Acad. Sci. U. S A. 89, 7247– 7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Identified phosphorylated peptides and phosphorylated sites along with further information ranging from evolutionary conservation to structural constraints and Swiss-Prot annotations were uploaded to Phosida, the phosphorylation site database (32). Besides the fly phosphoproteome, Phosida also contains phosphorylation sites of mouse, human, and various prokaryotes and now provides the described Drosophila-specific phosphorylation site predictor. Furthermore, the genome annotation section of Phosida links directly to the EnsEMBL database, which displays our proteomic data on the basis of the DAS source management. The setup of Phosida DAS layers for a given gene of interest in EnsEMBL is described in the “Background” section of Phosida and in EnsEMBL.
Moreover, the proteomic data (identified peptides) were uploaded to MAPU - Max-Planck Unified Proteome Database. The detected peptides, which have been assigned to gene entries as described in this study, are also available via the MAPU DAS source in the EnsEMBL database.