Abstract
Conventional LC-MS/MS data analysis matches each precursor ion and fragmentation pattern to their best fit within databases of theoretical spectra, yielding a peptide identification. Confidence is estimated by a score but can be validated by statistics, false discovery rates, and/or manual validation. A weakness is that each ion is evaluated independently, discarding potentially useful cross-correlations. In a classical approach to de novo sequence analysis, mixtures of peptides differing only in a carboxyl-terminal isotopic label yield fragmentation spectra with single, unlabeled b-type ions but pairs of isotope-labeled y-type ions, facilitating confident assignments. To apply this principle to identification by fragmentation pattern matching, we developed Validator, software that recognizes isotopic peptide pairs and compares their identifications and fragmentation patterns. Testing Validator 1 on a Mascot results file from FT-ICR LC-MS/MS of 16O/18O-labeled yeast cell lysate peptides yielded 2,775 peptide pairs sharing a common identification but differing in carboxyl-terminal label. Comparing observed b- and y-ions with the predicted fragmentation pattern improved the threshold Mascot score for 5% false discovery from 36 to 22, significantly increasing both sensitivity and specificity. Validator 2, which identifies pairs by precursor mass difference alone before comparing observed fragmentation with that predicted by Mascot, found 2,021 isotopic pairs, similarly achieving improved sensitivity and specificity. Finally Validator 3, which finds pairs based on mass difference alone and then deconvolutes fragmentation patterns independently of Mascot, found 964 predicted peptides. Validator 3 allowed raw mass spectrometry data to be mined not only to validate Mascot results but also to discover peptides missed by Mascot. Using standard desktop hardware, the Validator 1–3 software processed the 11,536 spectra in the 93-MB Mascot .DAT file in less than 6 min (32 spectra/s), revealing high confidence peptide identifications without regard to Mascot score, far faster than manual or other independent validation methods.
MS/MS combined with informatics analysis is now a uniquely powerful approach for identifying the components of complex protein samples (1–3). Although new technologies have dramatically enhanced the speed, sensitivity, and precision of LC-MS/MS instrumentation (4), data analysis has neither kept pace with nor taken full advantage of these advances. Determining peptide sequences from fragment ion spectra remains a difficult problem, and three main strategies have matured (5). In de novo sequencing, the peptide sequence is inferred directly from the fragment ion spectra, and many algorithms have been developed to automate this process, including Lutefisk (6), PepNovo (7), NovoHMM (8), Peptide Identification via Integer linear Optimization (PILOT) (9), and others (10–13). Incomplete fragmentation patterns and low signal to noise (10) make this method difficult to implement as an exclusive means of peptide identification.
The most commonly used method involves comparing experimental MS/MS spectra to theoretical peptide fragmentation patterns derived from protein sequence databases (4) and reporting the best peptide match, which is then propagated forward through the process of determining likely protein components. Several programs are commonly used, including SEQUEST (14, 15), Mascot (16), and X! Tandem (17, 18). What these algorithms share is the determination of a score for a spectrum-peptide match and subsequently a protein identification, and it is the way in which these scores are assigned and interpreted that distinguishes them (19).
The third method for spectrum-peptide matching is a hybrid of de novo and database searching (5) in which small lengths of sequence are generated directly from the fragment ion spectra, and these “sequence tags” (20) are used to corroborate spectrum-database matches. Popular implementations of this strategy include DirecTag (21), GutenTag (22), and MultiTag (23). The limitations to this method include the requirement for consecutive fragmentation ions and the reliance on de novo algorithms to identify sequence tags.
Database search is highly susceptible to both overreporting false positives (low specificity) and underreporting true positives (low sensitivity). The search engines provide different scoring systems that cannot be directly compared, as the rankings of spectral quality are often based on arbitrary cutoff values. Recent research has focused less on the sequence matching algorithms themselves but more on the statistics used to evaluate the resulting match scores (24). PeptideProphet was one of the first algorithms developed to evaluate match scores and assign probabilities by evaluating each match with respect to all other peptide assignments. By using machine learning techniques (an expectation-maximization algorithm), PeptideProphet was shown to have high discriminating power for database search results (25). Initially developed for SEQUEST search results, PeptideProphet has been subsequently adapted for use with database search results from Mascot and X! Tandem. These components are combined in Scaffold, a commercial software suite developed by Proteome Software. An alternative approach is to filter the primary data to exclude poor quality MS/MS scans prior to the database search (26), thereby enhancing the likely significance of each reported match.
Using a false discovery rate instead of a false-positive rate is now the standard statistical measure for reporting error rates in data sets with large numbers of features (e.g. proteomics or genomics data) (5, 27). Target-decoy searching as an estimate of false discovery rate (FDR)1 involves first constructing a database of decoy peptides (28, 29), and this strategy is being incorporated into PeptideProphet (30, 31). For each peptide-spectrum match, the target spectrum is queried against a second (decoy) database with characteristics similar to those of the first (e.g. a database of reversed or random peptides). Matches to the decoy database are considered false discoveries, and the number of matches above a particular cutoff score threshold is reported. The target-decoy search option is now available in the newest version (version 2.2) of the database search engine Mascot (Matrix Science).
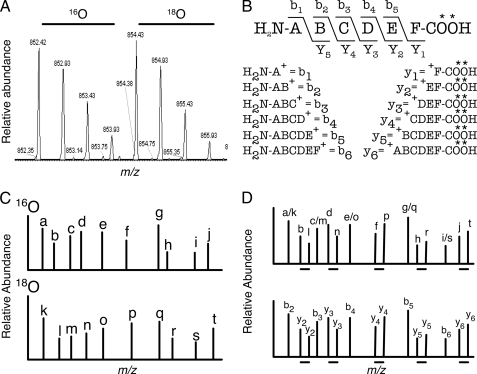
Despite these advances in mass spectrometry, database searching, and statistical approaches to validating matches, the process of analyzing mass spectrometry data remains time-consuming and computer processor-intensive, often requiring several steps and various data transformations (19). To overcome these limitations, we developed a fast and efficient method for peptide identification validation that minimizes the false discovery rate. Our algorithm relies on data from stable isotopic labeling, which is a standard method for quantifying relative protein abundance in complex mixtures (see Ref. 32 and references therein). Carboxyl-terminal labeling methods, including trypsin-catalyzed 18O exchange (33), result in a mixture of pairs of chemically identical but isotopically distinct peptides. The “light” and “heavy” peptides co-elute from HPLC but are readily distinguished by precursor mass (Fig. 1A). Each peptide also has an isotopic envelope comprised of isotopologues, molecules that are identical in composition except they can contain any number of isotopes. In the case of trypsin-catalyzed 18O exchange, two 18O atoms are substituted for the two carboxyl-terminal 16O atoms. Comparison of CID fragmentation patterns of carboxyl terminus-labeled light and heavy precursors (or isotopologues) distinguishes b-type and y-type ions (34, 35). The carboxyl-terminal fragments (y-ions) appear as light (16O) and heavy (18O-substituted) forms, but the amino-terminal fragments (b-ions) display a single shared mass (Fig. 1, B–D).
Fig. 1.
Peptide pair identification strategy. A, shown is an example of experimental spectra of a 16O/18O-peptide pair. Each peptide has an isotopic envelope comprised of three to four different isotopologues containing zero to three molecules of 13C, 15N, or other naturally occurring stable isotopes. The 18O envelope is shifted by about 2.0 Da, reflecting the difference in mass due to the substitution of two 18O atoms. Note that the difference of 2.0 Da is due to the peptide having a 2+ charge state. Peptide pairs with a 1+ charge would be separated by about 4.0 Da. B, the b-type and y-type ions from the collision-induced dissociation of a peptide are shown. Any carboxyl-terminal substitution (as in 18O, indicated by *) will affect the y-ions exclusively. C, idealized sample MS/MS spectra from the peptide and ions in B. The spectra from the 16O- and 18O-peptide forms have similar patterns, although the peak heights may be different. D, top, the two spectra from C are overlaid to demonstrate that the b-ions will have a nearly identical mass-to-charge ratio, whereas the y-ions will have a shift reflective of the stable isotope substitution. In the example given, peaks “a” and “k” from C are both b-ions and therefore overlap, whereas peaks “b” and “l” are y-ions with l being shifted due to the substitution of two 18O atoms. Shifted ions are indicated with a horizontal bar underneath. By observing which ions overlap and which have shifted, the identities of the b- and y-ions can be inferred (D, bottom).
The technique of using isotopic pairs to enhance peptide identification is not new, and several authors have recognized that isotopic labeling could be used to differentiate carboxyl-terminal from amino-terminal peptide fragments to facilitate peptide sequence analysis (2, 33, 35–38). This method has been productively applied to de novo analysis (12, 39–45) and peptide mass fingerprinting (46). In addition, analogous techniques have been applied to the analysis of mixtures of modified and unmodified peptides by probing for peptide mass differences that match known post-translational modifications (47); other groups have used MS/MS spectra information to corroborate these matches and remove noise (48, 49). Finally, isotopic labeling with 18O has been used for manual validation of peptide identifications by observing the predicted mass shift of y-ions (50). Nevertheless, this strategy has yet to be harnessed as a means for automated data analysis and peptide search validation.
The goal of this study was to develop a set of software tools designed to provide rapid and automatic validation of peptide assignments by Mascot and to determine the relative benefit of reducing false discovery and the magnitude of loss of bona fide identifications. We hypothesized that the characteristic shifting of y-type ions between fragmentation spectra of light and heavy precursors might provide a robust check for validity of peptide assignment by database search. Here we demonstrate the feasibility of quickly and efficiently analyzing searched mass spectrometry data, determining within minutes which peptide and protein assignments are likely valid. In its simplest form, Validator 1, identified isotopic pairs in a Mascot results file and improved the 5% FDR cutoff from a Mascot score of 36 to 22, thereby capturing many true identifications that would otherwise have been discarded. A more advanced algorithm, Validator 3, that considers only precursor ion mass, charge, and fragmentation spectral data to identify isotopic pairs independently of any peptide identifications, not only rapidly validated the Mascot results but also discovered peptides that Mascot had failed to match. Our software suite, Validator 1–3, provides new and robust tools for rapid validation of searched LC-MS/MS data obtained in stable isotope experiments, offering improved sensitivity and specificity over database searching alone.
EXPERIMENTAL PROCEDURES
Standardized and Normalized Data Sets
To provide normalized data for our analysis, we prepared a complex soluble protein sample from budding yeast cell lysate. The sample was subjected to proteolysis by trypsin. In detail, the proteins were mixed with 6 μl of Rapigest (Waters) and 10 mm tris(2-carboxyethyl)phosphine HCl, denatured at 37 °C for 30 min, alkylated with 10 μl of 50 mm iodoacetamide at room temperature in the dark for 40 min, and digested with 1:50 (w/w) trypsin in 50 mm ammonium bicarbonate, pH 8.9, at 37 °C overnight. The Rapigest was removed by adding 5 μl of 1% TFA. The sample was split and was exchanged in 100% [18O]water or 100% [16O]water using the 18O Proteome Profiler kit (Sigma-Aldrich). MALDI-TOF analysis was used to follow the reaction. Finally this sample was mixed in equal amounts to create a 1:1 16O:18O reference sample. The resulting peptide mixture was then subjected to reverse phase nanoelectrospray ionization LC-MS/MS on the LTQ-FT instrument (Thermo) using a standard gradient (Zorbax 300SB-C18 column, 150 mm × 75 μm; 0.1% formic acid in water with 5–60% acetonitrile; 0.5%/min gradient). The LTQ-FT instrument was run in positive ion mode at 50,000-ppm resolution MS for ICR. Parent ions were selected for fragmentation by data-dependent analysis using a cycle of one MS scan for ICR (m/z 400–2000) and up to five MS/MS scans in the LTQ (m/z 50–2000) of the most abundant ions using 120-s dynamic exclusion. A normalized collision energy of 35 was used for low energy CID MS/MS of peptide ions. Under these conditions, a high fraction of the most abundant peptides had both the 16O and 18O monoisotopic species subjected to CID based on our preliminary data. The data set was analyzed by Mascot (version 2.2, Matrix Science) and X! Tandem (version 2007.01.01.1, Global Proteome Machine Organization) to identify peptides and proteins from the MS/MS spectra. Mascot was set up to search the NCBInr_20060910 database (selected for Saccharomyces cerevisiae, 11,101 entries) assuming the digestion enzyme trypsin, a fragment ion mass tolerance of 1.0 Da, and a parent ion tolerance of 0.2 Da. Double 18O modification of carboxyl-terminal lysine or arginine, oxidation of methionine, N-formylation of the amino terminus, and iodoacetic acid derivative of cysteine were specified as variable modifications. X! Tandem was set to search the scd.fasta.pro database (selected for S. cerevisiae, 6,794 entries) also assuming trypsin with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10.0 ppm. Iodoacetamide derivative of cysteine was specified as a fixed modification. Double 18O modification, deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, sulfone of methionine, tryptophan oxidation to formyl, and acetylation of lysine and the amino terminus were specified as variable modifications. Scaffold (version Scaffold-01_06_00, Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications are accepted if they can be established at greater than 90.0% probability as specified by the PeptideProphet algorithm (51). Protein identifications are accepted at greater than 95.0% probability and contain at least one identified peptide with probabilities assigned by the ProteinProphet algorithm. Proteins that contain similar peptides and cannot be differentiated based on MS/MS analysis alone are grouped to satisfy the principles of parsimony.
Software Development
All software analysis was performed on searched Mascot data (e.g.“.DAT files”). Custom software was written in Python 2.6. Statistical analysis was performed using both Python scripting as well as Microsoft Excel. Charts and graphs were generated using both Python's Matplotlib library (SourceForge, Inc.) and GraphPad Prism. Software was run on standard desktop and laptop computers running both Windows XP (service pack 3) and Macintosh OS 10.5. Details about software development and implementation are included under “Results.”
RESULTS
The aim of this study is to describe a fast and efficient means for validating peptide identifications obtained by searching 18O-labeled MS/MS data with Mascot. Our approach is to mine the Mascot .DAT file to extract information not utilized by Mascot but potentially useful for automated validation. For the purposes of this study, we refer to a “query” as any precursor ion and its associated fragmentation ions, regardless of whether Mascot assigned a match, and to a “peptide” as any query to which Mascot assigned a match, regardless of Mascot score and without external validation. For each query, up to 10 possible peptides are assigned by Mascot, each with a probability score. For this study, we examined all query-peptide identifications as well as only the top scoring match suggested by Mascot. Using a 16O/18O-labeled data set from yeast cell lysate, analysis of the Mascot .DAT file revealed 20,759 queries and 17,200 peptide identifications, corresponding to 13,158 unique peptides and 5,962 unique proteins, using only the top suggested Mascot peptide identification (Table I). The FDR of 5% was achieved at a threshold Mascot peptide score of 36, and 2% was achieved at a cutoff score of 42.
Table I. Validator data.
For each version of Validator, the number of pairs, queries, and queries with peptides is shown. In addition, data are displayed after filtering the raw Mascot data for only those peptides with scores greater than 35. The precursor mass error range corresponds to the dotted (“all”) and solid (“>35”) lines in Fig. 3. NA, not applicable.
| Version | Raw | Raw >35 | 1 | 2 | 2e | 3 | 3e |
|---|---|---|---|---|---|---|---|
| Pairs identified | NA | NA | 2,775 | 3,209 | NA | 3,779 | 2,021 |
| Mascot queries | 20,759 | 2,308 | 2,345 | 3,185 | 1,782 | 3,615 | 2,310 |
| Queries with peptides | 17,200 | 2,308 | 2,345 | 3,177 | 1,782 | 3,545 | 2,289 |
| PME range (±) with 95%: all | 0.193 | 0.024 | 0.022 | 0.134 | 0.042 | 0.142 | 0.129 |
| PME range (±) with 95%: >35 | 0.024 | 0.024 | 0.017 | 0.011 | 0.011 | 0.011 | 0.013 |
| Unique peptides | 13,158 | 580 | 398 | 1,564 | 481 | 1,881 | 964 |
| Unique proteins | 5,962 | 186 | 125 | 1,150 | 234 | 1,391 | 696 |
| Score at FDR 5% | 36 | 36 | 22 | 36 | 29 | 37 | 37 |
| Score at FDR 2% | 42 | 42 | 32 | 41 | 34 | 43 | 43 |
| Percentage of queries with Mascot score >35 | 13.4 | 100 | 78.0 | 46.6 | 75.2 | 42.1 | 57.1 |
The majority of peptides have low Mascot scores (Fig. 2A). As expected, peptides with the highest Mascot scores tend to have a low precursor mass error (PME) (Fig. 3A). In fact, the search results represent two populations: peptides with high Mascot score/low PME and peptides with low Mascot score/high PME. A plot of the Mascot score versus the variance of the PME for all peptide matches above that score illustrates a steep fall in the variance, plateauing close to a Mascot score of 35 (supplemental Fig. 1), providing an approximate cutoff threshold separating the two populations. Of the 17,200 peptides identified by Mascot, 2,308 have scores greater than 35. The width of precursor mass error range that encompasses 95% of these peptides with high Mascot scores is 0.048 Da, whereas the interval that covers 95% of all peptides is 0.386 Da (Fig. 3).
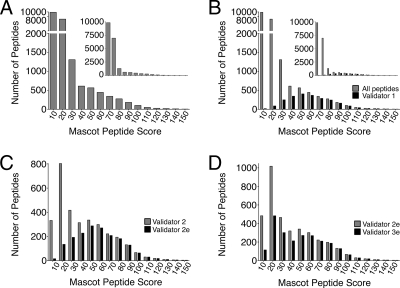
Fig. 2.
Distribution of Mascot scores. A, the raw Mascot data file was parsed, and the number of peptides in each score group was tallied. The vast majority of scores were less than 30. Note that the y axis has a break at 2,000. See the inset for the full-scale graph with identical x axis but no break in the y axis. B, Validator 1 finds 16O/18O pairs in the searched Mascot data file. The distribution of Validator 1-derived peptide scores (black) is seen against the raw distribution (gray) from A. Again, note the broken y axis and the inset showing the full y axis scale. At the low end of the scores, Validator 1 rejects most of the peptides while retaining most of the high scoring peptides. C, the Validator 2e-identified peptides with fragment ion tallies greater than 10 (black) are shown compared with the Validator 2 results (gray). At low scores, Validator 2e rejects most low scoring peptides while retaining most peptides with high Mascot scores. D, Validator 3e (black) performs similarly to Validator 2e (gray) despite not utilizing any Mascot search information.
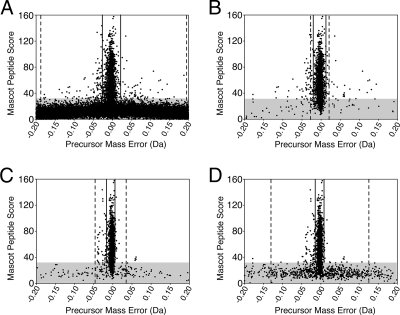
Fig. 3.
Precursor mass error versus Mascot score. Low Mascot peptide scores, as defined as a score less than 35, are shown in the shaded gray area. A, the raw data are separated into two distinct zones: the high Mascot score peptides, most with low precursor mass error, and the low Mascot score peptides, most with high precursor mass error. As the Mascot score increases from 0 to 35, the variance of the precursor mass errors of all peptide matches above this score falls dramatically (see also supplemental Fig. 1). We determined cutoffs for precursor mass error that would encompass 95% of all peptides (dashed lines) and 95% of peptides with Mascot peptide scores over 35 (solid lines). B, Validator 1 successfully removes most of the peptides with low Mascot peptide scores. Note the more narrow 95% range for all peptides (dashed lines) compared with A as well as the much tighter 95% interval for peptides with Mascot peptide scores greater than 35 (solid lines). C, Validator 2e-identified peptides with a fragment ion tally of 10 or more are shown. Note that although the interval encompassing 95% of the peptides (dashed lines) is wider than for Validator 1 it is much narrower than for the raw data. In addition, the 95% interval for peptides with Mascot peptide scores greater than 35 (solid lines) is narrower than for Validator 1-identified peptides. D, Validator 3e-identified peptides with a fragment ion tally of at least 10 are shown. Again the intervals encompassing 95% of the peptides (dashed lines) and 95% of peptides with Mascot scores greater than 35 (solid lines) are shown.
Validator 1
As a proof of concept, we first sought to find all 16O/18O pairs in the Mascot summary file (“.DAT file”). Here a 16O/18O pair refers to a peptide sequence identified in two distinct isotopic forms in the same Mascot file as an unlabeled 16O-peptide and as a peptide containing two 18O atoms. The 18O form of each peptide is 4.008491 Da heavier than its unlabeled 16O form (Unimod). Our first program, Validator 1, is designed to utilize the peptide identifications made by Mascot. Validator 1 first iterates through all queries looking for identical top scoring peptides found in both 16O and 18O forms (a “16O/18O pair”). As the 16O and 18O forms are expected to co-elute from reverse phase columns, we added a constraint that the MS/MS scans of the two peptides must occur within 200 scan units (∼2.25 min) of each other. With these criteria, Validator 1 identified 2,775 pairs representing 2,345 unique matched queries with peptides. These peptides represented 398 unique peptides and 125 unique proteins (Table I). This analysis required ∼10 s of calculation on a laptop computer. The precursor mass range width that encloses 95% of the peptides with Mascot scores greater than 35 was 0.034 Da, whereas the width of the range that encompasses 95% of all peptides decreased by 89% compared with Mascot alone, to 0.044 Da (Fig. 3, A versus B).
There were 223 unique peptides with Mascot scores over 35 that Validator 1 failed to discover as a member of a 16O/18O pair. Manual examination of the raw spectra for 10 of the highest scoring of these peptides revealed three scenarios. For six peptides, the 16O form was fragmented and yielded a high Mascot score, but the 18O form was not selected for MS/MS. In one case, the 18O form subjected to MS/MS was an isotopologue not accounted for by the Mascot search and thus was not correctly identified. In three cases, a candidate pair was flagged by Validator 1, but the data turned out to correspond to two peaks within the isotopic envelope of a single peptide.
On the other hand, Validator 1 did not reject all low scoring peptides, particularly where the Mascot identifications yielded low precursor mass errors. As seen in Fig. 3B, these peptides represent a “comet tail” in the data, stretching all the way down to Mascot scores as low as 10. A closer inspection of these peptides (data not shown) reveals that most were also found in other queries with high Mascot scores. Nevertheless, of the low scoring peptides found by Validator 1, there were 21 proteins represented that would not be identified if only high Mascot scoring peptides were being retained.
Therefore, Validator 1 was able to rapidly identify 16O/18O pairs within searched Mascot data. Using 16O/18O pairs as a criterion rather than a simple Mascot threshold retained most high scoring peptides and rejected most low scoring peptides but also rescued several low scoring but likely correct identifications.
Validator 2
Validator 1 relies on Mascot to identify both the 16O- and 18O-labeled peptides. We reasoned that additional 16O/18O pairs might be found in the Mascot .DAT file by searching for pairs of queries where the precursor masses were separated by a difference of 4.008491 Da without regard to any features of the MS/MS data or whether Mascot had assigned the same, different, or even any identifications. Thus, the Validator program was modified to start with a query identified as a 16O- or 18O-peptide and search the Mascot .DAT file for queries within a range of 200 scan units (2.25 min) with a precursor mass difference of 4.008491 Da and with a mass error limit of 3 ppm. Using these criteria, Validator 2 found 3,209 pairs representing 1,564 unique peptides and 1,150 unique proteins.
The most significant distinction between Validator 1 and 2 was the retention of considerably more low scoring peptides. Notably, of the 3,177 peptides retained by Validator 2, 1,696 had Mascot scores below 35, and many also displayed a high mass error, suggesting a low likelihood of correct identification. These results raised the question of whether using additional criteria based on the MS/MS data embedded in the Mascot data file might help reveal potentially correct peptide matches with low Mascot peptide scores while filtering out incorrect identifications.
Validator 2e
Given that fragmentation spectra are available for each member of a candidate 16O/18O-peptide pair identified by Validator 1 or 2, we hypothesized that these data could be mined to distinguish false identifications. As noted above, comparing the MS/MS fragmentation of the light and heavy forms will reveal identical sets of b-ions but distinct y-ions with pairs of fragments shifted by 4.008491 Da, reflecting the exchange of two 18O atoms for 16O at the carboxyl terminus (Fig. 1). We therefore extended our program, dubbed Validator 2e, to take advantage of the embedded carboxyl-terminal labeling information to distinguish the b-type and y-type ions, facilitating peptide validation.
As a first step, we confirmed that the MS/MS ions in each query correspond with a theoretical fragmentation table based on the sequence of the peptide match provided by Mascot. For each peptide identification in the Mascot data file, we calculated the fragmentation table and counted the number of observed ions that fell within a window of 2000 ppm from a predicted b- or y-ion. As expected, there is a positive correlation between the number of b- and y-ion matches and Mascot peptide score (r = 0.596, p < 0.0001; supplemental Fig. 2A). To validate Mascot identifications for 16O/18O pairs, we tested whether the following held true: when pairs of ions matched predicted b-type ions, they should be identical (non-shifting), whereas those matching y-ions should differ by 4.008491 Da (shifting). The number of matching pairs of non-shifting b-ions and shifting y-ions were thus tallied to generate a “fragment ion tally.” We hypothesized that a high fragment ion tally would characterize a correct peptide identification for a query member of a 16O/18O pair.
For each pair identified by Validator 2, we calculated the fragment ion tally for each query member based on comparison with predicted fragmentation tables for the highest scoring peptide match provided by Mascot. Fragment ion tally correlates with a high Mascot peptide score (r = 0.639, p < 0.0001; supplemental Fig. 2B) with a fragment ion tally of 10 corresponding to a Mascot score of 35. We therefore filtered the list generated by Validator 2 to retain only pairs that yielded a fragment ion tally of at least 10 with at least two matching shifting (y-type) ions. The requirement of two y-ion (shifting) matches will reject pairs of ions derived from the same isotopic envelope that are predicted to yield many matching b-ions but no matching y-ions. Calculating fragment ion tallies for the 3,209 pairs of queries found by Validator 2 yielded 1,782 queries with counts greater than or equal to 10 (Table I). These queries represent 481 unique peptides and 234 proteins. Notably, of the query-peptide matches with fragment ion tallies of 10 or greater, only 442 (24.8%) had Mascot scores less than 35. Compared with Validator 2, Validator 2e eliminates many of the low scoring/high mass error peptides but retains most of the high scoring/low mass error peptides (Fig. 2C). Limiting the plot to peptides evaluated with Validator 2e that yield a fragment ion tally of 10 or greater, 95% of high scoring peptides fell within a precursor mass error range of 0.022 Da versus a range of 0.084 Da for all peptides (Fig. 3C). Compared with Validator 1, Validator 2e found 219 queries, 163 peptides, and 135 proteins not found by Validator 1 (supplemental Table 1).
Validator 3/3e
As a next logical step, we sought to find candidate pairs based solely on their mass difference and ion lists from raw data without regard to any peptide sequence information provided by Mascot in the .DAT file. Validator 3 identifies pairs much like Validator 2 except for not requiring that one member of the pair be a Mascot-identified 16O- or 18O-peptide. The program iterates through all queries and searches for another query with the predicted 4.008491-Da mass difference, allowing an error of 3 ppm. From the reference data set, the program identified 3,779 pairs, representing 3,615 unique queries, of which 3,545 have Mascot-assigned peptide identifications. Examination of the data revealed that some Validator 1 pairs remained unidentified, as their difference in precursor mass lies outside the 3-ppm tolerance limit imposed by Validator 3 (data not shown). Validator 3 found 1,875 queries, 1,540 peptides, and 1,279 proteins not found by Validator 1 (supplemental Table 1).
As with Validator 2e, we extended Validator 3 to 3e by utilizing the expectation of non-shifting b-ions and shifting y-ions to perform an internal validation of the proposed pairs, without relying on the peptide identification(s) provided by Mascot. Therefore Validator 3 was modified to find pairs of shifting and non-shifting fragment ions for each pair based on comparing the two lists of MS/MS ions and finding non-shifting b-ions and shifting y-ions within a mass tolerance of 2,000 ppm. To decrease the influence of noise, only fragment ions with a peak height of at least 0.5% of the intensity of the strongest ion were evaluated. To be considered a shifting or non-shifting pair, the difference in intensity between the heavy and light forms of the candidate could be no more than 25%. Again a fragment ion tally was determined from the number of pairs of candidate b- (non-shifting) and y (shifting)-ions while requiring at least two y-ions. To validate the scoring scheme, the fragment ion tally and Mascot peptide scores were compared, and as with Validator 2e, we found a significant positive correlation (r = 0.395, p < 0.0001; supplemental Fig. 2C).
Because two complete sets of MS/MS ions are being compared without regard to a predicted fragmentation pattern, we expected to identify more pairs with higher fragment ion tallies. To facilitate comparison with Validator 2e, we filtered based on a fragment ion tally cutoff of 10, yielding 2,310 queries (Table I). These correspond to 964 peptides and 696 proteins identified. As expected, Validator 3e was less selective than Validator 2e in rejecting low scoring peptides (Fig. 2D) while retaining a higher proportion of high mass error peptides (Fig. 3D). The precursor mass error range containing 95% of peptides with scores greater than 35 was quite similar to that of Validator 2e, 0.026 versus 0.022 Da, but considerably wider for all peptides, 0.258 versus 0.084 Da. These data show that a strategy agnostic to Mascot-specific peptide information can be used to identify peptides highly likely to represent bona fide 16O/18O pairs, providing independent validation for Mascot identifications.
Comparison with Scaffold
The commercial proteomics software suite Scaffold (Proteome Software) uses the PeptideProphet algorithm (25) to generate lists of peptides and proteins with an associated probability. Many groups use Scaffold for downstream data analysis, and we feel that it is important to compare the performance of our software with that of this commonly used analysis tool. Using the same Mascot .DAT file, the data were analyzed in Scaffold using probability cutoffs for peptides and proteins of 90 and 95%, respectively. The list of proteins meeting these criteria along with the constituent peptides was compared with the peptide and protein lists generated by Validator versions 1–3e (Table II). Using the top scoring Mascot peptide identifications only, Validator 1 found 69.5% of the peptides and 91.9% of the proteins found by Scaffold. The performance of Validator 2e was similar, identifying 62.6 and 84.9% of the peptides and proteins, respectively. Validator 3e found 59.1% of the peptides and 88.4% of the proteins found by Scaffold. The seven proteins identified by Scaffold but not identified by Validator 1 were examined. Four proteins had peptide pairs with the MS mass difference outside of the Validator 3e tolerance of 3 ppm. One protein had a fragment ion tally below the cutoff limit of 10. Two proteins were identified solely from 16O-peptides with no 18O partner and would thus not be identified by any form of the Validator software.
Table II. Scaffold comparison.
Results are shown comparing the performance of Validator versions 1–3 with the peptide and protein output from the commercial software package Scaffold. In addition, data are displayed after filtering the raw Mascot data for only those peptides with scores greater than 35. The Scaffold filtering criteria were to include only peptides with a 90% confidence, proteins with a 95% confidence, and only those for which there were at least two unique peptides identified. For instance, using only the top peptide match from Mascot for each query, Validator 1 captured 69.5% of the peptides and 91.9% of the proteins as identified by Scaffold. Also shown are results when using all possible peptide and protein guesses by Mascot. ID'd, identified.
| Version | Raw | Raw >35 | 1 | 2 | 2e | 3 | 3e |
|---|---|---|---|---|---|---|---|
| Top Mascot query match | |||||||
| Percentage of Scaffold peptides ID'd | 99.6 | 99.4 | 69.5 | 66.1 | 62.6 | 67.1 | 59.1 |
| Percentage of Scaffold proteins ID'd | 100 | 100 | 91.9 | 93.0 | 84.9 | 94.2 | 88.4 |
| Percentage of peptides ID'd not in Scaffold | 96.4 | 18.8 | 18.6 | 80.4 | 39.7 | 83.4 | 71.7 |
| Percentage of proteins ID'd not in Scaffold | 97.5 | 56.8 | 47.6 | 90.2 | 64.8 | 91.6 | 84.7 |
| All Mascot query matches | |||||||
| Percentage of Scaffold peptides ID'd | 100 | 99.8 | 71.1 | 68.9 | 64.4 | 69.9 | 60.7 |
| Percentage of Scaffold proteins ID'd | 100 | 100 | 97.7 | 98.8 | 95.3 | 98.8 | 96.5 |
| Percentage of peptides ID'd not in Scaffold | 99.5 | 96.7 | 95.9 | 98.5 | 97.4 | 98.6 | 98.1 |
| Percentage of proteins ID'd not in Scaffold | 98.2 | 97.6 | 96.9 | 97.9 | 97.5 | 97.9 | 97.7 |
Corroboration of Validator 1-identified Peptide Pairs
Returning to the 16O/18O pairs identified by Validator 1, we sought to corroborate the pairs by analysis of shifting and non-shifting fragment ions. The Validator 3e program was extended to analyze all Validator 1-identified pairs, first by finding all shifting and non-shifting ions between the two MS/MS ion lists. Then the list of matches was compared with the predicted fragmentation table for the Mascot-identified peptide to calculate a fragment ion tally. To determine the significance of each potential match, the following algorithm was used: for each potential peptide pair, we randomly permuted the peptide sequence 30 times, each time computing the fragmentation table for the random peptide and determining a fragment ion tally. Based on the distribution of fragment ion tallies for the randomly permuted peptides, a 95% confidence interval was determined. Using a criterion that the fragment ion tally for the Mascot-identified peptide must fall outside this range, the fragment ion tallies for 2,626 (94.6%) of the 2,775 Validator 1-identified peptides were found to be significant. In other words, using internal pair validation based on matching shifting and non-shifting MS/MS ions, we were able to corroborate almost every 16O/18O pair found by Validator 1. This is highly significant as it both demonstrates the strength of using 16O/18O pair finding as a route to high confidence peptides and validates our method of peptide validation by matching MS/MS ions.
Statistical Analyses
We next sought to analyze our results by applying a conventional validation method of false discovery rate determination and receiver operating characteristic (ROC) curve plotting. Whenever a protein sequence from the target database is tested, a random sequence of equal length and similar amino composition is generated and tested (Matrix Science and Refs. 29 and 52). Any matches to the decoy database are assumed to be false positives, and this approach assumes that matches to the decoy peptides have the same distribution as false-positive matches to the original target data (5). For calculation of FDR at a given threshold score, we used the method described by Käll et al. (27, 29) of dividing the number of decoy peptides identified (with scores over the threshold) by the number of target peptides identified (with scores over the threshold score). In general, the identified decoy peptides have low Mascot peptide scores and high precursor mass errors (supplemental Fig. 3). Searching the data set with Mascot against the reference proteomes of 17,200 target peptides and 17,687 decoy peptides yielded an FDR of 5% at a Mascot peptide score of 36 (Fig. 4A). At this cutoff score, Mascot retains 2,250 target peptides and 106 decoy peptides. We were interested in comparing the features of decoy peptides as an independent means of estimating the ability of Validator to decrease FDR. We therefore applied this test to analyze the filtering ability of Validator versions 1–3 (Table I). As an example, recall that Validator 2e identifies pairs by first finding a pair member that Mascot has identified as having either a carboxyl-terminal 16O or 18O and then finding the other pair member by searching for a peptide with the appropriate difference in m/z. Using this Mascot-identified peptide for each pair member, the program identifies the b- and y-ions from the list of MS/MS ions. This list is searched against the list of MS/MS ions from the isotopic partner to determine the number of non-shifting (b-type) and shifting (y-type) ions, and the sum of these is the fragment ion tally. Peptide-spectrum matches with a fragment ion tally of 10 or greater are retained. Validator 2e retains 1,782 target but only 650 decoy peptides. The majority of decoy peptides have a low Mascot score so that an FDR of 5% is achieved at a cutoff score of 29 (Fig. 4B). At that score, the algorithm retains 1,457 target peptides and 62 decoy peptides.
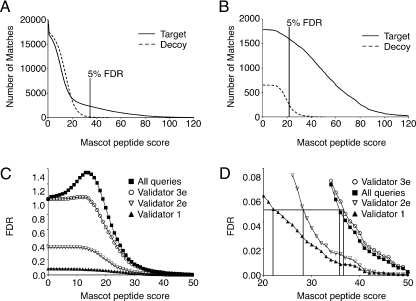
Fig. 4.
Analysis of FDRs. A, number of Mascot peptide-spectrum matches for target (solid) and decoy data (dotted). The total number of matches with peptide scores over the given Mascot cutoff score is shown, and the score threshold for an FDR of 5% is indicated. B, number of Validator 2e matches for target data (solid) and decoy data (dotted). Note the different y axis scale compared with A. C and D, false discovery rate for raw Mascot and data filtered by Validator versions 1, 2e, and 3e. False discovery rate is the number of decoy peptides divided by the number of target peptides with scores exceeding a given threshold. In D, the black lines mark the Mascot peptide score cutoffs to achieve an FDR of 5% for Mascot (35.6) and Validator 1 (22), 2e (29), and 3e (37).
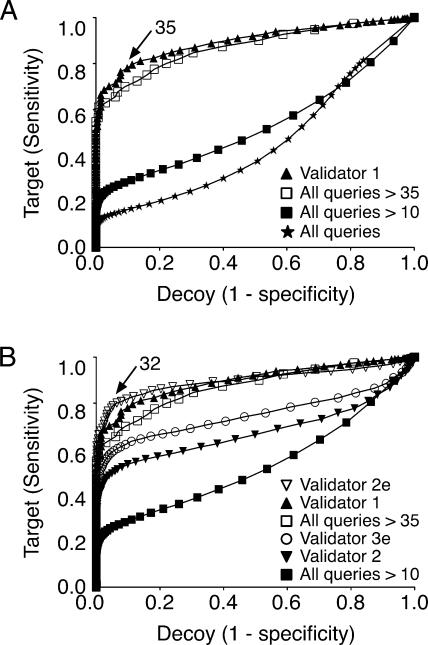
Receiver operating characteristic curves are a useful way to visualize the relationship between the sensitivity and specificity of a test. We used ROC analysis to probe the relationship between sensitivity and specificity for Mascot peptide scores over all data, prefiltered data, and Validator-filtered data. For a typical mass spectrometry experiment, a true ROC curve cannot be plotted because the true-positive rate is unknown. Typically the search results from the target and decoy data sets are used to approximate the sensitivity and specificity of the search engine filter (Matrix Science). Sensitivity is approximated by the ratio of the number of queries with peptide scores above a given value to the total number of queries. Likewise specificity is approximated by the ratio of the number of decoy queries with assigned peptides above a given score to the total number of decoy peptides. ROC analysis of the full set of Mascot-searched data demonstrates poor sensitivity and specificity throughout most of the range of score thresholds (Fig. 5A, stars). It is only at a very low threshold score that the sensitivity approaches 100% (capturing all correct identifications) while the specificity is close to zero (capturing all incorrect identifications). As expected, restricting the ROC analysis to peptides with Mascot scores above 10 or above 35 (Fig. 5A, solid and open squares) improves sensitivity and specificity. When the Validator 1 filtering algorithm is applied to the data (Fig. 5A, triangles), the ROC curve demonstrates a stronger relationship between sensitivity and specificity with a sensitivity of 80% and specificity of 89% at a threshold score of 35 (Fig. 5A, arrow). The performance of Validator versions 2, 2e, and 3e are similarly compared in Fig. 5B. Note that Validator 2e has the best ROC curve with a sensitivity of 80% and a specificity of 94% at a Mascot peptide score threshold of 32 (Fig. 5B, arrow).
Fig. 5.
ROC curves. For a given threshold Mascot peptide score, the sensitivity is the ratio of the number of identifications with scores greater than the cutoff score to the total number of queries, whereas the specificity is the ratio of the number of decoy peptide identifications over the cutoff score to the total number of decoy peptide identifications. A, ROC curves for Mascot-searched data and Validator 1-filtered peptides. Validator 1 (triangles) outperforms a simple score cutoff of 35 (open boxes). B, ROC curves for Validator versions 1–3. Both Validator 1 and 2e outperform using a simple Mascot score cutoff of 35 (open boxes).
Corroboration of Validator 3-identified Peptide Pairs
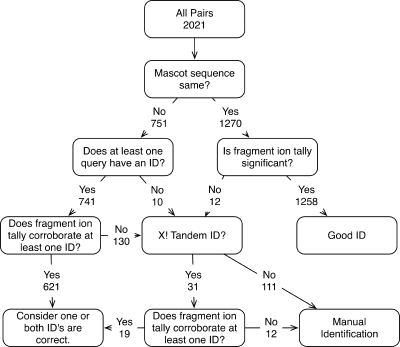
A schema for corroboration of Validator 3-identified peptide pairs is shown in Fig. 6. For the pairs identified by Validator 3e, we utilized the Mascot information, where available, to determine the significance of the match. If the Mascot identification was the same for both members of the pair, we determined the significance of the match using the corroboration strategy of determining fragment ion tallies after randomization of the candidate peptide. Of the 1,270 pairs where the peptide identifications were the same, the score was found to be significant in 1,258 pairs. For the 741 cases where the Mascot identifications were to different sequences, or only one member of a pair had an identification, the same technique was applied to determine the significance. In 621 cases, the corroboration score was significant for at least one matched peptide. For the 130 pairs where there was no corroboration or where neither peptide had a Mascot identification, 31 could be identified using X! Tandem. Of these, we were able to corroborate 19 using the randomization strategy. This left only 133 pairs that passed the fragment ion tally threshold of 10 but lacked any peptide identification to validate. Overall we were able to corroborate 1,898 of 2,021 Validator 3e pairs (93.9%).
Fig. 6.
Schema for corroborating Validator 3e-identified peptide pairs. The tallies reflect the results for the test data set. If the Mascot identification (ID) was the same, the shifting and non-shifting ions were matched against the fragmentation table. 1,258 of 1,270 pairs were corroborated this way. Of the remaining pairs, if at least one had a Mascot identification, the shifting and non-shifting ions were compared with the theoretical fragmentation table, and if one or both had a valid fragment ion tally, it was assumed correct. This was true for 621 pairs. Of the remaining pairs, a search was performed using X! Tandem, an alternate search engine, and if a peptide was identified, the corroboration was repeated. For 31 peptides, an identification was made using X! Tandem, and for 19 of these, the match was corroborated with the identified ions. For the remaining pairs (133 in this case), a manual review will need to be performed to determine the identity of the peptide and the validity of the match.
Performance
All versions of Validator are written in Python version 2.6 running on desktop and laptop hardware. Versions were tested both in Windows XP and Mac OS X environments. Our reference Mascot .DAT data file is 92.8 MB and 1.24 million lines, consisting of 11,536 scans, 20,759 queries, and their analysis. On standard hardware (e.g. Intel Core-2 Duo processors with 2–4 GB of RAM), Validator versions 1–3 run in sequence in less than 6 min (∼32 spectra/s), including a complete parsing of the .DAT file, pair finding, and corroboration and full FDR analysis. Validator 1 by itself runs from start to finish in 70 s. Most of this time is spent building the query dictionaries, and once loaded, Validator 1 is able to find all 16O/18O pairs in about 10 s, including decoy search and false discovery rate determination. This corresponds to processing >1,000 spectra/s. Once optimized and compiled, it is expected that Validator should be able to run several times faster. To facilitate further development, software will be available freely both as stand alone code as well as a Web-based tool (www.msvalidator.org).
DISCUSSION
We have developed Validator, a novel proteomics database search validation software that provides a direct and independent means to validate peptide identifications provided by Mascot analysis of tandem mass spectrometry data. Our algorithm is based on LC-MS/MS analysis of a mixture of carboxyl-terminal stable isotope-labeled and non-labeled peptides, a common sample in quantitative mass spectrometry (32, 53–57). We exploit the characteristic fragmentation of isotopically labeled peptides to enhance their identification, a well established principle that goes back to the period preceding the modern era of ESI and LC-MS/MS (36, 37) and has since been applied effectively by a number of investigators (e.g. Refs. 2, 5, 12, 14, 33, 35, 38–48, and 50). Where both the light (unlabeled) and heavy (labeled) forms of a peptide are selected for fragmentation, the resulting spectra can be compared, thereby distinguishing pairs of non-shifting b-ions from pairs of y-ions that display a shift determined by the isotopic label. These data are then used to test the validity of Mascot peptide identifications, comparing observed with predicted fragmentation patterns. We found that this approach allows rapid and efficient automated filtering of Mascot analysis of LC-MS/MS data to improve both the sensitivity and specificity of peptide identification while salvaging potentially useful low scoring peptides not captured by conventional validation strategies.
Our naive, first approach was to rapidly identify all Mascot-derived 16O/18O pairs from a Mascot .DAT file where both peptides received the same identification. Our data show that a majority of the highest scoring peptides are validated by this simple strategy, and this method was not only able to find 91% of the proteins identified by the commercial analysis package Scaffold but also to capture peptides where the Mascot scores would have fallen below any standard significance threshold. This analysis takes less than 10 s and results in a list of very high confidence peptide and protein identifications. The surprising performance of this simple approach probably reflects the high bar required for Mascot to independently match each of the fragmentation spectra to the 16O and 18O forms of the same peptide, even when the resulting scores fall below normal significance thresholds. In turn, this single criterion efficiently rejects most false identifications as from decoy data.
Validator 2 relaxes the requirement for Mascot to make the same identification for both spectra in a pair and simply seeks a partner for each 16O- or 18O-labeled peptide based on the expected difference in precursor mass. We have shown that this is also a fast and reliable way of identifying pairs, and we found many 16O/18O-labeled potential matches not identified by Validator 1. With Validator 2e, we extracted the b-type (non-shifting) and y-type (shifting) fragment ions from the MS/MS spectra of each pair and then compared these data with the theoretical peptide fragmentation table calculated from the Mascot peptide identifications. Validator 2e confirmed both low and high scoring Mascot identifications but also rejected many others, including nearly all high scoring matches to the decoy database. Thus, Validator 2e was able to achieve an FDR of 5% at a score of 29 versus 36 for Mascot alone. These data suggest that for any arbitrary level of significance running Validator can significantly increase confidence in peptide identifications independently of the Mascot score.
To develop a validation scheme agnostic to Mascot-derived information, we reasoned that peptide pairs could be found based only on the difference in precursor mass. Validator 3 was able to quickly find all Validator 2-identified pairs as well as many others. Here, even though in many pairs neither the light nor heavy forms were matched by Mascot, we again wanted to corroborate the peptides by matching shifting and non-shifting ions. By comparing the two MS/MS ion series directly, shifting and non-shifting ions were rapidly identified by Validator 3e, and we were able to confirm the majority of high Mascot scoring peptides by tallying the number of shifting and non-shifting ions and again efficiently reject Mascot decoy matches. In addition, Validator 3e validated many pairs that had received low Mascot scores and even determined fragmentation patterns for pairs of queries for which Mascot had made no assignments at all.
Using this fragment ion matching scheme, we were able to corroborate most of the 2,775 pairs found by Validator 1. To study Validator 3-identified peptides, we applied a more complicated but systematic approach and corroborated 94% of peptide pairs by combining multiple analysis methods including X! Tandem and manual validation. These results demonstrate that we can quickly (<5 min) parse a Mascot results file, returning a list of high confidence peptide pairs, many of which would be missed using conventional score cutoff techniques.
Because our software is designed to analyze data from samples that are a mixture of peptides labeled at the carboxyl terminus with either 16O or 18O, there is some concern that MS analysis of the mixture will result in fewer protein identifications than for an unlabeled sample due to an increase in fragmentation of “redundant” isotopologues at the expense of other peptides. Indeed when we analyzed 16O and 18O samples separately, we found that Mascot identified about 30% more peptides in either singly labeled sample than when the MS was performed on the 1:1 mixture. Thus, we modified Validator to allow for separate 16O and 18O fractions to be combined and analyzed as a single data set, and as expected, analysis of the combined fractions rescues the lost identifications (data not shown). Whether analyzed separately (requiring more MS time) or together (and potentially losing some protein identifications) Validator can accommodate the data analysis.
We intend to provide Validator versions 1–3 both as a downloadable, open source program and as a Web-based tool for parsing and analyzing searched Mascot data. In addition, this approach is readily applied to other labeling schemes used for quantitative analysis, such as stable isotope labeling by amino acids in cell culture (SILAC) or ICAT. Thus, we intend to adapt the software to accommodate other stable isotope tags. Analysis will also be extended to other search platforms such as SEQUEST or X! Tandem.
This study raises the possibility of implementing a new approach to proteomics data acquisition and analysis to speed up and enhance protein identification based on identifying peptides “on the fly” during the LC-MS/MS run. Our data suggest that peptides might be readily identified, even in a complex sample, based on detecting pairs of precursor ions with a characteristic mass difference. Then MS/MS could be performed on both the heavy and light forms followed by comparison to detect shifting and non-shifting fragment ions. The lists of precursor ion masses and b- and y-ions determined from such a match could be used to generate sequence tags as done by Mann and Wilm (20) to directly identify each peptide and thus the protein. With such a strategy, protein identification in real time during the LC-MS/MS run is entirely feasible from a computational perspective. Toward these ends, we anticipate pursuing rapid recognition of 16O/18O pairs in raw LC-MS/MS data and interrogating pairs of fragmentation patterns to search for matching shifting and non-shifting ions.
In its current incarnation, our Validator software offers a simple and powerful tool to filter searched tandem mass spectrometry proteomics data. By applying the techniques outlined above, a list of high confidence peptide and protein identifications can be obtained within minutes, thus reducing the complexity of downstream proteomics analyses.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM60443 and R01 HG003864 (to S. K.). This work was also supported by a Department of Defense Breast Cancer Research Program multidisciplinary postdoctoral award (to K. K.) and a grant from the Cancer Research Foundation (to S. V.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
1 The abbreviations used are:
- FDR
- false discovery rate
- ROC
- receiver operating characteristic
- LTQ
- linear trap quadrupole
- PME
- precursor mass error.
REFERENCES
- 1.Aebersold R., Mann M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198– 207 [DOI] [PubMed] [Google Scholar]
- 2.Hunt D. F., Yates J. R., 3rd, Shabanowitz J., Winston S., Hauer C. R. ( 1986) Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 83, 6233– 6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin D., Tabb D. L., Yates J. R., 3rd ( 2003) Large-scale protein identification using mass spectrometry. Biochim. Biophys. Acta 1646, 1– 10 [DOI] [PubMed] [Google Scholar]
- 4.Liu T., Belov M. E., Jaitly N., Qian W. J., Smith R. D. ( 2007) Accurate mass measurements in proteomics. Chem. Rev. 107, 3621– 3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nesvizhskii A. I., Vitek O., Aebersold R. ( 2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787– 797 [DOI] [PubMed] [Google Scholar]
- 6.Taylor J. A., Johnson R. S. ( 1997) Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 11, 1067– 1075 [DOI] [PubMed] [Google Scholar]
- 7.Frank A., Pevzner P. ( 2005) PepNovo: de novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964– 973 [DOI] [PubMed] [Google Scholar]
- 8.Fischer B., Roth V., Roos F., Grossmann J., Baginsky S., Widmayer P., Gruissem W., Buhmann J. M. ( 2005) NovoHMM: a hidden Markov model for de novo peptide sequencing. Anal. Chem. 77, 7265– 7273 [DOI] [PubMed] [Google Scholar]
- 9.DiMaggio P. A., Jr., Floudas C. A. ( 2007) De novo peptide identification via tandem mass spectrometry and integer linear optimization. Anal. Chem. 79, 1433– 1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank A. M., Savitski M. M., Nielsen M. L., Zubarev R. A., Pevzner P. A. ( 2007) De novo peptide sequencing and identification with precision mass spectrometry. J. Proteome Res. 6, 114– 123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu S., Chen X. ( 2005) Precise proteomic identification using mass spectrometry coupled with stable isotope labeling. Analyst 130, 1225– 1231 [DOI] [PubMed] [Google Scholar]
- 12.Shevchenko A., Chernushevich I., Ens W., Standing K. G., Thomson B., Wilm M., Mann M. ( 1997) Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 11, 1015– 1024 [DOI] [PubMed] [Google Scholar]
- 13.Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. ( 2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626– 4639 [DOI] [PubMed] [Google Scholar]
- 14.Eng J. K., McCormack A. L., Yates J. R., 3rd ( 1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976– 989 [DOI] [PubMed] [Google Scholar]
- 15.Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. ( 1995) Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67, 1426– 1436 [DOI] [PubMed] [Google Scholar]
- 16.Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. ( 1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551– 3567 [DOI] [PubMed] [Google Scholar]
- 17.Craig R., Beavis R. C. ( 2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466– 1467 [DOI] [PubMed] [Google Scholar]
- 18.Craig R., Beavis R. C. ( 2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17, 2310– 2316 [DOI] [PubMed] [Google Scholar]
- 19.Deutsch E. W., Lam H., Aebersold R. ( 2008) Data analysis and bioinformatics tools for tandem mass spectrometry in proteomics. Physiol. Genomics 33, 18– 25 [DOI] [PubMed] [Google Scholar]
- 20.Mann M., Wilm M. ( 1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66, 4390– 4399 [DOI] [PubMed] [Google Scholar]
- 21.Tabb D. L., Ma Z. Q., Martin D. B., Ham A. J., Chambers M. C. ( 2008) DirecTag: accurate sequence tags from peptide MS/MS through statistical scoring. J. Proteome Res. 7, 3838– 3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabb D. L., Saraf A., Yates J. R., 3rd ( 2003) GutenTag: high-throughput sequence tagging via an empirically derived fragmentation model. Anal. Chem. 75, 6415– 6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunyaev S., Liska A. J., Golod A., Shevchenko A., Shevchenko A. ( 2003) MultiTag: multiple error-tolerant sequence tag search for the sequence-similarity identification of proteins by mass spectrometry. Anal. Chem. 75, 1307– 1315 [DOI] [PubMed] [Google Scholar]
- 24.Tabb D. L. ( 2008) What's driving false discovery rates? J. Proteome Res. 7, 45– 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383– 5392 [DOI] [PubMed] [Google Scholar]
- 26.Savitski M., Nielsen M. L., Zubarev R. A. ( 2005) New data base-independent, sequence tag-based scoring of peptide MS/MS data validates Mowse scores, recovers below threshold data, singles out modified peptides, and assesses the quality of MS/MS techniques. Mol. Cell. Proteomics 4, 1180– 1188 [DOI] [PubMed] [Google Scholar]
- 27.Käll L., Storey J. D., MacCoss M. J., Noble W. S. ( 2008) Posterior error probabilities and false discovery rates: two sides of the same coin. J. Proteome Res. 7, 40– 44 [DOI] [PubMed] [Google Scholar]
- 28.Elias J. E., Gygi S. P. ( 2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207– 214 [DOI] [PubMed] [Google Scholar]
- 29.Käll L., Storey J. D., MacCoss M. J., Noble W. S. ( 2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29– 34 [DOI] [PubMed] [Google Scholar]
- 30.Choi H., Ghosh D., Nesvizhskii A. I. ( 2008) Statistical validation of peptide identifications in large-scale proteomics using the target-decoy database search strategy and flexible mixture modeling. J. Proteome Res. 7, 286– 292 [DOI] [PubMed] [Google Scholar]
- 31.Choi H., Nesvizhskii A. I. ( 2008) Semisupervised model-based validation of peptide identifications in mass spectrometry-based proteomics. J. Proteome Res. 7, 254– 265 [DOI] [PubMed] [Google Scholar]
- 32.Ong S. E., Mann M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252– 262 [DOI] [PubMed] [Google Scholar]
- 33.Schnölzer M., Jedrzejewski P., Lehmann W. D. ( 1996) Protease-catalyzed incorporation of 18O into peptide fragments and its application for protein sequencing by electrospray and matrix-assisted laser desorption/ionization mass spectrometry. Electrophoresis 17, 945– 953 [DOI] [PubMed] [Google Scholar]
- 34.Scoble H. A., Martin S. A. ( 1990) Characterization of recombinant proteins. Methods Enzymol. 193, 519– 536 [DOI] [PubMed] [Google Scholar]
- 35.Takao T., Hori H., Okamoto K., Harada A., Kamachi M., Shimonishi Y. ( 1991) Facile assignment of sequence ions of a peptide labelled with 18O at the carboxyl terminus. Rapid Commun. Mass Spectrom. 5, 312– 315 [DOI] [PubMed] [Google Scholar]
- 36.Gray W. R., Del Valle U. E. ( 1970) Application of mass spectrometry to protein chemistry. I. Method for amino-terminal sequence analysis of proteins. Biochemistry 9, 2134– 2137 [DOI] [PubMed] [Google Scholar]
- 37.Gray W. R., Wojcik L. H., Futrell J. H. ( 1970) Application of mass spectrometry to protein chemistry. II. Chemical ionization studies on acetylated permethylated peptides. Biochem. Biophys. Res. Commun. 41, 1111– 1119 [DOI] [PubMed] [Google Scholar]
- 38.Rose K., Simona M. G., Offord R. E., Prior C. P., Otto B., Thatcher D. R. ( 1983) A new mass-spectrometric C-terminal sequencing technique finds a similarity between gamma-interferon and alpha 2-interferon and identifies a proteolytically clipped gamma-interferon that retains full antiviral activity. Biochem. J. 215, 273– 277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cagney G., Emili A. ( 2002) De novo peptide sequencing and quantitative profiling of complex protein mixtures using mass-coded abundance tagging. Nat. Biotechnol. 20, 163– 170 [DOI] [PubMed] [Google Scholar]
- 40.Goodlett D. ( 2003) Stable isotopic labeling and mass spectrometry as a means to determine differences in protein expression. Trends Analyt. Chem. 22, 282– 290 [Google Scholar]
- 41.Goodlett D. R., Keller A., Watts J. D., Newitt R., Yi E. C., Purvine S., Eng J. K., von Haller P., Aebersold R., Kolker E. ( 2001) Differential stable isotope labeling of peptides for quantitation and de novo sequence derivation. Rapid Commun. Mass Spectrom. 15, 1214– 1221 [DOI] [PubMed] [Google Scholar]
- 42.Gu S., Pan S., Bradbury E. M., Chen X. ( 2003) Precise peptide sequencing and protein quantification in the human proteome through in vivo lysine-specific mass tagging. J. Am. Soc. Mass Spectrom. 14, 1– 7 [DOI] [PubMed] [Google Scholar]
- 43.Lee Y. H., Han H., Chang S. B., Lee S. W. ( 2004) Isotope-coded N-terminal sulfonation of peptides allows quantitative proteomic analysis with increased de novo peptide sequencing capability. Rapid Commun. Mass Spectrom. 18, 3019– 3027 [DOI] [PubMed] [Google Scholar]
- 44.Qin J., Herring C. J., Zhang X. ( 1998) De novo peptide sequencing in an ion trap mass spectrometer with 18O labeling. Rapid Commun. Mass Spectrom. 12, 209– 216 [DOI] [PubMed] [Google Scholar]
- 45.Zhang N., Aebersold R., Schwikowski B. ( 2002) ProbID: a probabilistic algorithm to identify peptides through sequence database searching using tandem mass spectral data. Proteomics 2, 1406– 1412 [DOI] [PubMed] [Google Scholar]
- 46.Pratt J. M., Robertson D. H., Gaskell S. J., Riba-Garcia I., Hubbard S. J., Sidhu K., Oliver S. G., Butler P., Hayes A., Petty J., Beynon R. J. ( 2002) Stable isotope labelling in vivo as an aid to protein identification in peptide mass fingerprinting. Proteomics 2, 157– 163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potthast F., Gerrits B., Häkkinen J., Rutishauser D., Ahrens C. H., Roschitzki B., Baerenfaller K., Munton R. P., Walther P., Gehrig P., Seif P., Seeberger P. H., Schlapbach R. ( 2007) The Mass Distance Fingerprint: a statistical framework for de novo detection of predominant modifications using high-accuracy mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 854, 173– 182 [DOI] [PubMed] [Google Scholar]
- 48.Bandeira N., Tsur D., Frank A., Pevzner P. A. ( 2007) Protein identification by spectral networks analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 6140– 6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savitski M. M., Nielsen M. L., Zubarev R. A. ( 2006) ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics 5, 935– 948 [DOI] [PubMed] [Google Scholar]
- 50.Heller M., Mattou H., Menzel C., Yao X. ( 2003) Trypsin catalyzed 16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J. Am. Soc. Mass Spectrom. 14, 704– 718 [DOI] [PubMed] [Google Scholar]
- 51.Kolker E., Hogan J. M., Higdon R., Kolker N., Landorf E., Yakunin A. F., Collart F. R., van Belle G. ( 2007) Development of BIATECH-54 standard mixtures for assessment of protein identification and relative expression. Proteomics 7, 3693– 3698 [DOI] [PubMed] [Google Scholar]
- 52.Elias J. E., Haas W., Faherty B. K., Gygi S. P. ( 2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667– 675 [DOI] [PubMed] [Google Scholar]
- 53.Mirgorodskaya O. A., Kozmin Y. P., Titov M. I., Körner R., Sönksen C. P., Roepstorff P. ( 2000) Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using (18)O-labeled internal standards. Rapid Commun. Mass Spectrom. 14, 1226– 1232 [DOI] [PubMed] [Google Scholar]
- 54.Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. ( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43– 50 [DOI] [PubMed] [Google Scholar]
- 55.Qian W. J., Monroe M. E., Liu T., Jacobs J. M., Anderson G. A., Shen Y., Moore R. J., Anderson D. J., Zhang R., Calvano S. E., Lowry S. F., Xiao W., Moldawer L. L., Davis R. W., Tompkins R. G., Camp D. G., 2nd, Smith R. D. ( 2005) Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell. Proteomics 4, 700– 709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakai J., Kojima S., Yanagi K., Kanaoka M. ( 2005) 18O-labeling quantitative proteomics using an ion trap mass spectrometer. Proteomics 5, 16– 23 [DOI] [PubMed] [Google Scholar]
- 57.Yao X., Freas A., Ramirez J., Demirev P. A., Fenselau C. ( 2001) Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal. Chem. 73, 2836– 2842 [DOI] [PubMed] [Google Scholar]