Abstract
Formalin-fixed paraffin-embedded (FFPE) tissue specimens comprise a potentially valuable resource for retrospective biomarker discovery studies, and recent work indicates the feasibility of using shotgun proteomics to characterize FFPE tissue proteins. A critical question in the field is whether proteomes characterized in FFPE specimens are equivalent to proteomes in corresponding fresh or frozen tissue specimens. Here we compared shotgun proteomic analyses of frozen and FFPE specimens prepared from the same colon adenoma tissues. Following deparaffinization, rehydration, and tryptic digestion under mild conditions, FFPE specimens corresponding to 200 μg of protein yielded ∼400 confident protein identifications in a one-dimensional reverse phase liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The major difference between frozen and FFPE proteomes was a decrease in the proportions of lysine C-terminal to arginine C-terminal peptides observed, but these differences had little effect on the proteins identified. No covalent peptide modifications attributable to formaldehyde chemistry were detected by analyses of the MS/MS datasets, which suggests that undetected, cross-linked peptides comprise the major class of modifications in FFPE tissues. Fixation of tissue for up to 2 days in neutral buffered formalin did not adversely impact protein identifications. Analysis of archival colon adenoma FFPE specimens indicated equivalent numbers of MS/MS spectral counts and protein group identifications from specimens stored for 1, 3, 5, and 10 years. Combination of peptide isoelectric focusing-based separation with reverse phase LC-MS/MS identified 2554 protein groups in 600 ng of protein from frozen tissue and 2302 protein groups from FFPE tissue with at least two distinct peptide identifications per protein. Analysis of the combined frozen and FFPE data showed a 92% overlap in the protein groups identified. Comparison of gene ontology categories of identified proteins revealed no bias in protein identification based on subcellular localization. Although the status of posttranslational modifications was not examined in this study, archival samples displayed a modest increase in methionine oxidation, from ∼17% after one year of storage to ∼25% after 10 years. These data demonstrate the equivalence of proteome inventories obtained from FFPE and frozen tissue specimens and provide support for retrospective proteomic analysis of FFPE tissues for biomarker discovery.
Formalin-fixed paraffin-embedded (FFPE)1 tissue samples are routinely prepared during the pathological characterization of clinical specimens and are abundantly available in pathology archives worldwide. The fixation process yields clinically relevant samples that can be stored at ambient temperature and are suitable for pathological examination by light microscopy even after years in storage. Given the wealth of clinical data associated with specimens collected over a span of decades, such as patient treatment regimens and outcomes, FFPE tissue represents a potentially valuable resource for biomarker discovery through retrospective analysis (1, 2).
However, fixation of tissue in formalin leads to significant cross-linking among proteins and other biomolecules, rendering the samples incompatible with many biochemical analyses. Immunohistochemical (IHC) analysis of FFPE tissue has been conducted since the 1970s using either proteolysis or protein denaturants to expose antigenic regions of proteins (3, 4). Since the 1990s, detection of antigens in FFPE tissue has been improved through the development of so-called antigen retrieval techniques (5, 6). These methods involve application of heat in the presence of any of a variety of buffers resulting in the cleavage of methylene bridges formed during the course of fixation (2).
Despite their utilization for IHC analysis, FFPE tissue samples have been largely overlooked in proteomics studies, due to the assumption that tissue fixation would make proteomic analysis intractable. Recent work appears to refute this notion. In 2005, Hood et al. (7) first described the successful application of shotgun proteome analysis to FFPE tissue. Using laser capture microdissected cells and an optimized extraction method, hundreds of proteins were identified from a cancerous prostate lesion and benign prostate hyperplasia, thus opening the door to comparative proteomic analyses of FFPE tissue. Moreover, the same study showed that the numbers and identities of proteins observed were remarkably similar when applying the method to frozen and FFPE mouse liver, thus lending support to the use of FFPE tissue in biomarker discovery studies. Since the initial demonstration of its feasibility, FFPE tissues from diverse origins including breast, liver, kidney, lymphoma, and bone successfully have been subjected to proteomic analyses (8–14).
Although this work suggests the feasibility of biomarker discovery from FFPE tissue, most of these previous studies have been performed on small amounts of material with one-dimensional reverse phase liquid chromatography-tandem mass spectrometry (LC-MS/MS) methods. The use of multidimensional peptide separations can extend the dynamic range of the LC-MS/MS analyses to detect lower abundance proteins. Recently, the use of capillary isotachophoresis as the first dimension in a multidimensional peptide separation strategy for analyzing FFPE tissue was described (8). In this study, thousands of proteins were identified out of <4 μg of digest from FFPE human liver sections. However, the apparatus used was an in-house, custom-designed system, not readily accessible to other laboratories. In several of these studies, proteins identified by a single peptide were accepted as valid identifications. Use of single peptide-based identifications elevates the probability of false positive protein identifications, and these identifications often constitute the majority of protein identifications (15).
The equivalence of fresh/frozen and FFPE tissue proteomes is a critical issue in evaluating the suitability of employing FFPE tissues for biomarker discovery by comparative proteomic analyses. Hood et al. (7) and Guo et al. (14) reported comparisons from analyses of paired fresh and frozen tissue specimens. Guo et al. (14) reported an apparent overlap of 83% in protein identifications between FFPE and frozen brain tissue specimens, whereas Hood et al. (7) did not report the degree of overlap, but found that FFPE mouse liver tissue yielded about 88% of the identifications determined for frozen mouse liver tissue. The majority of protein identifications in both studies were based on single peptide assignments. These investigations did not explicitly address the effect of formaldehyde-derived modifications on the inventories of identified peptides.
An unexplored question with FFPE tissue specimens is the extent to which normal variability in fixation process and storage duration affect the proteomes observed. The duration of tissue fixation is not highly standardized and may vary from hours to several days. One of the most attractive features of FFPE specimens is the opportunity for retrospective biomarker discovery, but the effects of storage for many years on tissue proteomes remains unknown.
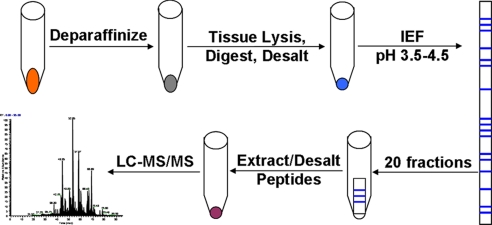
Here, we address these questions through detailed comparative studies of the analysis of fresh frozen and FFPE tissues by LC-MS/MS-based shotgun proteomics. We used the same fresh tissue specimens to prepare both frozen and FFPE samples for paired comparisons. We evaluated conditions for tissue lysis and digestion and the effects of fixation time and storage duration on the number of protein IDs obtained during shotgun proteomic analysis of FFPE tissue. We also characterized the differences in peptides observed between fixed and frozen specimens in an effort to understand the effect of fixation from a practical biomarker discovery standpoint. Furthermore, we compared analyses of fresh frozen and FFPE colon adenoma tissue by multidimensional LC-MS/MS using gel-based isoelectric focusing of peptides (Fig. 1). The results demonstrate a remarkable overlap in the number and identities of proteins between the fixed and frozen tissue and indicate that variations in duration of fixation and storage have a minimal effect on protein inventories obtained by shotgun proteomic analysis. The data indicate essential equivalence between protein inventories obtained from fresh frozen and FFPE tissue specimens by shotgun proteomics and validate the use of FFPE tissue specimens for biomarker discovery.
Fig. 1.
Strategy for multidimensional LC-MS/MS analysis of FFPE tissue.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Sub-X xylene substitute was obtained from Surgipath (Richmond, IL), and 10% buffered formalin solution was purchased from Starplex Scientific, Inc. (Etobicoke, Ontario, Canada). Iodoacetamide and pyridoxamine were from Sigma, tris-carboxyethylphosphine was from Pierce, sequencing grade trypsin was from Promega (Madison, WI), and trifluoroethanol and dithiothreitol were from Acros (Geel, Belgium). Trifluoroacetic acid, ammonium bicarbonate, and urea were purchased from Fisher Scientific, and EDTA was purchased from Invitrogen.
Tissue Digest
Fixed and frozen colon adenoma biopsies were obtained from the Cooperative Human Tissue Network-Western Division (Vanderbilt University, Nashville, TN). The de-identified samples and experimental protocol were subject to Institutional Review Board exempt approval (Institutional Review Board No. 080856). Slices of tissue (60 μm) were placed in separate centrifuge tubes. Paraffin was removed with three washes in 1 ml of Sub-X, and rehydration was achieved with three washes each in 1 ml of 100, 85, and 70% ethanol.
Adenoma slices were resuspended in 100 μl of ammonium bicarbonate (100 mm, pH 8.0) either alone or with 1 mm EDTA or 100 mm pyridoxamine as mentioned in the text. Samples were heated at 80 °C for 2 h. Tryptic digestion was done by an adaptation of the method of Wang et al. (16). Trifluoroethanol (100 μl) was then added, and the samples were sonicated for 20 s followed by 30 s incubation on ice. The sonication was repeated twice. The resulting homogenate was heated for 1 h at 60 °C followed by a second series of sonication steps, as stated above. The homogenate was reduced with carboxyethylphosphine (10 mm) and dithiothreitol (25 mm) at 60 °C for 30 min followed by alkylation with iodoacetamide (50 mm) in the dark at ambient temperature for 20 min. The reduced and alkylated homogenate was diluted to 1 ml with ammonium bicarbonate (50 mm, pH 8.0) followed by addition of trypsin at 1:50 (w/w). The digest was incubated overnight at 37 °C followed by freezing at −80 °C and lyophilization. Samples were resuspended in 1 ml of high pressure liquid chromatography water, desalted over 1 cc (100 mg) Sep-Pak Vac C18 cartridges (Waters Corp., Milford, MA) and evaporated in vacuo with a SpeedVac sample concentrator (ThermoFisher, Waltham, MA).
Isoelectric Focusing of Peptides
Isoelectric focusing (IEF) of tryptic peptides was adapted from the method of Cargile et al. (17). Adenoma tryptic peptides (200 μg) were resuspended in 500 μl of 6 m urea and loaded in an IPGphor rehydration tray. Immobiline immobilized pH gradient strips (24 cm, pH 3.5–4.5) were placed over the samples and allowed to rehydrate overnight at ambient temperature. The loaded strips were focused at 21 °C on an Ettan IPGPhor-3 IEF system (GE Healthcare) using the following program: step at 300 V for 900 volt-hours; gradient to 1000 V for 3900 volt-hours; gradient to 8000 V for 13500 volt-hours; step to 8000 V for 93700 volt-hours. The strips were then cut into 20 (1.2 cm) pieces and placed in separate wells of a 96-well enzyme-linked immunosorbent assay (ELISA) plate. Peptides were eluted from the strips as follows: 200 μl of 0.1% formic acid (FA) for 15 min; 200 μl of 50% acetonitrile (ACN)/0.1% FA for 15 min; 200 μl of 100% ACN/0.1% FA for 15 min. Solutions of extracted peptides were evaporated in vacuo, resuspended in 1 ml of 0.1% trifluoroacetic acid and desalted over a 96-well C18 Oasis hydrophilic-lipophilic balance plate 30 μm (10 mg) (Waters Corp.). Peptide solutions were evaporated in vacuo, resuspended in 100 μl of 0.1% FA, and placed in sample vials for LC-MS/MS analysis.
Reverse Phase LC-MS/MS
LC-MS/MS analyses were performed on an LTQ-XL mass spectrometer (Thermo Electron, San Jose, CA) equipped with an Eksigent nanoLC (Dublin, CA) and Thermo Survey or micro-autosampler. Peptides were resolved on a 100 μm × 11 cm fused silica capillary column (Polymicro Technologies, LLC., Phoenix, AZ) packed with 5 μm, 300 Å Jupiter C18 (Phenomenex, Torrance, CA). Liquid chromatography was carried out at ambient temperature at a flow rate of 0.6 μl min−1 using a gradient mixture of 0.1% (v/v) formic acid in water (solvent A) and 0.1% (v/v) formic acid in ACN (solvent B). Peptides eluting from the capillary tip were introduced into the LTQ source in micro-electrospray mode with a capillary voltage of ∼2 kV. A full scan was obtained for eluting peptides in the range of 400–2000 amu followed by three data-dependent MS/MS scans. MS/MS spectra were recorded using dynamic exclusion of previously analyzed precursors for 60 s with a repeat of 1 and a repeat duration of 1. MS/MS spectra were generated by collision-induced dissociation of the peptide ions at normalized collision energy of 35% to generate a series of b- and y-ions as major fragments.
Data Analysis
The “ScanSifter” algorithm read tandem mass spectra stored as centroided peak lists from Thermo RAW files and transcoded them to mzData v1.05 files. Only MS/MS scans were written to the mzData files; MS scans were excluded. If 90% of the intensity of a tandem mass spectrum appeared at a lower m/z than that of the precursor ion, a single precursor charge was assumed; otherwise the spectrum was processed under both double and triple precursor charge assumptions. Tandem mass spectra were assigned to peptides from the IPI Human database version 3.33 (September 13, 2007; 67837 sequences) by the MyriMatch algorithm, version 1.1.0 (18). The sequence database was doubled to contain each sequence in both normal and reversed orientations, enabling false discovery rate estimation. MyriMatch was configured to expect all cysteines to bear carboxamidomethyl modifications and to allow for the possibility of oxidation on methionines. Candidate peptides were required to feature trypsin cleavages or protein termini at both ends, though any number of missed cleavages was permitted. A precursor error of 1.25 m/z was allowed, but fragment ions were required to match within 0.5 m/z. The IDPicker algorithm v1.53.3 (19) filtered the identifications for each reverse phase liquid chromatography run to include the largest set for which a 5% identification false discovery rate could be maintained, as described by Qian et al. (20). Indistinguishable proteins were recognized and grouped, and parsimony rules were applied to generate a minimal list of proteins that explained all of the peptides that passed our entry criteria (19). This approach uses bipartite graph analysis to derive a minimal list of protein identifications with shared clusters of peptides. These identifications were pooled for each IEF sample set. Proteins were required to have at least two different peptide sequences observed within an IEF sample set. False discovery rates (FDR) for peptide identifications were computed by the formula (15): FDR = (2 × reverse)/(forward + reverse). The algorithm reported the number of spectra and number of distinct sequences observed for each protein and protein group in each sample set. Each sample set consisted of multiple technical replicates at the tissue lysis step, and the results for each replicate are reported in the supplemental Tables S1–S7, denoted as “Runs”.
Statistical Analysis
Numbers of protein group IDs obtained and log-transformed lysine to arginine ratios were compared using a one-way ANOVA test with Bonferroni post-test in cases of multiple comparisons, and 95% confidence intervals were determined. Previous results have shown that spectral counting provides a rough measure of protein levels in complex protein mixtures, especially for more abundant proteins (21). We performed regression analysis and permutation testing using a quasi-likelihood model based on Poisson distribution commonly used for count data (22) with a correction for FDR (23). We compared protein group and peptide identifications between the FFPE and frozen tissue groups and generated quasi p values and ratios of model-predicted frequencies to identify those proteins and peptides that had an unequal distribution between these two groups.
RESULTS
Correction Factor for Protein Concentration Estimation in FFPE Tissue
To facilitate quantitative analyses of proteins identified between runs, equal sample loading is imperative. Yet, estimation of protein concentration from FFPE tissue lysates using, for example, the bicinchoninic acid assay, is not straightforward because those amino acids that contribute to the reduction of copper are also susceptible to reactions with formaldehyde. Thus, the protein concentration estimated from an FFPE tissue lysate will be lower than the true concentration by an unknown magnitude. To account for this effect, serial 60-μm cross-sections of a frozen colon adenoma were prepared. The adenoma was cylindrical in shape, thus variability in protein amounts due to differences in tissue area were kept to a minimum. Four slices were randomly selected and subjected to fixation in 10% formalin for 24 h at ambient temperature. The formalin was decanted, and the deparaffinization and trifluoroethanol/ammonium bicarbonate lysis procedure was applied to the four fixed and four additional frozen tissue slices. Protein concentration in the lysates was estimated using the bicinchoninic acid assay. The fixed sections consistently gave a protein concentration estimate that was 56% of the measured value for frozen tissue (supplemental Fig. S1). Assuming equivalent amounts of protein in each tissue slice, this result provides a correction factor permitting a more accurate assessment of protein concentration in FFPE colon adenoma lysates.
Buffer Optimization for Tryptic Digestion of FFPE Tissue Samples
To ensure deep coverage of the FFPE tissue proteome, various buffers were evaluated in an effort to maximize the number of confident protein IDs obtained during shotgun analysis of tryptic digests from 60-μm tissue slices. The buffers were chosen on the basis of their suitability for mass spectrometry following a solid phase extraction step and included: 100 mm ammonium bicarbonate, pH 8.0; 1 mm EDTA in 100 mm ammonium bicarbonate, pH 8.0; and 100 mm pyridoxamine in 100 mm ammonium bicarbonate, pH 8.0. EDTA was chosen because it has been reported that calcium ions can stabilize interactions between methylol groups that are generated as methylene bridges between cross-linked proteins are cleaved during heating (24, 25). This inhibits antibody recognition of certain antigens and also may affect trypsin access to substrates cleavage sites. Pyridoxamine was chosen due to its nucleophilicity, which may enhance cross-link cleavage through the scavenging of reactive aldehydes. Trifluoroethanol was added to each buffer prior to sonication, to a final concentration of 50% (v/v).
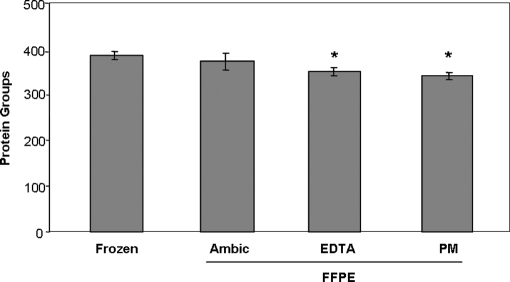
ANOVA testing of the numbers of protein groups identified from shotgun analysis of the various extracts indicated a significant difference between the groups (ANOVA, p = 0.0065). Applying a statistical test correcting for multiple comparisons revealed a significant reduction in numbers of protein groups identified in both the EDTA and pyridoxamine FFPE extracts versus the ammonium bicarbonate-only frozen extract (Bonferroni post-test p < 0.05; Fig. 2, supplemental Table S1). No significant difference in the number of protein groups identified was observed when comparing frozen versus FFPE tissue extracted with ammonium bicarbonate alone, nor when comparing the FFPE tissue extracts to one another. These findings are consistent with previous reports that the ionic strength and buffer composition have relatively little effect on protein extraction efficiency, but that heating at temperatures >60 °C is critical (2).
Fig. 2.
Protein group identifications obtained from reverse phase LC-MS/MS of 600 ng of protein from frozen and FFPE tissue. FFPE tissues were prepared by tryptic digestion with or without prior treatment with EDTA and pyridoximaine HCl as described under “Experimental Procedures.” Error bars represent standard deviation (n = 3). *, significantly different from results with frozen tissue (ANOVA, p = 0.0065; Bonferroni post-test versus frozen: *, p < 0.05).
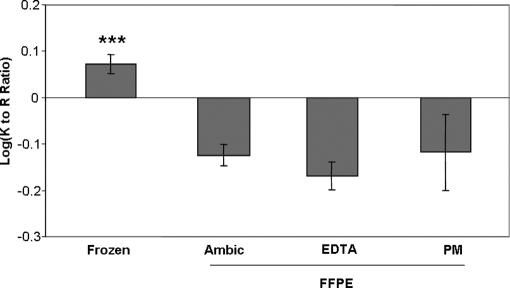
Inter- and intramolecular cross-linking reactions caused by formaldehyde are known to involve the primary amines of lysine side chains (26). Thus, one might expect C-terminal lysine-containing peptides to be under-represented upon shotgun proteomic analysis of a tryptic digest of FFPE tissue. Comparison of the ratios of C-terminal lysine to arginine observed during shotgun analysis of FFPE and frozen tissue digests confirmed this expectation. The log-transformed ratios of Lys- to Arg-terminal peptides revealed a significant reduction of this ratio in the FFPE dataset, suggesting a disproportionate loss of lysine-terminal peptides in the fixed samples (ANOVA, p < 0.0001; Fig. 3, supplemental Table S1). This finding is consistent with the known chemistry of formaldehyde and with previous reports that lysine-terminal peptides are under-represented in proteomic analyses of FFPE tryptic digests (7).
Fig. 3.
Comparison of log-transformed ratios of C-terminal lysine versus arginine peptides observed from frozen and FFPE tissue. Error bars represent 95% confidence interval (n = 3). *** indicates significantly different from results with FFPE tissue (ANOVA, p < 0.0001; Bonferroni post-test versus each FFPE group: ***, p < 0.001).
Another possible explanation for this shift in K to R ratio could be that modification of amino acid side chains with formaldehyde would preclude their identification during database searching. Candidate modifications previously reported include +12 Da (imine) on N termini and lysine and tryptophan side chains and +30 Da (methylol) on cysteine, histidine, lysine, and arginine (26). However, searching the spectra against the IPI database and allowing for these potential variable modifications did not yield any additional peptide IDs that could be verified through manual spectral evaluation. We also conducted an analysis using the P-Mod software tool, which allows unbiased discovery of peptide modifications among proteins known to be represented in a dataset, without the need to specify the mass shifts of interest (27). This approach detects unanticipated mass shifts and modifications and avoids the bias introduced by doing multiple rounds of database searches with amino acid mass shifts incorporated as dynamic modifications. P-Mod searches of the sequences of the 20 most abundant proteins (based on observed spectral counts from the original database searches) against the entire MS/MS datasets for frozen and FFPE tissues failed to detect any modifications due to formaldehyde chemistry. The results collectively suggest that most amino acid modifications induced by formalin fixation of tissue ultimately result in cross-links, or that these modifications may affect the ionization efficiency and fragmentation of the target peptides. The algorithms commonly used for peptide sequence determination from MS/MS spectra are unable to account for the complexity of spectra resulting from the fragmentation of cross-linked peptides. Algorithms designed especially for this objective have been described (28, 29) but require knowledge of the proteins involved in the cross-linking. Absent a targeted enrichment step, the complexity of FFPE tissue samples precludes identification of cross-linked species at this time.
Effect of Fixation Time on FFPE Tissue Shotgun Proteomics
An important potential source of variability in peptide and protein identifications from FFPE samples is fixation time. Fixation protocols can vary among pathology laboratories, and specimens may be left in formalin for varying amounts of time, thus possibly affecting the extent of fixation and protein cross-linking.
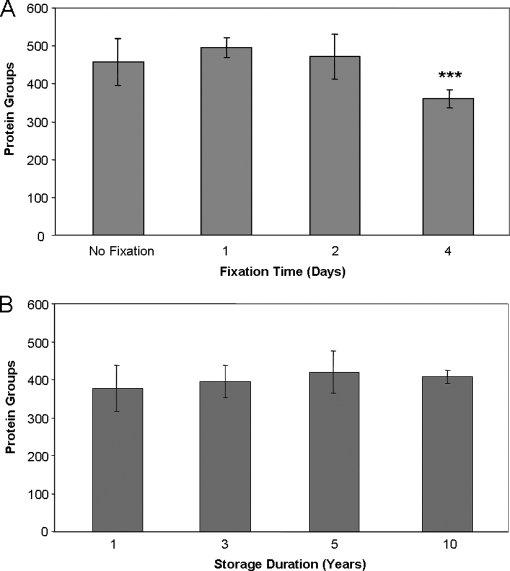
To address this issue, we cut a frozen colon adenocarcinoma into four pieces of equal size. One piece was left frozen, and the other three were fixed in a 10% buffered formalin solution for 1, 2, or 4 days. Fixed tissue was embedded in paraffin and serial 60-μm slices of tissue from each treatment were prepared, and nine slices were analyzed for each time point. ANOVA testing indicated a significant difference in protein groups identified among the fixation time points (ANOVA, p < 0.0001). Comparisons between the individual groups revealed no significant difference in the numbers of protein groups identified among the frozen, 1 day fixation, and 2 day fixation samples (Fig. 4A, supplemental Table S2). However, the 4-day fixation sample yielded fewer protein group IDs with a mean number of IDs that was statistically different from all the other groups (Bonferroni post-test, p < 0.001). The numbers of spectral counts derived from these samples were also compared and found to bear significant differences (ANOVA, p = 0.0001). The post-test indicated a significant reduction in the number of observed spectra for both the 2 day and 4 day fixation time points compared with the frozen sample (Bonferroni post-test, p < 0.05 and p < 0.001 respectively; supplemental Fig. S2A). This suggests that increased duration of fixation has a negative impact on the compatibility of the samples with thorough proteomic characterization. Also, fewer lysine-terminal peptide spectral counts were observed at all the fixation time points compared with the frozen sample, consistent with the observations from the analysis of the effect of various buffers on protein identifications (supplemental Fig. S2B). These results would appear to be in conflict with a recent report in which similar numbers of IDs were obtained between frozen tissue and tissue fixed for up to 14 days (8). It is notable, however, that in the previous study fixation was modeled in a system using entire mouse livers with no paraffin embedding. The efficiency of fixation when applied to an intact organ is likely to be less than what would occur in our process, where a frozen polyp was quartered and fixed prior to embedding in paraffin. Thus, as anticipated, the duration and extent of fixation are likely to have some impact on sample suitability for unfractionated shotgun proteomic analysis, although our data likely represents a worst-case scenario.
Fig. 4.
Assessment of FFPE sample processing and storage variability on protein identifications by shotgun LC-MS/MS. A, effect of fixation time on efficiency of shotgun proteomic analysis. Error bars represent standard deviation (n = 9). B, effect of storage duration on efficiency of shotgun proteomic analysis on FFPE samples. Error bars represent standard deviation (n = 9). *** indicates significantly different from all other groups (ANOVA, p < 0.0001, Bonferroni post-test versus frozen, 1 day and 2 day fixation: ***, p < 0.001).
Effect of Storage Duration on FFPE Tissue Shotgun Proteomics
FFPE tissue specimens may be retained for decades. Therefore, another possible source of variability in analyses of archival FFPE tissue is the length of time of sample storage. To assess this effect, we conducted shotgun proteomic analysis on FFPE colon adenoma tissue samples that had been in storage for 1, 3, 5, or 10 years. The samples were selected by an experienced gastroenterology pathologist (Mary Kay Washington) on the basis of similar sample size, tissue characteristics and diagnosis. Triplicate 60-μm slices from each of three different FFPE colon adenomas at each time point were processed as described under “Experimental Procedures” section. No significant difference was observed in the number of protein IDs obtained from tissues that had been stored for up to a decade (Fig. 4B and supplemental Table S3). In addition, we compared the total spectral counts leading to the positive identification of a peptide sequence among the runs as a measure of the quality of spectra obtained from each sample. We observed no significant difference in the number of confidently identified spectra among all of the time points considered (supplemental Fig. S3A). Furthermore, there was no statistical difference in the number of lysine-terminal or arginine-terminal peptide spectral counts observed for the various storage time points, suggesting that the duration of storage has no discernable effect on the extent of cross-linking (supplemental Fig. S3B and S3C).
Storage duration may impact other modifications among proteins, such that one might expect oxidative modifications (such as methionine oxidation) to be more prevalent in older samples. We did observe a modest increase in the percentage of oxidized methionine residues among methionine-containing peptides, which correlated with storage duration (16.8%, 17.8% 18.2%, and 25.2% for the 1, 3, 5, and 10 year storage time points, respectively) (supplemental Table S3). Methionine-oxidized peptides were detected because this oxidation was specified as a variable modification in our database searches. Additional analyses with P-Mod were done to detect spectra corresponding to modified cysteine-containing peptides from high-abundance proteins (albumin, HSP70, and filamin A) in three fresh-frozen and three 10-year-old FFPE samples. These analyses detected only S-carboxamidomethylated cysteines but did not detect cysteic acid modifications. These results suggest that a very modest degree of protein oxidation occurs during FFPE tissue storage, mainly on methionine residues.
These results are encouraging because they indicate that long-term storage of up to 10 years duration should not be a severe impediment to retrospective proteomic analysis. Also, since it is unlikely that these samples were fixed in exactly the same manner, having been collected and processed over the span of a decade, these results suggest that routine variability in fixation procedure can be accommodated during shotgun proteomic analysis.
IEF of FFPE Tryptic Digests
Multidimensional separation strategies have been widely employed at the protein and peptide level to enhance proteomic coverage by presenting multiple peptide fractions for LC-MS/MS analysis (21). However, the utility of gel-based IEF on immobilized pH gradient strips for analysis of FFPE samples has yet to be demonstrated. To assess this separation method, 200 μg of tryptic digest derived from each of three 60-μm FFPE and frozen colon adenoma tissue slices were analyzed using a multidimensional peptide separation platform, shown schematically in Fig. 1. A narrow pI range (pI 3.5–4.5) separation contains tryptic peptides from a majority of proteins and was chosen based on previous studies (17, 30).
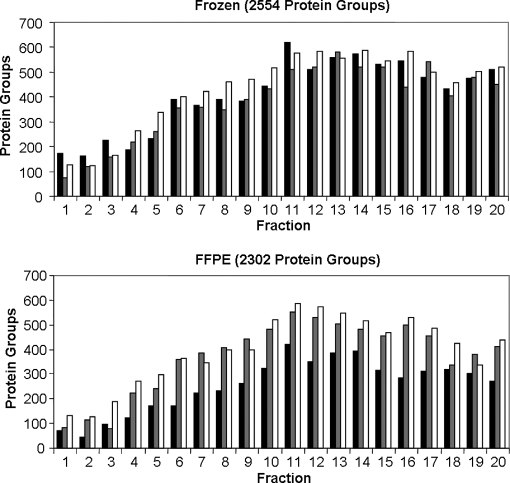
This fractionation strategy yielded a >6-fold increase in protein group identifications compared with single dimension, reverse phase LC-MS/MS analysis. Moreover, the total number of protein groups identified in the FFPE samples was 90% of that identified in frozen tissue and a similar distribution of identifications over the 20 fractions was observed for both sample types (Fig. 5 and supplemental Table S4). The total number of unique peptides identified was 10349 from FFPE samples and 12265 from the frozen samples, whereas the number of spectral counts observed was 27004 and 34482, respectively.
Fig. 5.
Protein group identifications observed in IEF fractions from frozen tissue (top panel) and FFPE tissue (bottom panel). Lysis, digestion and IEF runs were performed in triplicate as described under “Experimental Procedures.”
Equivalence of Identifications for FFPE and Frozen Samples
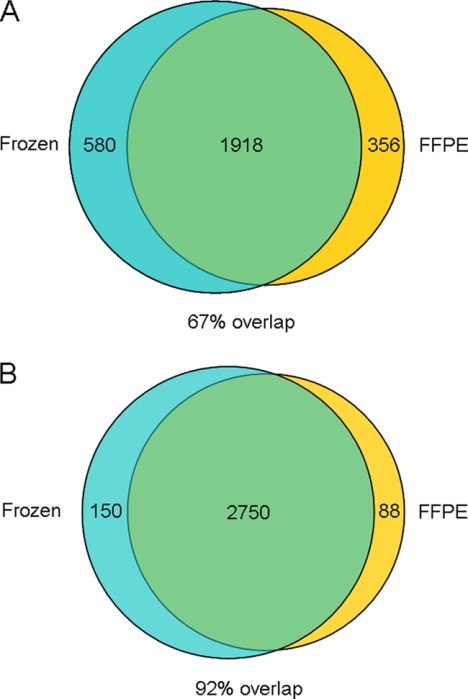
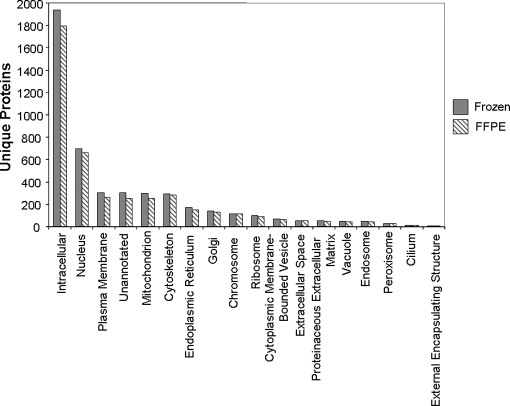
The resulting protein IDs were analyzed using the WebGestalt gene annotation tool, which allows facile comparison of large protein datasets (31). This analysis involves conversion of IPI protein accession numbers generated by our analyses to Entrez gene accession numbers, resulting in slight differences in the numbers of proteins represented in this analysis compared with the IDPicker dataset. The apparent overlap in protein identifications between the two datasets was ∼67%, suggesting differences in the pools of proteins detectable between frozen and FFPE samples (Fig. 6A). This degree of divergence is surprising because both the FFPE and frozen polyps were derived from the same patient. The distribution of proteins among subcellular compartments was nearly identical between the FFPE and frozen tissue datasets (Fig. 7), suggesting that the apparent differences in proteins observed in the two datasets were not biased toward a specific cell compartment.
Fig. 6.
A, overlap of protein groups identified between frozen and FFPE samples searched separately. B, overlap of protein groups identified between frozen and FFPE samples when datasets are combined for database search. See text for discussion.
Fig. 7.
Annotated subcellular distribution of unique proteins identified from frozen and FFPE tissue. Results were compiled using the WebGestalt gene analysis tool.
A possible explanation for the apparent divergence in specific proteins identified in the frozen and FFPE datasets is that modification and cross-linking of peptides containing lysine residues might shift the sampling of peptides to favor those terminating in arginine. To address this possibility, we examined the ratio of lysine- to arginine-terminal peptides for the IEF-fractionated samples. In the frozen samples, this ratio remained essentially identical to the relative prevalence of lysine and arginine among proteins (supplemental Fig. S4) (32). The Lys/Arg ratio was lower for the IEF-fractionated FFPE samples, and the difference between the frozen and FFPE ratios was significant using a two-tailed t test (p = 0.0005; supplemental Fig. S4). (We did note, however, that the Lys/Arg peptide ratio was higher in the IEF-fractionated FFPE samples than in the unfractionated FFPE samples (see Fig. 3, above). This result may indicate that the acidic peptides (pI 3.5–4.5) targeted in our fractionation strategy are less susceptible to modification and cross-linking by formaldehyde.) Consistent with the observed difference in Lys/Arg ratios for frozen and FFPE samples, the number of proteins identified only by lysine-terminal peptides was greater in the frozen samples than in the FFPE samples (345 versus 270, respectively), whereas the number identified only by arginine-terminal peptides was less in the frozen than FFPE (289 versus 359). Despite these differences in peptide characteristics, only 166 of the proteins in these categories were uniquely identified in the FFPE dataset (62 lysine only, 104 arginine only) representing less than half of the total IDs apparently unique to the FFPE samples (Fig. 6A).
The best explanation for the apparent divergence in protein groups observed emerges upon examination of the numbers of peptides identified by database searching with the combined FFPE and frozen datasets. When analyzed in this manner using IDPicker and WebGestalt, a total of 2988 protein groups were identified, of which only 88 were unique to the FFPE dataset and 150 were unique to the frozen, representing an overlap of 92% (Fig. 6B and supplemental Table S5). Since our analysis required at least two distinct peptides to define a protein hit, those proteins with a single unique peptide observed in the FFPE dataset were not identified when searching the frozen and FFPE datasets separately. When the datasets are considered together, nonidentical, single peptide identifications from each combine to yield additional confident identifications. The combined dataset benefits from enhanced coverage, and the overlap in identifications is maximized.
We also compared numbers of spectral counts observed from proteins identified in the combined dataset. Only three proteins were found for which there was a confident difference in spectral counts of 3-fold or greater between the frozen and FFPE sets. These proteins were histone H2A, collagen, and a U4/U5/U6 tri-small nuclear ribonuclear protein associated protein. Thus, despite an unavoidable loss of sensitivity associated with the use of FFPE tissue, we found no indication of a qualitative difference in the proteins observable through shotgun proteomic analysis of FFPE and frozen tissue samples. These results suggest that the collection of proteins observed using our multidimensional peptide separation platform on FFPE tissue samples accurately reflects the proteome observable in frozen tissue.
False Differences between Frozen and FFPE Proteomes Derived from Single Peptide-based Protein Identifications
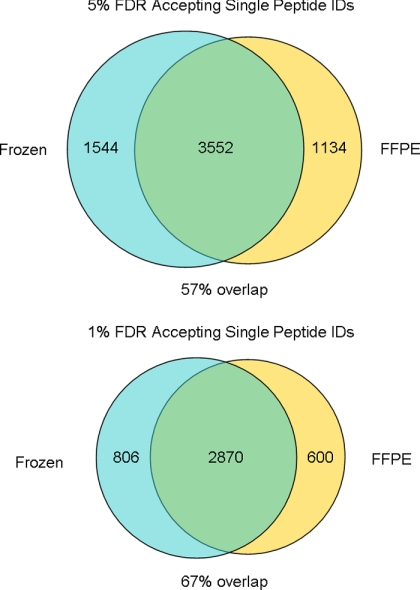
The data obtained from LC-MS/MS analysis of the three FFPE and three frozen IEF-fractionated colon adenoma samples described above were combined and subjected to IDPicker analysis at 5 and 1% FDR, whereas allowing single peptide identifications to specify a protein identification. A large apparent divergence in identified proteins between frozen and FFPE proteomes was observed when proteins identified by single peptides were accepted as valid (Fig. 8 and supplemental Tables S6 and S7). This occurs despite combining the FFPE and frozen data in order to leverage parsimonious protein identification and to reduce spurious unique IDs. Since both the FFPE and frozen specimens were derived from the same individual, we would expect complete overlap in the identities of proteins. However, this is clearly not the case here and stands in stark contrast to the 92% overlap obtained when combining the datasets and requiring at least two unique peptides per protein (Fig. 6B).
Fig. 8.
Divergence in protein identifications when accepting single peptide IDs. Datasets were combined and analyzed using IDPicker to achieve the most parsimonious protein set. Numbers represent protein group identifications.
Given the stochastic nature of the sampling of low abundance peptides inherent in LC-MS/MS and the fact that a significant majority (>70%) of the single hit proteins observed were identified by only a single spectrum (at either a 5% or 1% FDR), the apparent proteome diversity observed through such an analysis alone cannot be taken as proof of differential expression. Since single peptide identifications can account for as many as half of protein identifications in studies accepting them as valid, extreme caution should be exercised when considering the results of such analyses.
DISCUSSION
Recent work demonstrating the feasibility of employing FFPE tissue specimens for shotgun proteome analyses offers the prospect of retrospective biomarker discovery (7–13). Although this body of work indicates that FFPE tissue samples yield protein identifications, these studies do not indicate whether shotgun proteome inventories accurately represent the collection of proteins identified in similar analyses of fresh frozen tissues. Our data indicate that shotgun proteome analyses of FFPE tissues yield proteome inventories that are greater than 90% equivalent to those generated from frozen tissues, both in numbers of proteins identified and in identities of the protein inventories.
Proteins in FFPE tissues have undergone extensive modifications due to formaldehyde reaction with lysine ε-amino groups and protein N termini. Nevertheless, after deparaffinization and rehydration, these cross-linked proteins are still efficiently digested with trypsin under mild conditions typically used for fresh tissues and without need for additional specialized reagents. Our results indicate that proteome inventories of fresh and FFPE tissues encompass virtually the same proteins, despite a decreased representation of C-terminal lysine-containing proteins (see below). Comparison of identifications based on gene ontology categories representing cellular localization and function indicates no organelle- or function-related bias in identifications from FFPE tissues. Thus, the key conclusion of our work is that proteome analyses of FFPE and frozen tissues afford the same proteomic view of the biology of the system under study. It is this criterion, more than simply an ability to generate protein identifications that establishes the validity of proteomic analysis of FFPE tissues for biomarker discovery studies.
The widespread reaction of formaldehyde with proteins would be expected to produce several protein modifications, including lysine cross-links, and the effects of fixation are clearly manifested in our datasets. FFPE peptide inventories exhibited a disproportionate loss of C-terminal lysine peptides (Fig. 3 and supplemental Fig. S4). FFPE samples also yielded fewer spectral counts overall, fewer proteins identified by 8 or more peptides in the IEF dataset (278 for FFPE, 361 for frozen) and fewer peptides overall than observed with frozen samples. These differences result in an apparent disparity in protein group identifications when the FFPE and frozen datasets are processed separately in IDPicker. Surprisingly, we did not directly observe any peptide modifications consistent with known lysine-formaldehyde chemistry. We observed no MS/MS evidence for formaldehyde-derived modifications in the FFPE samples. We searched for modifications in both a targeted manner (i.e. specifying modifications as variable modifications to lysine in database searches) and in an unbiased manner (i.e. P-Mod search of data for proteins identified in the samples). We note that P-Mod detects spectra that correspond to modified variants of peptides specified as search sequences. Presumably, the majority of intermediates formed from amino acids reacting with formaldehyde go on to form cross-linked products that are not amenable to identification using current database searching algorithms or P-Mod. However, we should note that, although protein input was standardized between FFPE and frozen samples, the exact quantity of peptides loaded onto the IEF strips was not known. A disproportionate loss of material during the desalting of peptides from the FFPE samples cannot be ruled out, but appears unlikely.
Our analyses demonstrate that proteome inventories for frozen and FFPE samples are equivalent at the level of global protein expression. However, we did not attempt to analyze the posttranslational modification (PTM) status of proteins in FFPE specimens beyond our limited screen using P-Mod for modifications among the 20 most abundant proteins. These analyses only established that formaldehyde-derived modifications were not detected. Successful analysis of biologically derived PTMs typically requires affinity enrichment for the modification of interest, which we did not attempt. Even in our analyses of frozen tissues, PTMs would have been detected only sporadically, due to the low abundance of modified peptides relative to unmodified peptides. Although we cannot draw any conclusions about the analyses of PTM in FFPE tissues from our data, it seems reasonable to suspect that PTM-directed analyses in FFPE would be complicated by dynamic changes during the fixation process and due to complications of affinity enrichment and the difficulty of verification using Western blotting with FFPE samples.
An important lesson learned in our studies is the impact of methods for data analysis on the degree of overlap between frozen and FFPE protein identifications. Our initial analysis of the data involved separate database searches of the data from frozen and FFPE tissue. Comparison of the identification lists indicated a significant disparity in protein group identifications (>30% divergence; Fig. 6A), which suggested that application of a common analysis platform to the two sample types yielded different proteome inventories. However, this apparent disparity was an artifact caused by searching the FFPE and frozen tissue datasets separately and then comparing the identified protein lists. A search restricted to either FFPE or frozen tissue datasets imposes a limit on protein identifications due to the requirement for two peptide identifications per protein identification. For example, in separate searches of the FFPE and frozen tissue data, if a protein is identified by two peptides in the frozen tissue dataset, but only by one peptide in the FFPE tissue dataset, then that protein is assigned only to the frozen tissue when the protein lists are compared. When the combined datasets are searched together, single peptide identifications of proteins from one set (e.g. FFPE) can be “rescued” by identification of one or more additional peptides in the other set (e.g. frozen). The combined search recognizes evidence for the same protein in both datasets and more correctly represents the overlap of protein identifications between frozen and FFPE tissues at over 90% (Fig. 6B). This rationale is distinct from the issue of whether to accept single peptide-based protein identifications. If the requirement for protein identification by at least two peptide identifications is relaxed, then the overlap between FFPE and frozen tissues decreases dramatically, even when the FDR for peptide identification is made more stringent (Fig. 8).
Another important aspect of our work with FFPE tissues is the standardization of protein input. We empirically determined a correction factor for protein analysis of FFPE tissues using the bicinchoninic acid assay and used this approach to measure the amount of protein analyzed. In our experience, a colon adenoma FFPE sample of 60-μm thickness and 5-mm diameter typically yielded 300–400 μg of protein. We analyzed FFPE samples corresponding to 200 μg of protein, which is equivalent to ∼2 mg (wet weight) of fresh or frozen tissue. These samples typically yielded about 400 protein group identifications in reverse phase LC-MS/MS analyses and about 2300 protein group identifications in multidimensional IEF-reverse phase LC-MS/MS analyses. In work with laser capture microdissected prostate tissue, Hood et al. (7) identified ∼200–300 proteins with at least two peptide identifications from 100,000 cells. Their evaluation of gene ontology categories represented by their datasets from FFPE and similar frozen tissue specimens also suggested little or no selectivity bias in analysis of FFPE tissue. Thus, we expect that our conclusions regarding the equivalence of FFPE and frozen tissues apply to laser capture microdissected specimens.
An important advantage of working with FFPE tissues is the linkage of these specimens with information about clinical outcomes related to disease course or response to therapy. Because these outcomes may describe events occurring long after specimen collection, the proteins in archival FFPE samples must be sufficiently stable so that later analyses provide an accurate representation of the proteome at the time the tissue was obtained. Our results indicate that long-term storage of FFPE colon adenoma samples did not compromise the identification of proteins in these specimens. On the other hand, extended fixation for greater than 2 days did compromise protein identifications. These results provide evidence that use of archival FFPE specimens for retrospective studies is possible. Despite these encouraging results, we recognize that factors affecting the stability and utility of archival FFPE specimens will require much more detailed and systematic evaluation, particularly with respect to PTM. Our work builds upon the previous cited work on shotgun proteomics with FFPE tissue specimens and clearly establishes the identity of FFPE samples with corresponding fresh frozen tissue. This demonstration is a critical element in justifying their use in retrospective biomarker discovery studies.
Acknowledgments
We thank Sarah Stuart in the Ayers Institute for expert technical assistance.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health, NCI (the National Cancer Institute Clinical Proteomic Technologies Assessment for Cancer program Grant 1U24CA126479 and by the National Cancer Institute Specialized Program of Research Excellence in Gastrointestinal Cancer Grant P50CA95103).
 The on-line version of this article (available at http://www.mcp.org) contains supplemental material.
The on-line version of this article (available at http://www.mcp.org) contains supplemental material.
1 The abbreviations used are:
- FFPE
- formalin-fixed paraffin-embedded
- IHC
- immunohistochemistry
- PM
- pyridoxamine
- ID
- identification
- IEF
- isoelectric focusing
- FA
- formic acid
- ACN
- acetonitrile
- FDR
- false discovery rates
- ANOVA
- analysis of variance
- PTM
- posttranslational modification.
REFERENCES
- 1.Hood B. L., Conrads T. P., Veenstra T. D. ( 2006) Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics 6, 4106– 4114 [DOI] [PubMed] [Google Scholar]
- 2.Yamashita S. ( 2007) Heat-induced antigen retrieval: mechanisms and application to histochemistry. Prog. Histochem. Cytochem. 41, 141– 200 [DOI] [PubMed] [Google Scholar]
- 3.Huang S. N., Minassian H., More J. D. ( 1976) Application of immunofluorescent staining on paraffin sections improved by trypsin digestion. Lab. Invest. 35, 383– 390 [PubMed] [Google Scholar]
- 4.Shiozawa M., Aiso S., Hoshiai O., Tahara H., Yamashita S., Yasuda K. ( 1986) Localization of gamma-glutamyl transpeptidase in the proliferative state of liver. An enzyme histochemical and immunohistochemical study. Okajimas Folia Anat. Jpn. 63, 209– 221 [DOI] [PubMed] [Google Scholar]
- 5.Shi S. R., Key M. E., Kalra K. L. ( 1991) Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. 39, 741– 748 [DOI] [PubMed] [Google Scholar]
- 6.Namimatsu S., Ghazizadeh M., Sugisaki Y. ( 2005) Reversing the effects of formalin fixation with citraconic anhydride and heat: a universal antigen retrieval method. J. Histochem. Cytochem. 53, 3– 11 [DOI] [PubMed] [Google Scholar]
- 7.Hood B. L., Darfler M. M., Guiel T. G., Furusato B., Lucas D. A., Ringeisen B. R., Sesterhenn I. A., Conrads T. P., Veenstra T. D., Krizman D. B. ( 2005) Proteomic analysis of formalin-fixed prostate cancer tissue. Mol. Cell. Proteomics 4, 1741– 1753 [DOI] [PubMed] [Google Scholar]
- 8.Xu H., Yang L., Wang W., Shi S. R., Liu C., Liu Y., Fang X., Taylor C. R., Lee C. S., Balgley B. M. ( 2008) Antigen retrieval for proteomic characterization of formalin-fixed and paraffin-embedded tissues. J. Proteome Res. 7, 1098– 1108 [DOI] [PubMed] [Google Scholar]
- 9.Crockett D. K., Lin Z., Vaughn C. P., Lim M. S., Elenitoba-Johnson K. S. ( 2005) Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab. Invest. 85, 1405– 1415 [DOI] [PubMed] [Google Scholar]
- 10.Hwang S. I., Thumar J., Lundgren D. H., Rezaul K., Mayya V., Wu L., Eng J., Wright M. E., Han D. K. ( 2007) Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene 26, 65– 76 [DOI] [PubMed] [Google Scholar]
- 11.Jiang X., Jiang X., Feng S., Tian R., Ye M., Zou H. ( 2007) Development of efficient protein extraction methods for shotgun proteome analysis of formalin-fixed tissues. J. Proteome Res. 6, 1038– 1047 [DOI] [PubMed] [Google Scholar]
- 12.Palmer-Toy D. E., Krastins B., Sarracino D. A., Nadol J. B., Jr., Merchant S. N. ( 2005) Efficient method for the proteomic analysis of fixed and embedded tissues. J. Proteome Res. 4, 2404– 2411 [DOI] [PubMed] [Google Scholar]
- 13.Shi S. R., Liu C., Balgley B. M., Lee C., Taylor C. R. ( 2006) Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J. Histochem. Cytochem. 54, 739– 743 [DOI] [PubMed] [Google Scholar]
- 14.Guo T., Wang W., Rudnick P. A., Song T., Li J., Zhuang Z., Weil R. J., DeVoe D. L., Lee C. S., Balgley B. M. ( 2007) Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J. Histochem. Cytochem. 55, 763– 772 [DOI] [PubMed] [Google Scholar]
- 15.Elias J. E., Gygi S. P. ( 2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207– 214 [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Qian W. J., Mottaz H. M., Clauss T. R., Anderson D. J., Moore R. J., Camp D. G., 2nd, Khan A. H., Sforza D. M., Pallavicini M., Smith D. J., Smith R. D. ( 2005) Development and evaluation of a micro- and nanoscale proteomic sample preparation method. J. Proteome Res. 4, 2397– 2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cargile B. J., Sevinsky J. R., Essader A. S., Stephenson J. L., Jr., Bundy J. L. ( 2005) Immobilized pH gradient isoelectric focusing as a first-dimension separation in shotgun proteomics. J. Biomol. Tech. 16, 181– 189 [PMC free article] [PubMed] [Google Scholar]
- 18.Tabb D. L., Fernando C. G., Chambers M. C. ( 2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6, 654– 661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B., Chambers M. C., Tabb D. L. ( 2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6, 3549– 3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian W. J., Liu T., Monroe M. E., Strittmatter E. F., Jacobs J. M., Kangas L. J., Petritis K., Camp D. G., 2nd, Smith R. D. ( 2005) Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 4, 53– 62 [DOI] [PubMed] [Google Scholar]
- 21.Liu H., Sadygov R. G., Yates J. R., 3rd ( 2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193– 4201 [DOI] [PubMed] [Google Scholar]
- 22.Breslow N. ( 1990) Further studies in the variability of pock counts. Stat. Med. 9, 615– 626 [DOI] [PubMed] [Google Scholar]
- 23.Benjamini Y., Hochberg Y. ( 1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289– 300 [Google Scholar]
- 24.Shi S. R., Cote R. J., Hawes D., Thu S., Shi Y., Young L. L., Taylor C. R. ( 1999) Calcium-induced modification of protein conformation demonstrated by immunohistochemistry: What is the signal? J. Histochem. Cytochem. 47, 463– 470 [DOI] [PubMed] [Google Scholar]
- 25.Morgan J. M., Navabi H., Schmid K. W., Jasani B. ( 1994) Possible role of tissue-bound calcium ions in citrate-mediated high-temperature antigen retrieval. J. Pathol. 174, 301– 307 [DOI] [PubMed] [Google Scholar]
- 26.Metz B., Kersten G. F., Baart G. J., de Jong A., Meiring H., ten Hove J., van Steenbergen M. J., Hennink W. E., Crommelin D. J., Jiskoot W. ( 2006) Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjug. Chem. 17, 815– 822 [DOI] [PubMed] [Google Scholar]
- 27.Hansen B. T., Davey S. W., Ham A. J., Liebler D. C. ( 2005) P-Mod: an algorithm and software to map modifications to peptide sequences using tandem MS data. J. Proteome Res. 4, 358– 368 [DOI] [PubMed] [Google Scholar]
- 28.Gao Q., Xue S., Doneanu C. E., Shaffer S. A., Goodlett D. R., Nelson S. D. ( 2006) Pro-CrossLink. Software tool for protein cross-linking and mass spectrometry. Anal. Chem. 78, 2145– 2149 [DOI] [PubMed] [Google Scholar]
- 29.Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. ( 2008) Identification of cross-linked peptides from large sequence databases. Nature methods 5, 315– 318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slebos R. J. C., Brock J. W. C., Winters N. F., Stuart S. R., Martinez M. A., Li M., Chambers M. C., Zimmerman L. J., Ham A. J., Tabb D. L., Liebler D. C. ( 2008) Evaluation of strong cation exchange versus isoelectric focusing of peptides for multidimensional liquid chromatography-tandem mass spectrometry. J. Proteome Res. 7, 5286– 5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B., Kirov S., Snoddy J. ( 2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741– 748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voet D., Voet J. G. ( 1995) Biochemistry, 2nd Ed., p. 56, John Wiley and Sons, Inc., New York [Google Scholar]