Abstract
Parkinson's disease is a common neurodegenerative disorder in which familial-linked genes have provided novel insights into the pathogenesis of this disorder. Mutations in Parkin, a ring-finger-containing protein of unknown function, are implicated in the pathogenesis of autosomal recessive familial Parkinson's disease. Here, we show that Parkin binds to the E2 ubiquitin-conjugating human enzyme 8 (UbcH8) through its C-terminal ring-finger. Parkin has ubiquitin–protein ligase activity in the presence of UbcH8. Parkin also ubiquitinates itself and promotes its own degradation. We also identify and show that the synaptic vesicle-associated protein, CDCrel-1, interacts with Parkin through its ring-finger domains. Furthermore, Parkin ubiquitinates and promotes the degradation of CDCrel-1. Familial-linked mutations disrupt the ubiquitin–protein ligase function of Parkin and impair Parkin and CDCrel-1 degradation. These results suggest that Parkin functions as an E3 ubiquitin–protein ligase through its ring domains and that it may control protein levels via ubiquitination. The loss of Parkin's ubiquitin–protein ligase function in familial-linked mutations suggests that this may be the cause of familial autosomal recessive Parkinson's disease.
Parkinson's disease (PD) is the second most prevalent neurodegenerative disorder, but the etiology remains poorly understood (1, 2). Patients with PD suffer from rigidity, slowness of movement, tremor, and disturbances of balance (1, 2). PD is characterized by the progressive loss of dopamine-containing neurons in the substantia nigra pars compacta (3, 4) and the accumulation of Lewy bodies, proteinaceous intracytoplasmic accumulations of eosinophilic material that stain for ubiquitin (5). Novel insights into the molecular mechanisms of the pathogenesis of PD have come from the identification of genes associated with rare forms of familial PD (6). Mutations in α-synuclein (A53T and A30P) are linked to autosomal dominant PD (7, 8). This led to the discovery that α-synuclein is a major component of Lewy bodies and suggests that derangements in α-synuclein could be a major cause or contributor to the pathogenesis of sporadic PD (9, 10). Consistent with this notion are observations that overexpression of α-synuclein in transgenic fruit flies and mice causes a Parkinsonian phenotype and replicates many of the pathological features of PD (11–13).
Other autosomal dominant genes or loci linked to PD have been described and include a mutation (I93M) in ubiquitin carboxyl-terminal–hydrolase-L1 (14), a member of the ubiquitin C-terminal hydrolase family that hydrolyzes small C-terminal adducts of ubiquitin to generate ubiquitin monomers and is involved in facilitating the degradation and processing of proteins through the 26 S proteasome. Thus, derangements in ubiquitin processing may be linked to the pathogenesis of PD. Linkages to chromosome 2P and 4P have been described, as well as yet to be identified loci, but the genes await identification (15–17).
Mutations in the Parkin gene are responsible for autosomal recessive PD (18). Several Parkin-associated pedigrees have been described with both deletions and point mutations, as well as compound heterozygosity causing autosomal recessive PD (19–21). Recent studies suggest that mutations in Parkin are the major cause of autosomal recessive familial PD (19); thus, understanding the function of Parkin and how mutations interfere with the function of Parkin may provide novel insights into the pathogenesis of PD. The function of the Parkin protein remains unknown. However, Parkin shows mild homology to ubiquitin at the N terminus and contains two ring-finger motifs and an in-between ring-finger (IBR) domain at the C terminus (22). Recently, a few proteins with ring-finger motifs similar to Parkin were shown to be involved in E2-dependent ubiquitination (23–26). Ubiquitination requires the ATP-dependent activation of ubiquitin by the ubiquitin-activating enzyme E1. Ubiquitin is transferred to an E2 ubiquitin-conjugating enzyme, which works in conjunction with an E3 ubiquitin–protein ligase to ubiquitinate substrate proteins (23, 24). The existence of two ring-finger motifs and the N-terminal homology to ubiquitin suggests that Parkin may be involved in the ubiquitination pathway. Interestingly, several of the point mutations identified in Parkin fall within the two ring-finger domains, whereas the majority of the nonpathologic polymorphisms are located outside the ring-finger domains, thus implicating the importance of the ring-finger motifs in Parkin function (21). These recent findings prompted us to investigate whether the two ring-finger motifs and IBR domain of Parkin may provide an E2-binding interface and whether Parkin may function as an E3 ubiquitin–protein ligase. We show here that Parkin interacts with the E2 ubiquitin-conjugating enzyme, UbcH8, and has E3 ubiquitin–protein ligase activity. Parkin is self-ubiquitinated, and the self-ubiquitination promotes its own degradation. In an attempt to identify potential substrates of Parkin, we performed yeast two-hybrid screening and identified a synaptic protein, CDCrel-1. Parkin interacts with and ubiquitinates CDCrel-1 and promotes its degradation, whereas familial-linked Parkin mutants are defective in the degradation of CDCrel-1.
Materials and Methods
Database Searching.
Full-length Parkin protein sequence was entered as a query for a PSI-Blast search (http://www.ncbi.nlm.nih.gov/blast/psiblast.cgi) using default settings (27) and four iterations.
Generation of Plasmids.
Full-length Parkin cDNA was cloned into pRK5-myc vector and pDBleu vector between SalI and NotI sites. The cDNA sequences encoding amino acids 220–465, 220–403, 32–465, 220–318, 304–404, and 395–465 of Parkin were cloned into pRK5-myc vector between SalI and NotI sites to generate pRK5-myc-R1-IBR-R2, pRK5-myc-R1-IBR, pRK5-myc-IBR-R2, pRK5-myc-R1, pRK5-myc-IBR, and pRK5-myc-R2, respectively. The mutants pRK5-myc-ParkinThr240Arg, pRK5-myc-ParkinThr415Asn, pRK5-myc-ParkinCys421Ala, pRK5-myc-ParkinCys431Ala, pRK5-myc-ParkinGln311stop, and pRK5-myc-ParkinTrp453Stop were generated by PCR-mediated site-directed mutagenesis (28). Full-length mouse CDCrel-1 cDNA was cloned into pRK5-myc and pRK5-hemagglutinin (HA) vector to generate pRK5-myc-CDCrel-1 and pRK5-HA-CDCrel-1, respectively. Full-length UbcH5, UbcH7, and UbcH8 cDNA were cloned into pRK5-HA between SalI and NotI sites to generate pRK5-HA-UbcH5, pRK5-HA-UbcH7, and pRK5-HA-UbcH8. The integrity of all of the constructs was confirmed by sequencing.
Coimmunoprecipitation Assays.
HEK 293 cells were transfected with 2 μg of each plasmid using Lipofectamine (GIBCO). Thirty-six hours later, cells were washed with cold PBS and harvested in IP buffer (1% Triton X-100/2 μg/ml aprotinin/100 μg/ml PMSF in PBS). The lysate was then rotated at 4°C for 1 h followed by centrifugation at 16,000 × g for 15 min. The supernatants were combined with 50 μl of protein G Sepharose (Amersham Pharmacia) preincubated with anti-HA (Roche Molecular Biochemicals) or anti-myc antibody (Roche Molecular Biochemicals) followed by rotating at 4°C for 2 h. The protein G Sepharose was then spun down and washed thoroughly three times using IP buffer or IP buffer with additional 500 mM NaCl followed by three additional washes. The precipitates were resolved on SDS/PAGE gel and subjected to Western blot analysis.
In Vitro Ubiquitination Assays.
Parkin was in vitro translated by TnT using an in vitro translation kit according to the manufacturer's instructions (Promega). Translation reactions of Parkin (5 μl) were evaluated for ubiquitination in ubiquitination reaction buffer, which contains 50 mM Tris⋅HCl, pH 7.4/5 mM MgCl2/2 mM ATP/2 mM DTT/100 ng of E1 (Calbiochem)/5 μl of lysate of Escherichia coli overexpressing UbcH5, UbcH7, or UbcH8/10 μg of Ub (Sigma). Reactions were conducted for 1.5 h at 30°C. The reactions were terminated by adding SDS sample buffer and boiling for 5 min. The reactions were then resolved in 10% SDS/PAGE gel and subjected to Western blotting with anti-ubiquitin antibody.
In Vivo Ubiquitination Assays.
To determine the self-ubiquitination of Parkin, HEK 293 cells were transfected with 2 μg of pRK5-myc-Parkin or Parkin mutants and 2 μg of pMT123-HA-ubiquitin plasmids using Lipofectamine. Thirty-six hours later, immunoprecipitation was performed with an anti-myc antibody. The precipitates were analyzed via Western blot with anti-HA antibody. To determine ubiquitination of CDCrel-1, Myc-tagged CDCrel-1 and HA-tagged ubiquitin were cotransfected with pCMV-Parkin or control plasmid. 36 h later, cells were treated with the specific proteasome inhibitor clasto-Lactacystin β-lactone at 1 and 2 μM for indicated time periods. Immunoprecipitation was performed with an anti-myc antibody. The precipitates were analyzed via Western blot with anti-HA antibody.
Yeast Two-Hybrid Screening.
Saccharomyces cerevisiae Mav203 was transformed with PDBleu-Parkin, and 3 × 106 stable transformants were further transformed with 15 μg of pPC86 adult mouse brain library cDNA (CLONTECH). Transformants were selected and confirmed according to the manufacturer's instructions.
[35S]Methionine Pulse–Chase Experiments.
HEK 293 cells were transfected with corresponding plasmids as indicated in the figure legends using Lipofectamine. Twenty-four hours after transfection, the cells were washed and incubated with methionine-free medium for 1 h. The cells were then pulsed with 100 μCi of [35S]methionine for 3 h and then washed and chased in normal medium for 16 h. At 30 min, 1.5 h, 3 h, 6 h, and 16 h cells were harvested for immunoprecipitation with anti-myc antibody. The immunoprecipitates were resolved on a 10% SDS/PAGE gel and visualized with a phosphoimager. The amount of radioactivity was quantitated for each protein of interest (Parkin or CDCrel-1) by using the image analysis software of the Molecular Dynamics Phosphoimager and plotted versus time.
Results
Parkin Interacts with the E2 Ubiquitin-Conjugating Enzyme UbcH8.
Based on several missense mutations in the ring-finger domains of the Parkin gene being implicated in the pathogenesis of autosomal recessive PD, we conducted homology searches for proteins with similar ring-finger structures using PSI-BLAST. PSI-BLAST searches for proteins with common functional domains and can be used to identify proteins with similar putative function (27). We identified three proteins that share sequence similarity with Parkin including the human homologue of Drosophila ariadne (HHARI, NP005735), UbcH7-associated protein (H7-AP1, CAB45870), and UbcM4-interacting protein 28 (UIP28, AAD24572) (Fig. 1A). All three proteins share sequence similarities with Parkin by the presence of two ring fingers and an IBR domain. HHARI and H7-AP1 were recently shown to interact with the E2 ubiquitin-conjugating enzymes UbcH7 and UbcH8 (29). UIP28 was also recently shown to interact with the E2 ubiquitin-conjugating enzyme UbcM4, which is the mouse homologue of human UbcH8 (30).
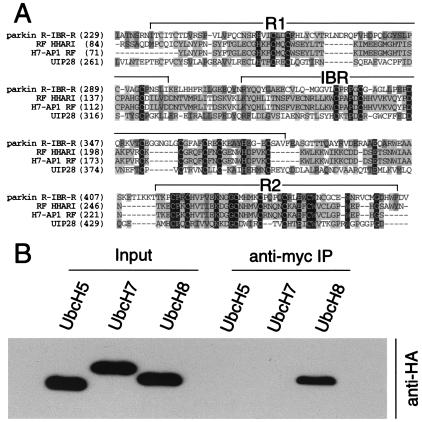
Figure 1.
Parkin interacts with the E2 ubiquitin-conjugating enzyme, UbcH8. (A) Multiple sequence alignment of ring-finger domains of Parkin (BAA25751), HHARI (NP005735), H7-AP1 (CAB45870), and UIP28 (AAD24572). Conserved residues are highlighted, and the ring fingers (R1 and R2) and the IBR domain are indicated. Abbreviations: HHARI, ariadne homologue (Homo sapiens); H7-AP1, UbcH7-binding protein; UIP28, UbcM4 interacting protein 28. (B) Lysates prepared from HEK 293 cells transfected with pRK5-myc-Parkin and pRK5-HAUbcH5, pRK5-HAUbcH7, or pRK5-HAUbcH8 were subjected to immunoprecipitation (IP) with anti-myc antibody followed by anti-HA immunoblotting. This experiment was replicated three times with similar results.
Because Parkin shares sequence similarities with three proteins that either interact with the E2 ubiquitin-conjugating enzymes UbcH7 and UbcH8 or the mouse UbcH8 homologue, we reasoned that Parkin might interact with UbcH7 or UbcH8. Accordingly, we monitored the ability of Parkin to interact with UbcH7, UbcH8, and the unrelated E2 UbcH5 (Fig. 1B). Cotransfection of Myc-tagged Parkin and HA-tagged UbcH5, UbcH7, and UbcH8 followed by coimmunoprecipitation reveals that Parkin selectively interacts with UbcH8 (Fig. 1B). Interestingly, UbcH8 is enriched in the central nervous system (31).
Parkin Binds UbcH8 Through Its R2 Ring-Finger Domain, and Familial-Linked and Ring-Finger Parkin Mutants Have Defective E2 Binding.
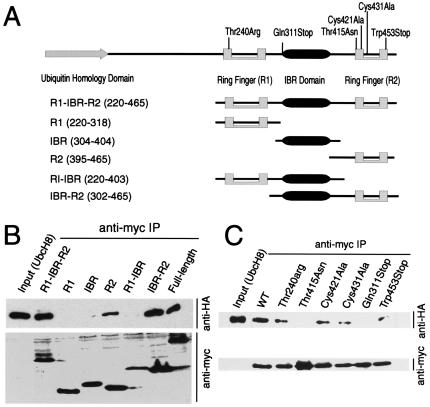
A series of constructs containing the ring fingers and IBR domains of Parkin were generated to map the UbcH8, E2-binding domain (Fig. 2B). A coimmunoprecipitation experiment reveals that the C-terminal ring-finger (R2) domain of Parkin is the minimal region required for UbcH8 binding and that UbcH8 does not bind to the N-terminal ring-finger (R1) domain (Fig. 2B). The combined R2 and IBR domains bind UbcH8 stronger than R2 domain alone (Fig. 2B).
Figure 2.
UbcH8 interacts preferentially with the C-terminal R2 ring-finger domain of Parkin. (A) Schematic representation of putative functional domains of Parkin used in the mapping experiments. (B) Lysates prepared from HEK 293 cells transfected with pRK5-HA-UbcH8 and various Myc-tagged Parkin domain constructs were subjected to immunoprecipitation (IP) with anti-myc antibody followed by anti-HA immunoblotting (Upper). The blot was stripped and reprobed with the anti-myc antibody (Lower) to illustrate that equivalent levels of the Parkin constructs were present in the extracts. This experiment was replicated three times with similar results. (C) Lysates prepared from HEK 293 cells transfected with pRK5-HA-UbcH8 and various Myc-tagged Parkin mutant constructs (see text for details) were subjected to immunoprecipitation (IP) with anti-myc antibody followed by anti-HA immunoblotting (Upper). The blot was stripped and reprobed with the anti-Myc antibody (Lower) to illustrate that equivalent levels of the Parkin constructs were present in the extracts. This experiment was replicated three times with similar results.
Because other ring-finger-containing E3 ubiquitin-protein ligases require active cysteines for a functional ring-finger domain (26) and the identification of several missense mutations in the ring-finger domains of Parkin cause PD, we assessed the requirements for cysteine residues and the effects of a variety of familial-linked Parkin mutations on the ability of Parkin to bind to UbcH8 (Fig. 2C). We elected to focus our attention on the R2 ring-finger domain as Parkin binds UbcH8 with its second ring finger. Initial experiments indicate that Parkin and the Parkin mutants interact with relatively high affinity with UbcH8, as we were only able to discern differences in binding to UbcH8 when the coimmunoprecipitates were washed with 500 mM NaCl (Fig. 2C). In this assay, the familial-linked missense mutation Thr415Asn within the R2 ring-finger domain, the familial-linked mutation Gln311Stop that results in a truncated protein without the IBR and R2 ring-finger domain, completely eliminates the ability of Parkin to interact with UbcH8 (Fig. 2C). In contrast, the familial-linked mutation Trp453Stop that results in a truncated protein without the last 12 amino acids impairs, but does not eliminate, the interaction between Parkin and UbcH8. The familial-linked R1 ring-finger mutant Thr240Arg partially reduces UbcH8 binding, suggesting that an intact R1 ring-finger domain may be required for full-length Parkin to interact with UbcH8. We further explored the role for the ring fingers in regulating the interaction between UbcH8 and Parkin by mutating cysteines within the second ring finger (Fig. 2C). Mutations of either the first or second cysteine of the R2 ring-finger domain (Cys421Ala and Cys431Ala, respectively) reduces the interaction between Parkin and UbcH8 (Fig. 2C). These results, taken together, indicate that the ring-finger R2 domain is critical for the interaction between Parkin and UbcH8 and suggest that this interaction may be important for Parkin-mediated ubiquitination.
Parkin Has E3 Ubiquitin–Protein Ligase Activity, and Familial-Linked Parkin Mutants Are Defective in E3 Ubiquitin–Protein Ligase Activity.
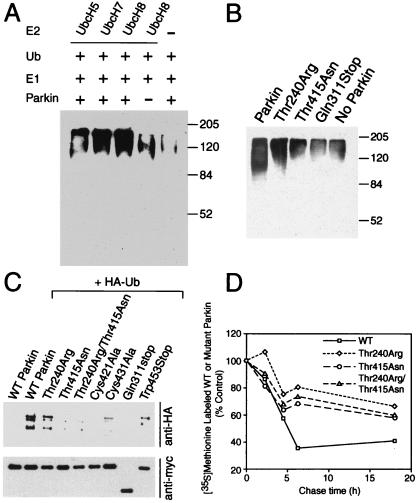
To ascertain whether Parkin might have a role in ubiquitination, we determined whether Parkin has E2-dependent E3 ubiquitin–protein ligase activity in vitro (Fig. 3A). Free ubiquitin and the ubiquitin-activating enzyme (E1) were incubated with bacterial lysates expressing UbcH8, UbcH7, UbcH5, or control bacteria lysate, in the presence and absence of in vitro translated Parkin in the ubiquitination reaction buffer (see Materials and Methods). Ubiquitinated species were monitored by Western blot analysis with an anti-ubiquitin antibody. Significantly more ubiquitination occurs in the presence of UbcH8 and Parkin and also, to a lesser extent, in the presence of Parkin and the closely related E2, UbcH7. The ubiquitination observed in the absence of Parkin is probably caused by intrinsic E3 ubiquitin–protein ligase activity that is contained within reticulocyte lysates. The unrelated UbcH5 also appears to partially support Parkin-dependent ubiquitination, which may be related to the high concentration of the E2s contained within the bacterial lysates. Thus, Parkin appears to be an E2-dependent ubiquitin ligase (Fig. 3A), and UbcH8 and UbcH7 can both support Parkin-mediated ubiquitination, whereas UbcH8 is more efficient.
Figure 3.
Parkin is involved in E2-dependent self-ubiquitination both in vitro and in vivo. (A) In vitro translated Parkin (5 μl) was incubated with 100 ng of E1, 10 μg of ubiquitin (Ub) with 5 μl of bacterially expressed UbcH8, UbcH7, UbH5, or control bacteria lysate in the ubiquitination reaction buffer for 1.5 h at 30°C. The reaction products were immunoblotted with anti-ubiquitin antibody. Molecular weight markers are indicated. This experiment was replicated three times with similar results. (B) In vitro translated Parkin (5 μl) or Parkin mutants were incubated with 100 ng of E1, 10 μg of Ub with 5 μl of bacterially expressed UbcH8 in the ubiquitination reaction buffer for 1.5 h at 30°C. The reaction products were immunoblotted with anti-ubiquitin antibody. Molecular weight markers are indicated. This experiment was replicated three times with similar results. (C) Lysates prepared from HEK 293 cells transfected with pRK5-myc-Parkin or myc-tagged mutant Parkin constructs and pMT123-HA-ubiquitin were subjected to immunoprecipitation (IP) with anti-myc antibody followed by anti-HA immunoblotting (Upper). The blot was stripped and reprobed with the anti-myc antibody (Lower) to illustrate that equivalent levels of the Parkin constructs were present in the extracts. This experiment was replicated three times with similar results. (D) Familial-linked Parkin mutants impair Parkin degradation. HEK 293 cells were transfected with pRK5-myc-Parkin, pRK5-myc-ParkinThr240Arg, pRK5-myc-Parkin-Thr415Asn, and pRK5-myc-ParkinThr240Arg/Thr415Asn plasmids using Lipofectamine. 24 h after transfection, cells were washed and incubated with methionine-free medium for 1 h. The cells were pulsed-chased with 100 μCi of [35S]methionine and were harvested at the indicated times for immunoprecipitation with anti-myc antibody. The immunoprecipitates were resolved on a 10% SDS/PAGE gel and visualized and quantitated with a phosphoimager. This experiment was replicated with similar results.
To determine whether the mutations in Parkin that prevent or alter the interaction with UbcH8 affect the ability of Parkin-mediated ubiquitination, we monitored the in vitro ubiquitination activity of mutant Parkin in the presence of UbcH8, E1, and ubiquitin (Fig. 3B). The familial-linked mutation Thr415Asn, which occurs in the second ring finger, and the Gln311Stop mutation, which truncates the IBR and R2 ring-finger domains of Parkin, prevent Parkin-mediated ubiquitination (Fig. 3B). The familial-linked Thr240Arg mutation, which occurs in the first ring-finger domain, has decreased E3 ubiquitin–protein ligase activity. These results taken together suggest that Parkin functions as an E2-dependent E3 ubiquitin–protein ligase and that the second ring-finger (R2) of Parkin is essential and the first ring finger (R1) may be required for E3 ubiquitin–protein ligase activity of Parkin.
Parkin Ubiquitinates Itself Leading to Its Degradation, and Familial-Linked and Ring-Finger Parkin Mutants Impair Parkin Degradation Through Defective Self-Ubiquitination.
Given the precedence that Mdm2, a ring-finger protein with E3 ubiquitin–protein ligase activity, ubiquitinates its target, p53, and also itself (32), we tested whether Parkin is also self-ubiquitinated in intact cells. In this assay, HEK 293 cells were transfected with pRK5-myc-Parkin and pMT123-HA-ubiquitin plasmids using Lipofectamine. After 36 h, immunoprecipitation was performed with an anti-myc antibody. The precipitates were stringently and extensively washed and then submitted to Western blot analysis with anti-HA antibody. The immunoprecipitated Parkin shows significant anti-HA immunoreactivity, consistent with Parkin self-ubiquitination (Fig. 3C).
We then tested whether familial-linked Parkin mutations affects the self-ubiquitination of Parkin using the same assay (Fig. 3C). The familial-linked R2 ring-finger mutation Thr415Asn markedly reduces Parkin self-ubiquitination. The familial-linked truncation mutant, Gln311Stop, which eliminates the IBR and R2 ring-finger domains, has no detectable self-ubiquitination. In contrast, the familial-linked mutation Trp453Stop that results in a truncated protein without the last 12 amino acids impairs, but does not eliminate, Parkin self-ubiquitination, which is consistent with it partially binding to the E2, UbcH8. The familial-linked R1 ring-finger mutant, Thr240Arg, also decreases the self-ubiquitination, consistent with the in vitro ubiquitination and E2 binding assays. We further tested whether disruption of the second ring-finger domain affects the self-ubiquitination of Parkin by examining the R2 ring-finger cysteine mutants. Both Cys421Ala and Cys431Ala mutations significantly decrease the self-ubiquitination of Parkin (Fig. 3C).
We next tested whether the self-ubiquitination of Parkin regulates its own degradation and whether the familial-linked mutants have different degradation kinetics. We metabolically labeled Parkin with [35S]methionine and chased it for 18 h by harvesting cells at different time points and measuring the amount of [35S]methionine labeled immunoprecipitated Parkin (Fig. 3D). Pulse–chase experiments show that the familial-linked mutants, Thr240Arg, Thr415Asn, and a synthetic mutant harboring both of mutations, have significantly slower degradation rates than wild-type Parkin (Fig. 3D). Thus, Parkin ubiquitinates itself and thereby promotes its own degradation, and familial-linked mutations impair this process.
Parkin Interacts with and Ubiquitinates and Promotes the Turnover of CDCrel-1.
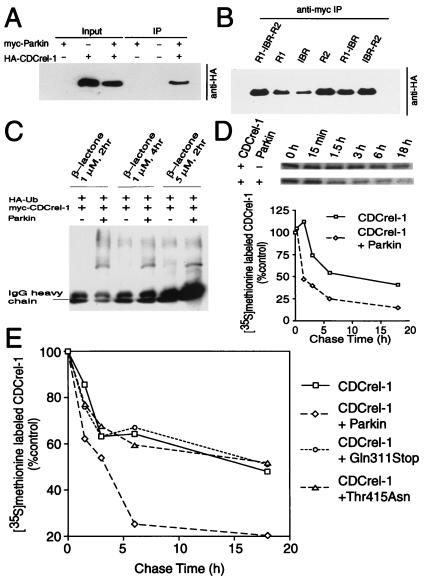
The ability of Parkin to interact with UbcH8 and ubiquitinate itself suggests that Parkin may target one or more proteins for ubiquitin-mediated degradation through the 26 S proteasome. To identify a potential protein, which Parkin might target for ubiquitin-dependent degradation, we conducted yeast two-hybrid screening (data not shown). We screened an adult mouse brain cDNA library with full-length Parkin as bait and identified and confirmed one positive interacting clone, CDCrel-1, a synaptic vesicle-enriched septin GTPase of 44 kDa implicated in the inhibition of exocytosis through its interactions with syntaxin (33). To confirm the yeast two-hybrid results, we performed cotransfection experiments with myc-tagged Parkin and HA-tagged CDCrel-1 followed by coimmunoprecipitation. CDCrel-1 coimmunoprecipitates with Parkin (Fig. 4A). We next evaluated whether Parkin interacts with CDCrel-1 through its ring-finger domains by cotransfecting myc-tagged Parkin domain constructs and HA-tagged CDCrel-1 followed by coimmunoprecipitation. Our results show that Parkin interacts with CDCrel-1 through the entire ring-finger domain with preferential binding to the R2 ring-finger domain (Fig. 4B). Familial-linked Parkin mutants do not disrupt the interaction with CDCrel-1 (data not shown).
Figure 4.
Parkin interacts with, ubiquitinates and promotes the degradation of CDCrel-1. (A) Lysates prepared from HEK 293 cells transfected with pRK5-HA-CDCrel-1 and pRK5-myc-Parkin constructs were subjected to immunoprecipitation (IP) with anti-myc antibody followed by anti-HA immunoblotting. This experiment was replicated three times with similar results. (B) The ring-finger domains of Parkin interact with CDCrel-1. Lysates prepared from HEK 293 cells transfected with pRK5-HA-CDCrel-1 and myc-tagged Parkin domain constructs were subjected to immunoprecipitation (IP) with anti-myc antibody followed by anti-HA immunoblotting. This experiment was replicated three times with similar results. (C) Parkin ubiquitinates CDCrel-1. HEK 293 cells were transfected with pRK5-Myc-CDCrel-1, pMT123-HA-ubiquitin and pCMV-Parkin or control plasmid. Cells were then treated with 1 or 2 μM clasto-Lactacystin β-lactone for indicated times. Lysates were prepared from these cells and subjected to immunoprecipitation (IP) with anti-myc antibody followed by anti-HA immunoblotting. This experiment was replicated with similar results. (D) Parkin promotes the degradation of CDCrel-1. HEK 293 cells were transfected pRK5-myc-CDCrel-1 and pRK5-myc-Parkin or empty vector. 24 h after transfection, cells were washed and incubated with methionine-free medium for 1 h. The cells were pulsed-chased with 100 μCi of [35S]methionine and were harvested at the indicated times for immunoprecipitation with anti-myc antibody. The immunoprecipitates were resolved on a 10% SDS/PAGE gel and visualized and quantitated with a phosphoimager. This experiment was replicated with similar results. (E) Familial-linked Parkin mutants fail to degrade CDCrel-1. The same experimental paradigm as described in Fig. 4D were performed with the familial-linked Parkin mutant pRK5-myc-Parkin-Thr415Asn or pRK5-myc-ParkinGln311 and compared with wild-type Parkin. This experiment was replicated with similar results.
We next evaluated whether Parkin ubiquitinates CDCrel-1 by an in vivo ubiquitination assay in the presence of the specific proteasome inhibitor clasto-Lactacystin β-lactone. Ubiquitination of CDCrel-1 is significantly increased in the presence of Parkin (Fig. 4C).
To investigate whether Parkin regulates CDCrel-1 degradation, we analyzed CDCrel-1 turnover in the presence or absence of Parkin or familial-linked Parkin mutants by pulse–chase experiments. CDCrel-1 levels are significantly reduced at all of the time points during the chase process in the presence of Parkin (Fig. 4D). The familial-linked mutations Thr415Asn and Gln311Stop do not significantly affect CDCrel-1 turnover (Fig. 4E). Taken together, these data suggest that CDCrel-1 is a target of Parkin and that Parkin regulates CDCrel-1 turnover.
Discussion
Our findings indicate that Parkin is a ring-finger containing E2-dependent E3 ubiquitin–protein ligase. Parkin preferentially interacts with the E2, UbcH8, through its C-terminal R2 ring-finger domain. Parkin ubiquitinates itself in an E2-dependent manner leading to its own degradation. Furthermore, Parkin interacts with, ubiquitinates, and regulates the turnover of the synaptic vesicle-associated protein, CDCrel-1. Familial-linked and ring-finger-containing cysteine mutations, particularly within the R2 ring-finger domain, disrupt the interaction of Parkin with UbcH8 and impair the ability of Parkin self-ubiquitination and retard Parkin-mediated degradation of itself and CDCrel-1. Thus, Parkin joins a growing family of ring-finger-containing proteins that function as E3 ubiquitin–protein ligases (23–26), and disruption of the E3 ubiquitin-protein ligase activity of Parkin is likely to be the cause of autosomal recessive PD.
While this work was in preparation, Shimura and colleagues reported that Parkin functions as a ubiquitin–protein ligase by preferentially binding to the E2, UbcH7, as well as UbcH8, although to a lesser degree (34). In contrast, we show that Parkin preferentially binds UbcH8. The difference in Parkin's E2 binding preference between the two studies may be related to methodologic differences. Because our in vitro ubiquitination assays indicate that UbcH7 can support Parkin-mediated ubiquitination (see Fig. 3), it is likely that Parkin interacts with both E2s, and the cellular and methodologic context may determine Parkin's preference for UbcH7, UbcH8, or as yet to be identified E2s. As UbcH8 is abundantly expressed in the central nervous system (31), whereas UbcH7 has barely detectable levels (35), we believe that UbcH8 plays a more prominent role in the function of Parkin in the central nervous system.
CDCrel-1 is a member of the septin family, comprising GTPases required for the completion of cytokinesis in diverse organisms (36, 37). Members of the septin family of proteins may function in synaptic vesicle transport, fusion, or recycling events in the brain. CDCrel-1 is predominantly expressed in the nervous system and is associated with membrane fractions (33). A significant fraction of the protein copurifies and coprecipitates with synaptic vesicles. CDCrel-1 directly binds to syntaxin via the SNARE interaction domain. Transfection of HIT-T15 cells with wild-type CDCrel-1 inhibits secretion, whereas GTPase dominant-negative mutants enhance secretion; thus, CDCrel-1 may regulate synaptic vesicle dynamics through interactions with syntaxin (33). Whether CDCrel-1 regulates dopamine release awaits further clarification, but failure of CDCrel-1 to be degraded by familial-linked Parkin mutations could increase the steady-state level of CDCrel-1 and thus potentially inhibit dopamine release and contribute to a Parkinsonian state. The mechanism underlying the degradation and turnover of synaptic vesicle-associated proteins has not been clarified, despite the identification of ubiquitin in the postsynaptic density and synaptic terminals (38–40). However, it is likely that Parkin or other nervous system-enriched E3s may play a prominent role in regulation of synaptic vesicle-associated protein levels. It is also not clear whether alterations in the turnover of CDCrel-1 contribute to the degeneration of dopamine neurons. However, three other members of the human septin family, Nedd5, H5, and Diff6, were identified in neurofibrillary tangles, neuropil threads, and dystrophic neurites in senile plaques in brains affected by Alzheimer's disease (41). These findings and our data suggest that septins, such as CDCrel-1, could be involved in neurodegeneration.
The E3 ubiquitin–protein ligase function of Parkin is likely to play a prominent role in maintaining “normal” dopaminergic function as patient mutations in Parkin that cause familial-linked PD disrupt the ubiquitin–protein ligase function of Parkin. Parkin seems to function as an adapter between the E2, UbcH8, and its target CDCrel-1. The disruption of Parkin's ability to ubiquitinate and target proteins for degradation could have profound effects on normal central nervous system physiology. Whether the inability to degrade CDCrel-1 is important in the pathogenesis of autosomal recessive PD awaits further study. It is likely that Parkin interacts with other yet to be identified substrate proteins, which may be important in the pathogenesis of PD. PD is characterized pathologically by the presence of cytoplasmic proteinaceous inclusions (Lewy bodies), which stain abundantly with ubiquitin (42). Patients with mutations in Parkin do not have Lewy bodies (43–45). Our observations that Parkin functions in the ubiquitin proteasome degradation pathway coupled with the postmortem observations suggest that Parkin function might contribute to the formation of Lewy bodies and that Lewy body formation may not be required for the development of PD. Further study of Parkin's role in the development of Parkinson's disease pathology, especially Parkin's interaction with proteins in the Lewy body and Parkin's role in Lewy body formation, will greatly enhance our knowledge and understanding of the pathophysiology of PD.
Acknowledgments
We thank Drs. P. Howley, J. Huibregtse, and M. Scheffner for providing UbcH5, UbcH7, and UbcH8 plasmids and other ubiquitin constructs and thank Dr. C. Pickart for providing the HA-ubiquitin plasmid. We also thank Bing Ye and Haining Zhong for technical help and Ann Schmidt for manuscript preparation. This work was supported by United States Public Health Service Grant NS38377 and the Edward O. and Anna Mitchell Family Foundation.
Abbreviations
- PD
Parkinson's disease
- IBR
in-between ring-finger
- HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240347797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240347797
References
- 1.Lang A E, Lozano A M. N Engl J Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- 2.Lang A E, Lozano A M. N Engl J Med. 1998;339:1130–1143. doi: 10.1056/NEJM199810153391607. [DOI] [PubMed] [Google Scholar]
- 3.Jenner P, Olanow C W. Ann Neurol. 1998;44:S72–S84. doi: 10.1002/ana.410440712. [DOI] [PubMed] [Google Scholar]
- 4.Dunnett S B, Bjorklund A. Nature (London) 1999;399:A32–A39. doi: 10.1038/399a032. [DOI] [PubMed] [Google Scholar]
- 5.Pollanen M S, Dickson D W, Bergeron C. J Neuropathol Exp Neurol. 1993;52:183–91. doi: 10.1097/00005072-199305000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Dawson V L, Dawson T M. Neurobiol Dis. 2000;7:240–250. doi: 10.1006/nbdi.2000.0319. [DOI] [PubMed] [Google Scholar]
- 7.Polymeropoulos M H, Lavedan C, Leroy E, Ide S E, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 8.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, Przuntek H, Epplen J T, Schols L, Riess O. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 9.Spillantini M G, Schmidt M L, Lee V M, Trojanowski J Q, Jakes R, Goedert M. Nature (London) 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 10.Trojanowski J Q, Goedert M, Iwatsubo T, Lee V M. Cell Death Differ. 1998;5:832–837. doi: 10.1038/sj.cdd.4400432. [DOI] [PubMed] [Google Scholar]
- 11.Masliah E, Rockenstein E, Veinbergs I, Mallory M, Hashimoto M, Takeda A, Sagara Y, Sisk A, Mucke L. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- 12.Feany M B, Bender W W. Nature (London) 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 13.Dawson T M. Cell. 2000;101:115–118. doi: 10.1016/S0092-8674(00)80629-7. [DOI] [PubMed] [Google Scholar]
- 14.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein M J, Jonnalagada S, Chernova T, et al. Nature (London) 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 15.Farrer M, Gwinn-Hardy K, Muenter M, DeVrieze F W, Crook R, Perez-Tur J, Lincoln S, Maraganore D, Adler C, Newman S, et al. Hum Mol Genet. 1999;8:81–85. doi: 10.1093/hmg/8.1.81. [DOI] [PubMed] [Google Scholar]
- 16.Gasser T, Muller-Myhsok B, Wszolek Z K, Oehlmann R, Calne D B, Bonifati V, Bereznai B, Fabrizio E, Vieregge P, Horstmann R D. Nat Genet. 1998;18:262–265. doi: 10.1038/ng0398-262. [DOI] [PubMed] [Google Scholar]
- 17.Gwinn-Hardy K A, Crook R, Lincoln S, Adler C H, Caviness J N, Hardy J, Farrer M. Neurology. 2000;54:504–507. doi: 10.1212/wnl.54.2.504. [DOI] [PubMed] [Google Scholar]
- 18.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Nature (London) 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 19.Lucking C B, Durr A, Bonifati V, Vaughan J R, De Michele G, Gasser T, Harhangi B S, Meco G, Denefle P, Wood N W, et al. N Engl J Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 20.Lucking C B, Abbas N, Durr A, Bonifati V, Bonnet A M, de Broucker T, De Michele G, Wood N W, Agid Y, Brice A. Lancet. 1998;352:1355–1356. doi: 10.1016/s0140-6736(05)60746-5. [DOI] [PubMed] [Google Scholar]
- 21.Abbas N, Lucking C B, Ricard S, Durr A, Bonifati V, De Michele G, Bouley S, Vaughan J R, Gasser T, Marconi R, et al. Hum Mol Genet. 1999;8:567–574. doi: 10.1093/hmg/8.4.567. [DOI] [PubMed] [Google Scholar]
- 22.Morett E, Bork P. Trends Biochem Sci. 1999;24:229–231. doi: 10.1016/s0968-0004(99)01381-x. [DOI] [PubMed] [Google Scholar]
- 23.Kornitzer D, Ciechanover A. J Cell Physiol. 2000;182:1–11. doi: 10.1002/(SICI)1097-4652(200001)182:1<1::AID-JCP1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 24.Ciechanover A, Orian A, Schwartz A L. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 25.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 26.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- 29.Moynihan T P, Ardley H C, Nuber U, Rose S A, Jones P F, Markham A F, Scheffner M, Robinson P A. J Biol Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Noel G, Niedenthal R, Tamura T, Harbers K. FEBS Lett. 1999;454:257–261. doi: 10.1016/s0014-5793(99)00823-6. [DOI] [PubMed] [Google Scholar]
- 31.Kimura M, Hattori T, Matsuda Y, Yoshioka T, Sumi N, Umeda Y, Nakashima S, Okano Y. Cytogenet Cell Genet. 1997;78:107–111. doi: 10.1159/000134639. [DOI] [PubMed] [Google Scholar]
- 32.Fang S, Jensen J P, Ludwig R L, Vousden K H, Weissman A M. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 33.Beites C L, Xie H, Bowser R, Trimble W S. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 34.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, et al. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 35.Katsanis N, Fisher E M. Genomics. 1998;51:128–131. doi: 10.1006/geno.1998.5263. [DOI] [PubMed] [Google Scholar]
- 36.Trimble W S. J Membr Biol. 1999;169:75–81. doi: 10.1007/s002329900519. [DOI] [PubMed] [Google Scholar]
- 37.Field C M, Kellogg D. Trends Cell Biol. 1999;9:387–394. doi: 10.1016/s0962-8924(99)01632-3. [DOI] [PubMed] [Google Scholar]
- 38.Beesley P W, Mummery R, Tibaldi J, Chapman A P, Smith S J, Rider C C. Biochem Soc Trans. 1995;23:59–64. doi: 10.1042/bst0230059. [DOI] [PubMed] [Google Scholar]
- 39.Chapman A P, Smith S J, Rider C C, Beesley P W. Neurosci Lett. 1994;168:238–242. doi: 10.1016/0304-3940(94)90459-6. [DOI] [PubMed] [Google Scholar]
- 40.Chapman A P, Courtney S C, Smith S J, Rider C C, Beesley P W. Biochem Soc Trans. 1992;20:155S. doi: 10.1042/bst020155s. [DOI] [PubMed] [Google Scholar]
- 41.Kinoshita A, Kinoshita M, Akiyama H, Tomimoto H, Akiguchi I, Kumar S, Noda M, Kimura J. Am J Pathol. 1998;153:1551–1560. doi: 10.1016/S0002-9440(10)65743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galloway P G, Mulvihill P, Perry G. Am J Pathol. 1992;140:809–822. [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, Morita T, Tsuji S, Ikuta F. Neurology. 1994;44:437–441. doi: 10.1212/wnl.44.3_part_1.437. [DOI] [PubMed] [Google Scholar]
- 44.Mori H, Kondo T, Yokochi M, Matsumine H, Nakagawa-Hattori Y, Miyake T, Suda K, Mizuno Y. Neurology. 1998;51:890–892. doi: 10.1212/wnl.51.3.890. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa A, Takahashi H. J Neurol. 1998;245:4–9. [Google Scholar]