Abstract
Glycerol nucleic acid (GNA) is an interesting base-pairing system with an acyclic, three-carbon backbone. In the present study, GNA analogues with N2′→P3′ phosphoramidate linkages (npGNA) have been synthesized and their base-pairing properties examined. Thermal denaturation and circular dichroism studies show that npGNA can form stable duplexes with itself and with GNA. Furthermore, we show that npGNA can be assembled by template-directed ligation of 3′-imidazole-activated-2′-amino GNA dinucleotides. These results suggest that npGNA is a potential candidate for a self-replicating system based upon phosphoramidate linkages.
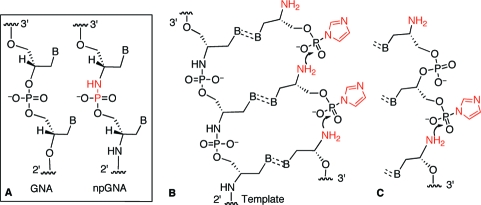
Glycerol nucleic acid (GNA), which is based upon an acyclic, three-carbon backbone (Figure 1A), is a promising starting point for the construction of an artificial genetic system owing to its ability to form stable, antiparallel duplexes.1,2 As proposed by Orgel,(3) we imagine that a nonenzymatic, template-directed polymerization process mediated replication during the evolution of early genetic systems,4,5 in place of enzymatic replication as in the modern genetic system. To recapitulate this process, our group has focused on the chemistry of polymerization facilitated by an imidazole-activated phosphate group and an amino group as the nucleophile (Figure 1B and 1C, highlighted in red).(5) This approach requires the newly synthesized strand with N→P phosphoramidate linkages to form stable, Watson−Crick base pairs with the template strand. The base-pairing properties of the GNA analogue with N2′→P3′ phosphoramidate linkages (npGNA, Figure 1A) have not been fully explored.(6) In this study, we demonstrate that npGNA can form stable duplexes with itself and with GNA. We also show that GNA oligonucleotides containing N2′→P3′ phosphoramidate linkages can be assembled via a nonenzymatic, template-directed ligation of 3′-imidazole-activated-2′-amino GNA dinucleotides (Figure 1C). These results suggest that npGNA is a potential candidate for a self-replicating system based on phosphoramidate linkages.
Figure 1.
GNA-based genetic system. (A) Structures of GNA and npGNA. (B and C) Chemistry of templated synthesis of npGNA using imidazole-activated monomer (B) or dimer (C). The nucleophile and leaving group are highlighted in red.
npGNA oligomers were synthesized via a solid-phase, oxidative amination coupling method7,8 using 2′-amino glycerol nucleosides as the monomers (1t−1g, Scheme 1). Compared with the lengthy synthesis (7−9 steps) of phosphoramidites used in the previously reported amine-exchange method,(6)1t−1g can be prepared in 2−3 steps from the readily available 3′-dimethoxytrityl (DMT) protected glycerol nucleosides 2t−2g (Scheme 1).(9) Each synthetic cycle of monomer addition consists of the following steps: detritylation, phosphitylation of the 3′-hydroxyl, hydrolysis, and oxidation followed by coupling as previously described by Gryaznov et al. (Scheme 2) (see Supporting Information for detailed procedures).7,8 Controlled-pore glass (CPG) resins premodified with thymidine were used in the synthesis (Scheme 2), which led to an extra thymidine at the 2′-terminus in all npGNA sequences synthesized in this study (Table S1).
Scheme 1. Synthesis of 2′-Amino Glycerol Nucleoside Monomers.
For 1t−4t, B = thymine; for 1c−4c, B = N4-benzoylcytosine; for 1a−4a, B = N6-benzoyladenine; for 1g−4g, B = N2-isobutyrylguanine. For 3t, R1 = O, R2 = CH3; for 3c, R1 = NBz, R2 = H. Conditions and yields: (a) DAST, pyridine/CH2Cl2; 3t, not isolated; 3c, 54%. (b) NaN3, 1:1 DMF/HMPA, 110 °C; 4t, 42%; 4c, 46%. (c) (i) MesCl, Et3N/CH2Cl2 (ii) NaN3, 1:1 DMF/HMPA, 100 °C. 4a, 71%; 4g, 96%. (d) For 4t and 4g, H2, Pd/C, 78% and 77%, respectively; for 4c and 4a, H2S, 15% Et3N in Pyridine, 0 °C, 83% and 91%, respectively.
Scheme 2. Synthesis of npGNA Using Solid-Phase, Oxidative Amination Coupling.
Conditions: (a) 3% dichloroacetic acid in CH2Cl2; (b) 0.4 M 2-cyanoethyl-N,N′-diisopropylchlorophosphite/0.4 M diisopropylethylamine in CH2Cl2; (c) 0.4 M tetrazole in 9:1 acetonitrile/water; (d) 0.2 M 1t, 1c, 1a, or 1g and 0.2 M triethylamine in 1:1 CCl4/acetonitrile. Functional groups involved in each transformation are highlighted in red.
This method was validated by the synthesis of dinucleotide model compounds (5t−5g, Figure S1). Only one diastereomer was observed for all 4 dinucleotides in 1H and 31P NMR spectra, indicating a clean SN2 attack by azide leading to 4t−4g. In addition, the dinucleotides had 31P chemical shifts of ∼7 to 8 ppm (Supporting Information, Figure S1) and underwent acid-catalyzed decomposition, both characteristic of a phosphoramidate linkage. The coupling efficiency of each cycle is ∼40% for 1a and 70−80% for 1t, 1c, and 1g. The decreased coupling efficiency compared with phosphoramidate DNA synthesis is thought to be related to the decreased nucleophilicity of the 2′-NH2 due to the presence of the 1′-nucleobase substitution.(10) Nevertheless, we were able to isolate several npGNA oligomers (up to 10-mer) in amounts sufficient for base-pairing studies. The unreacted monomers (1t−1g) were quantitatively recovered after each coupling step. Crude npGNA products were further purified by denaturing polyacrylamide gel electrophoresis or by anion-exchange HPLC (Figure S2) and characterized by MALDI-TOF MS (Table S1). All subsequent UV and CD experiments were performed on gel- or HPLC-purified npGNA oligomers.
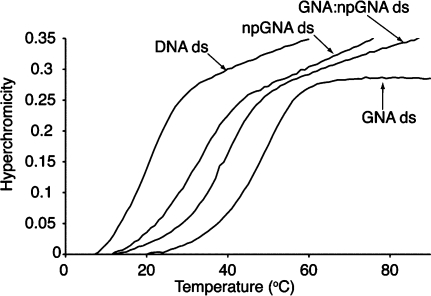
UV thermal denaturation studies on A9:T10 npGNA showed that npGNA formed a stable duplex (Figure 2). The 1:1 stoichiometry of each strand was established by a Job plot (Figure S3). Stable duplex formation was also observed in several other npGNA sequences (Table 1). The Tm values suggested that npGNA duplexes are more stable than DNA duplexes, but weaker than GNA duplexes of the same sequence (Table 1). Formation of duplex npGNA was confirmed by circular dichroism (CD) spectroscopy, which revealed a temperature-dependent conformational change upon denaturation of npGNA duplexes (Figures 3 and S4). The transition temperatures measured by CD experiments were consistent with those obtained in UV thermal denaturation studies (Table 1).
Figure 2.
Thermal denaturation curve of a 1:1 mixture of A9 and T10 npGNA compared with A10:T10 duplexes of DNA, GNA, and GNA:npGNA.
Table 1. Summary of Thermal Denaturation Studies.
| sequence | top:bottom strand | Tm (°C) |
|---|---|---|
| 3′-AAA AAA AAA A-2′ | npGNA:npGNAa | 34.5(32.5b) |
| 2′-TTT TT T TT T T-3′ | GNA:npGNA | 37.6(37.5b) |
| GNA:GNA | 49.5 | |
| DNA:DNA | 20.1 | |
| 3′-CGT ACG ACA T-2′ | npGNA:npGNA | 50.0(52.5b) |
| 2′-GCA TGC TGT A-3′ | GNA:npGNA | 64.1 |
| GNA:GNA | 74.0 | |
| DNA:DNA | 45.0 | |
| 3′-(CGA ATT CG)2-2′ | npGNA:npGNA | 32.0 |
| GNA:GNA | 40c | |
| TNA:TNAd | 29.8d | |
| DNA:DNA | 26c |
Figure 3.
Temperature-dependent CD studies on a 1:1 A9:T10 npGNA complex. The temperature range is between 10 and 55 at 5 °C intervals. The inset shows temperature-dependent CD signal change monitored at 273 nm.
Thermal denaturation (Figure 2) and CD (Figure S5−S7) studies also revealed stable heteroduplex formation between GNA and npGNA. Comparison of the Tm values in the same sequence context suggests that thermostability increases in the order of npGNA, npGNA:GNA, and GNA duplexes (Table 1). Interestingly, the reverse trend of thermostability was reported for N3′−P5′ phosphoramidate DNA (npDNA), which increases in the order of DNA, npDNA:DNA, and npDNA duplexes.(7) Previous X-ray structural studies and molecular dynamics simulations revealed that npDNA adopts an A-form conformation and its backbone is more rigid than that of DNA.(12) We found that GNA, GNA:npGNA, and npGNA duplexes share similar CD features (Figure S5), suggesting that all three share a similar conformation in solution.
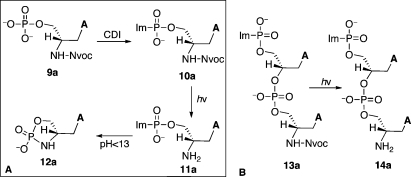
We then investigated the template-dependent synthesis of npGNA oligomers using 3′-imidazole-activated precursors (Figure 1B). A photolytic protecting group (6-nitroveratryl carbamate or Nvoc)(13) was used to protect the 2′-NH2 before activation of the 3′-phosphate group with carbonyl diimidazole (10a, Scheme 3A). After photolysis, however, we found that the imidazolide 11a underwent rapid intramolecular cyclization at pH < 13 to yield 12a (Scheme 3A). The rate of cyclization was too fast to be accurately measured by either NMR or HPLC analysis under the conditions for templated polymerization (at 4 °C, pH 8.4). However, we found that an activated dinucleotide (14a), which can be generated by photodeprotection of its precursor (13a, Scheme 3B), was much more stable than 12a with a half-life of 53.7 h at 4 °C and pH 8.4 as measured by HPLC (Figure S8). The breakdown products of 14a were cyclic and linear dinucleotides with a ratio of ∼3.5:1 (Figure S8).
Scheme 3. Synthesis of Imidazole-Activated Nucleotides for Templated Polymerization.
CDI, carbonyl diimidazole; Im, imidazole; Nvoc, 6-nitroveratryl carbamate.
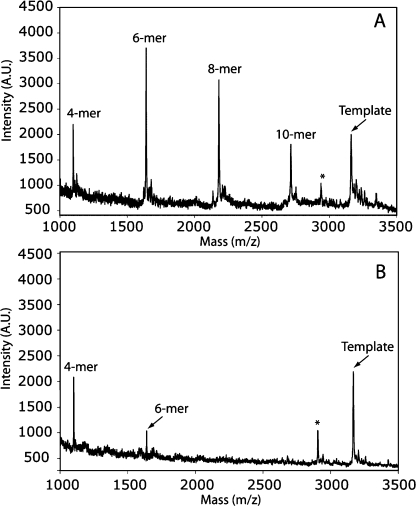
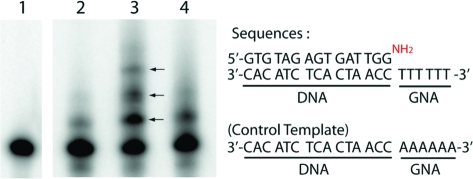
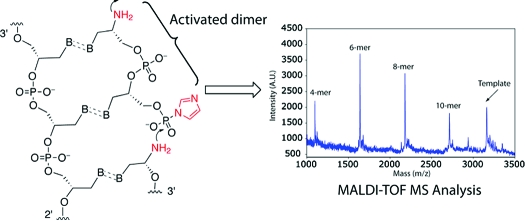
We envisioned that 13a could be used directly in templated polymerization reactions by generating 14a in situ via photolysis. Because GNA forms a stable duplex with npGNA and can be conveniently synthesized in a large quantity, GNA-(T)10 was used as the template for studying polymerization of 14a. As shown in Figure 4A, we observed polymerization of 14a by MALDI-TOF MS analysis. The full-length product (10-mer) was detected together with shorter oligomers after photolysis (Figure 4A). In the absence of the GNA template, only the 4-mer and a small amount of 6-mer products were observed with no full-length or 8-mer product (Figure 4B), suggesting that the template is necessary for efficient polymerization of 14a. We also investigated incorporation of 14a into a primer-template complex. Instead of using a GNA or npGNA primer-template complex, we used a well-characterized, 5′-[32P]-labeled, 3′-NH2-terminated DNA primer5,14 and a 3′-DNA-GNA hybrid template (Figure 5) for the preliminary primer extension studies on 14a. As shown in Figure 5, we observed multiple extension products after photolysis (lane 3, arrows). Extension products were significantly reduced without the template (lane 2) or with a mismatched template (lane 4), suggesting that polymerization of 14a was template dependent. The relatively low primer utilization (∼35%) compared with previous chemical extension studies (up to 100%)5,15 suggests that the B-form conformation in the primer-template pairing region is not favorable for primer-extension with 14a on the GNA template, which would lead to a duplex-GNA-like conformation.(1) In fact, primer-independent formation of 4, 6, and 8-mer was observed by MALDI-TOF MS analysis of the primer extension reactions. Future studies will be focused on the kinetics and fidelity of polymerization of short npGNA oligomers in an all-GNA or npGNA system to avoid unfavorable conformational transitions.
Figure 4.
Template-dependent polymerization of the activated dinucleotide 14a on T-3′-TTT TTT TTT T-2′-T analyzed by MALDI-TOF MS. (The underlined denotes the GNA 10-mer sequence): (A) with the template; (B) without the template. In B, the GNA template was cospotted with the sample as an internal control. The polymerization products were identified by comparing the signals to their expected masses. [M + H]+ for 4-mer: calcd, 1101.733; obsd, 1100.01. For 6-mer: calcd, 1642.349; obsd, 1640.67. For 8-mer: calcd, 2184.443; obsd, 2182.59. For 10-mer: calcd, 2725.798; obsd, 2723.68. For the template: calcd, 3168.994; obsd, 3167.08. (∗ denotes the n − 1 synthetic byproduct of the template).
Figure 5.
Template-dependent polymerization of 14a analyzed by primer extension: (lane 1) primer only as control; (lane 2) without template; (lane 3) with template; (lane 4) with control template. The reaction mixture of 10 μL contained 1 μM 5′-[32P]-primer/template, 2 mM 13a, 20 mM MgCl2, 0.2 M NaCl, 0.1 M HEPBS, pH 8.4. The reaction was initiated by photolysis at 4 °C for 6 h and, after another 12 h incubation at 4 °C, was analyzed by 20% denaturing polyacrylamide gel electrophoresis.
In summary, we have demonstrated that npGNA can form stable homo- and heteroduplexes with a conformation similar to that of GNA. In addition, N2′→P3′ linkages can be formed by template-dependent polymerization, which suggests that sequence information transfer can be achieved nonenzymatically in a genetic system based on GNA oligonucleotides.
Acknowledgments
J.J.C. was supported by a Fellowship from the Harvard Origins Initiative. This work was supported by NSF grant CHE 0434507 to J.W.S. J.W.S is an Investigator of the Howard Hughes Medical Institute. The authors thank Dr. A. Ricardo, Dr. M. Krishnamurthy, Dr. D. Treco, and Dr. B. Heuberger for helpful discussions.
Supporting Information Available
Procedures for synthesis and characterization of 1t−1g, 5t−5g, and 6a−14a, and solid-phase synthesis of npGNA, HPLC profile of the npGNA-(T)10 crude product, MALDI-TOF analysis of GNA and npGNA oligonucleotides in this study, UV melting curves, and additional CD spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Schlegel M. K.; Essen L. O.; Meggers E. J. Am. Chem. Soc. 2008, 130, 8158–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Peritz A.; Meggers E. J. Am. Chem. Soc. 2005, 127, 4174–5. [DOI] [PubMed] [Google Scholar]
- Orgel L. E. Nature 1992, 358, 203–9. [DOI] [PubMed] [Google Scholar]
- Szostak J. W.; Bartel D. P.; Luisi P. L. Nature 2001, 409, 387–90. [DOI] [PubMed] [Google Scholar]
- Mansy S. S.; Schrum J. P.; Krishnamurthy M.; Tobe S.; Treco D. A.; Szostak J. W. Nature 2008, 454, 122–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D.; Froeyen M.; Rozenski J.; Van Aerschot A.; Herdewijn P. Chem. Biodivers. 2007, 4, 740–61. [DOI] [PubMed] [Google Scholar]; Zhou D.; Lagoja I. M.; Rozenski J.; Busson R.; Van Aerschot A.; Herdewijn P. ChemBioChem 2005, 6, 2298–304. [DOI] [PubMed] [Google Scholar]
- Chen J. K.; Schultz R. G.; Lloyd D. H.; Gryaznov S. M. Nucleic Acids Res. 1995, 23, 2661–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryaznov S.; Chen J. K. J. Am. Chem. Soc. 1994, 116, 3143–4. [Google Scholar]
- Zhang L.; Peritz A. E.; Carrikk P. J.; Meggers E. Synthesis 2005, 4, 645–53. [Google Scholar]
- Schultz R. G.; Gryaznov S. M. Nucleic Acids Res. 1996, 24, 2966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoning K.; Scholz P.; Guntha S.; Wu X.; Krishnamurthy R.; Eschenmoser A. Science 2000, 290, 1347–51. [DOI] [PubMed] [Google Scholar]
- Cieplak P.; Cheatham T. E.; Kollman P. A. J. Am. Chem. Soc. 1997, 119, 6722–30. [Google Scholar]; Tereshko V.; Gryaznov S.; Egli M. J. Am. Chem. Soc. 1998, 120, 269–83. [Google Scholar]
- Amit B.; Zehavi U.; Patchorn A. J. Org. Chem. 1974, 39, 192–6. [Google Scholar]
- Stutz J. A. R.; Richert C. J. Am. Chem. Soc. 2001, 123, 12718–9. [DOI] [PubMed] [Google Scholar]
- Hagenbuch P.; Kervio E.; Hochgesand A.; Plutowski U.; Richert C. Angew. Chem., Int. Ed. 2005, 44, 6588–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.