Summary
Background
Although the molecules involved in mitosis are becoming better characterized, we still lack an understanding of the emergent mechanical properties of the mitotic spindle. For example, we cannot explain how spindle length is determined. To gain insight into how forces are generated and responded to in the spindle, we developed a method to apply controlled mechanical compression to metaphase mitotic spindles in living mammalian cells, while monitoring microtubules and kinetochores by fluorescence microscopy.
Results
Compression caused reversible spindle widening and lengthening to a new steady-state. Widening was a passive mechanical response, and lengthening an active mechanochemical process requiring microtubule polymerization but not kinesin-5 activity. Spindle morphology during lengthening and drug perturbations suggested that kinetochore fibers are pushed outwards by pole-directed forces generated within the spindle. Lengthening of kinetochore fibers occurred by inhibition of microtubule depolymerization at poles, with no change in sliding velocity, inter-kinetochore stretching, or kinetochore dynamics.
Conclusions
We propose that spindle length is controlled by a mechanochemical switch at the poles that regulates the depolymerization rate of kinetochore-fibers in response to compression, and discuss models for how this switch is controlled. Poleward force appears to be exerted along kinetochore fibers by some mechanism other than kinesin-5 activity, and we speculate that it may arise from polymerization pressure from growing plus-ends of interpolar microtubules whose minus-ends are anchored in the fiber. These insights provide a framework for conceptualizing mechanical integration within the spindle.
Introduction
Cell division can be viewed as a primarily mechanical problem. The mitotic spindle generates mechanical forces for tasks ranging from chromosome movement [1] to regulation of anaphase entry [2]. Although force-producing mechanisms at the molecular level are becoming well characterized (e.g. microtubule dynamics [1] and motors [3]), we have a poor understanding of how these forces are integrated. Most spindle forces give rise to a steady-state in position of some object at metaphase - chromosomes, poles, and the spindle within the cell - which means that forces must be position-dependent. Our understanding of these dependencies of force on position is rudimentary, and in most cases we do not know the principle by which a steady-state in position is achieved. Here we address the problem of spindle length determination, i.e. positioning of the poles relative to each other, or to metaphase chromosomes. Proposed spindle length determination mechanisms include a balance of external forces generated at the cell cortex [3], a balance of pushing and pulling forces internal to the spindle (using motors [3], microtubule dynamics [1, 4], or a scaffold matrix [5, 6]), and length-setting by thresholds of a spatial gradient of diffusible morphogens [7]. This profusion of models shows that the problem of spindle length determination is far from solved, and that perhaps more than one mechanism is involved.
Before the molecular era, students of the mitotic spindle developed an impressive battery of physical perturbations for probing spindle mechanics. For example, microneedle manipulation revealed the mechanical properties of spindle-chromosome attachments [8, 9], and quantified forces exerted by the spindle on chromosomes [10]. Most physical manipulation experiments were performed on large spindles in meiotic or early mitotic systems (with few exceptions, e.g. [11]) that are not easily accessible to molecular methods. Recently, physical manipulations were applied to the Xenopus egg extract spindle [12, 13], but length determining mechanisms in this anastral meiotic spindle may differ considerably from astral spindles in mammalian somatic cells.
To integrate mechanical and molecular models in a mammalian somatic spindle, it would be useful to develop controlled and informative mechanical perturbations in a cell type that is tractable for high resolution imaging and molecular manipulations. In this spirit, we developed a method to mechanically perturb spindles in mitotic Ptk2 (Potorous tridactylis) cells by compressing them in a controlled and reversible manner. These cells are relatively flat in mitosis, have a large spindle with few large chromosomes, and are amenable to molecular perturbations [14]. Using compression to mechanically perturb spindles, we pose two interdependent questions. First, what are the material properties of the metaphase spindle and how do they relate to its function? Second, what does the spindle’s response to compression tell us about the mechanical and chemical processes that determine its shape and size? Our results allow us to propose a new conceptual framework for mechanical integration within the spindle, where pushing forces are generated along the length of kinetochore fibers, and spindle length is regulated by a mechanical feedback that controls microtubule depolymerization at poles.
Results
Cell compression apparatus
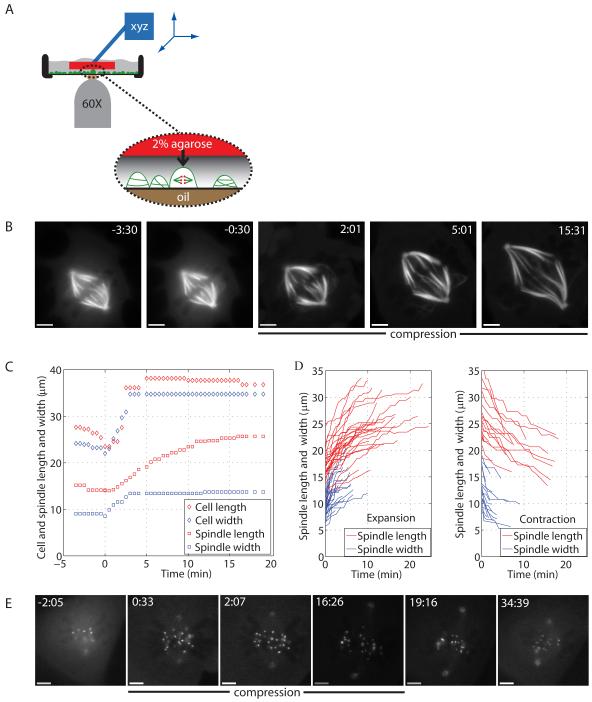
Our method is an extension of cell flattening methods reported previously [15]. We located mitotic cells by phase-contrast imaging, gently lowered an agarose pad saturated with medium (and drug in some experiments) on top of them, and then applied controlled and reversible downwards force with a metal rod held in a micromanipulator (Fig. 1A). The extent of compression was judged by phase contrast imaging of the cell’s response, and was generally completed within ∼10 s to provide a mechanical change that was step-like on the timescale of spindle dynamics. The agarose pad was kept in position for 10-90 min, and then raised again over ∼10 s. The response of the spindle and kinetochores was monitored by phase contrast and fluorescence from stably expressed EGFP-α-tubulin [16] or EYFP-cdc20 (gift from Jagesh Shah, Harvard Medical School) cells. Cdc20 localizes strongly to kinetochores and weakly to poles throughout metaphase and provides a convenient marker for measurement of spindle length and kinetochore dynamics.
Figure 1.
A compressed mitotic spindle expands asynchronously and reversibly.
(A) Method developed to mechanically perturb spindles in vivo. (B) Fluorescence imaging of a compressed spindle in a Ptk2 EGFP-α-tubulin cell. (C) Time courses of cell and spindle length and width changes upon compression for the cell in (B). (D) Time courses of spindle length and width changes during expansion (31 cells) and contraction (17 cells). For clarity, steady-state time points are not displayed. (E) Fluorescence imaging of a compressed spindle in a Ptk2 EYFP-cdc20 cell, with compression released at 16:30. (B-E), compression started at 0:00 (min:s) and scale bar corresponds to 5 μm.
Compression induces spindle widening and elongation asynchronously and reversibly
When metaphase cells were compressed, the cell and spindle expanded (Table 1, Fig. 1B, Movie S1). Spindle width and length plateaued after compression (Fig. 1C), and both changes were reversible (within limits, discussed below; Table 1, Fig. 1D-E, Movie S2), indicating that compression induces a new steady-state in spindle dimensions. Spindle length was defined as the pole-to-pole distance, where poles were defined either as a point of convergence of kinetochore fibers (k-fibers) in tubulin imaging, or as polar dots in cdc20 imaging. Spindle width was defined as the largest distance over which k-fibers or kinetochores spread out, and cell length and width were measured along the same axes as the spindle axes.
Table 1.
A compressed spindle expands asynchronously and reversibly. Expansion and contraction timescale (Δt), extent (Δ) and velocity (v), for spindle length (L; Lo, pre-compression value), spindle width (W), cell length (CL), and cell width (CW). Data was indiscriminately averaged over all experiments. Mean ± standard deviation
| Phase (# traces) |
ΔtL (min) | ΔtW (min) | ΔtCL (min) | ΔtCW (min) |
|---|---|---|---|---|
| Expansion (31) |
12.1±5.3 | 3.4±2.3 | 3.4±3.6 | 4.0±3.5 |
| Contraction (17) |
10.1±6.2 | 3.4±2.3 | 5.9±5.8 | 5.6±5.1 |
| Phase (# traces) |
ΔL (μm) [L0] |
ΔW (μm) [W0] |
ΔCL (μm) [CL0] |
ΔCW (μm) [CW0] |
| Expansion (31) |
8.0±3.5 [17.1] |
4.4±2.1 [8.7] |
8.0±8.3 [39.3] |
10.9±6.7 [27.3] |
| Contraction (17) |
-6.9±4.1 [24.8] |
-3.4±1.9 [13.0] |
-6.3±5.4 [46.2] |
-6.6±4.5 [37.7] |
|
Phase (# traces) |
vL (μm/min) | vW (μm/min) | vCL (μm/min) | vCW (μm/min) |
| Expansion (31) |
0.7±0.4 | 2.3±2.6 | 5.2±6.7 | 5.2±6.6 |
| Contraction (17) |
-0.9±0.8 | -1.5±1.5 | -1.3±0.7 | -2.5±3.0 |
Following compression, spindle width increased from ∼9 to ∼13 μm, spindle length from ∼17 to ∼24 μm, cell width from ∼27 to ∼38 μm, and cell length from ∼39 to ∼47 μm, on average (n=31, Table 1). The expansion parameters of the spindle and cell were correlated (Table S1). Cell width and length, and spindle width, all increased over similar, short timescales (∼3-4 min). In contrast, spindle length increased over a significantly longer timescale (∼12 min) in all cases (Fig. 1C). The observations that the expansion timescales of cell and spindle width are similar, and that the spindle never (0/17) widened more than the cell (in absolute terms, i.e. number of μm), suggest that spindle width expansion is a passive, purely mechanical, process. Spindle length, in contrast, increased over a period four times longer than cell length, and in 18/31 cases the spindle elongated more than the cell. Thus, spindles appear to lengthen by a different, and more complex, mechanism.
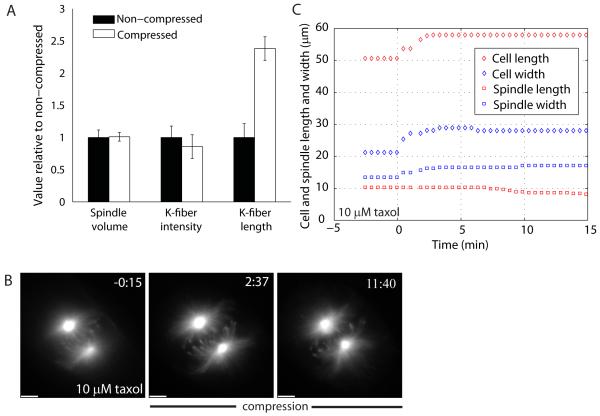
To better understand the spindle as a material, we followed its global response in three dimensions and calculated its volume. We measured spindle thickness by taking z-stacks of the spindle and finding the top and bottom planes with k-fibers or kinetochores in focus: on average, control spindles were 5.3±1.3 μm thick (n=22), while compressed spindles were 3.3±0.9 μm thick (n=7). Compressed spindles were 0.25-0.90-fold the thickness of their control counterparts. Surprisingly, estimated spindle volumes in the pre- and post-compression steady-states were, on average, the same, ∼400 μm3 (assuming an ellipsoid spindle shape; Fig. 2A). Because of the asynchrony in changes in spindle width and length, spindle volume decreased transiently upon compression, and increased transiently upon release, before returning to the steady-state volume.
Figure 2.
Tubulin polymerization is required for spindle elongation. (A) Histogram comparing non-compressed and compressed spindle steady-states: spindle volume (n=7), k-fiber cross-sectional intensity (n=19) and length of k-fibers (cell in Fig. S1B), all normalized to the non-compressed value. (B) Fluorescence imaging of a Ptk2 EGFP-α-tubulin bipolar spindle being compressed in 10 μM taxol, with its (C) time course of cell and spindle length and width changes. Compression started at 0:00 (min:s) and scale bar corresponds to 5 μm.
Spindle widening and elongation have different molecular requirements
Spindle width increased at 2.3±2.6 μm/min on average. In the most dramatic cases, spindle width doubled (Fig. S1, Movies S3A-S3B-S4). Imaging of both tubulin and kinetochores showed that k-fibers pivoted outward around the spindle poles during spindle widening. K-fibers still converged at the poles but now radiated more broadly (Fig. S1). This pivoting often occurred before k-fibers had appreciably increased in length. Interestingly, outward pivoting of k-fibers appeared to be physically limited by the edge of the spindle: when cell blebs sucked chromosome arms away from the spindle, the attached k-fibers did not pass the spindle edge (data not shown). We suspect that a physical barrier lies at the edge of the spindle, perhaps an envelope of ER membranes [17].
Spindle length increased at 0.7±0.4 μm/min on average, or 0.35 μm/min per k-fiber, which is similar to the rate of polewards tubulin sliding (“polewards flux”) in k-fibers in unperturbed metaphase cells [18]. Spindle elongation tended to slow down at the end of the response, before the new steady-state was reached. In the most dramatic cases, spindle length doubled, with imaged k-fibers elongating from ∼7 μm to ∼14 μm (Fig. 2A, Fig. S1, Movie S4). This length change cannot be accounted for by change in the angle of k-fibers relative to the optical axis; if we force the longest k-fiber flat, we can only account for <2 μm of this elongation by angle change. Therefore we conclude that the k-fibers elongated, which could occur by addition of new tubulin, or sliding apart of individual microtubules within the fiber.
K-fibers include at least 25 kinetochore microtubules (K-MTs) in Ptk cells, that bundle together with a similar number of non-K-MTs [19]. To test whether the elongation of k-fibers under compression was due to microtubules sliding apart within them, we measured their cross-sectional intensity in EGFP-α-tubulin images. This remained constant after background subtraction (Fig. 2A), implying lack of sliding apart. Rather, total tubulin polymer in k-fibers increased by microtubule lengthening, while the number of microtubules per fiber remained approximately constant. To confirm this new polymerization, we integrated EGFP-α-tubulin intensity over all z-stacks, and found that it indeed increased during compression in all examples tested. We do not quote a value for total polymer from this method, because our estimates were sensitive to potentially unreliable estimates of background fluorescence. The steady-state compressed spindle thus had the same volume, but more total polymer, because k-fibers were longer and contained similar numbers of microtubules. Thus, compression caused an increase in tubulin polymerization, either by promoting assembly or by inhibiting disassembly. Previous mechanical perturbations also caused correlated changes in polymer mass and spindle length [20, 21].
To test a requirement for polymerization dynamics in spindle elongation, we briefly pre-treated spindles with 10 μM taxol and then compressed them with taxol in the pad. Prior to compression, this treatment caused decreased inter-kinetochore distances, inhibition of kinetochore movement, and spindle shortening over time, as previously reported [22]. Since taxol strongly promotes polymerization, it is likely that the concentration of free tubulin dimers drops to a very low value after pre-incubation in drug, making further microtubule elongation difficult. Compression of spindles in 10 μM taxol revealed that they still widened, but k-fibers did not elongate significantly (Fig. 2B-C, Movie S5). The cell still expanded (Fig. 2C), so taxol treatment effectively uncouples change in cell shape from spindle shape in the length axis, but not in the width axis. K-fiber elongation upon compression thus requires microtubule dynamics, while outward pivoting does not.
Spindle elongation is driven by forces intrinsic to the spindle
Because the whole cell elongates following compression, we first hypothesized that the force causing spindle elongation came from pulling on astral microtubules attached to the outward-moving cell cortex. This model was previously proposed by Inoué for spindle expansion following egg compression [21], and it is consistent with the widely held view that pulling forces from the cortex serve to position spindles [23], and also to elongate them [3]. Unexpectedly, four lines of evidence, listed below, argued against cortical pulling in our system. Inoué’s proposal largely stemmed from the observation that the spindle narrowed as it elongated in compressed egg fragments [21]. In Ptk2 cells, we never observed a spindle narrow during elongation (n=31). The difference may arise from spindle-to-cell size ratios. Compressing a Ptk2 cell entails directly compressing its spindle, which is not the case in larger egg fragments.
Arguments that spindle elongation does not occur by pulling from the cortex
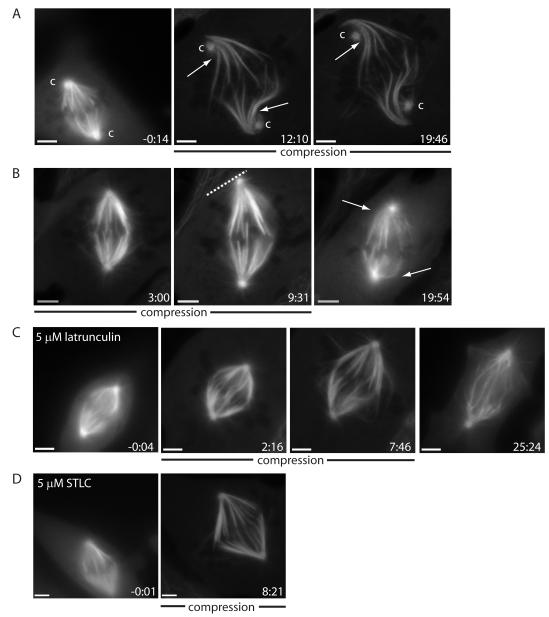
1) Centrosome detachment
In 3/16 cases where we imaged EGFP-α-tubulin, the spindle poles detached from the centrosomes during expansion, and then moved outward, passing the centrosomes (Fig. 3A). These image sequences are not compatible with spindle elongation by pulling from astral microtubules attached to centrosomes. They are consistent with the hypothesis that transient separation of centrosomes and spindle poles provides a signal (although not the force) for spindle elongation [24].
Figure 3.
Spindle elongation is driven by forces internal to the spindle that are kinesin-5-independent. (A) Example of extreme spindle compression where spindle poles disconnect from and grow passed centrosomes (marked ‘c’, with arrows marking k-fiber bends). (B) Example of a spindle elongating until it reaches the cell cortex, with cortex marked by a dashed line using the phase image (release at 10:00; arrows mark k-fiber bends post-release, suggesting that k-fibers impede shortening). Spindle being compressed in (C) 5 μM latrunculin (note bent interpolar microtubules at 7:46; release at 15:27) and in (D) 5 μM STLC. Ptk2 EGFP-α-tubulin cells, compression started at 0:00 (min:s) and scale bar corresponds to 5 μm.
2) K-fiber shape
In most cases, k-fibers remained straight during spindle elongation, but in some cases k-fibers appeared to bend or buckle, typically nearer the pole. This was clearest in cases where the centrosome detached (Fig. 3A), perhaps because the centrosome exerts a drag force on the outward-moving spindle poles. These bends strongly suggest that k-fibers push during spindle expansion, which implies they must be under compression near the poles, even though they are still under tension at kinetochores (as revealed by the distance between sister k-fibers at kinetochores). Bending at the poles was even more pronounced during shortening induced by decompression (discussed below). Like centrosome detachment, k-fiber bending suggests that pushing forces exerted on or by k-fibers drive spindle elongation.
3) Spindle versus cell length change
In a few cases, where neighbors prevented the cell from expanding along the spindle length axis, the spindle grew until it touched the cortex (Fig. 3B). Also, spindle length increased (in absolute terms) by more than cell length in 18/31 expansion experiments, and decreased by more than cell length in 10/17 contraction experiments. These results are inconsistent with the idea that cortical pulling is the sole mechanism responsible for spindle elongation.
4) Actin drugs
To test if actin is required for spindle elongation we compressed spindles in 5 μM latrunculin (Fig. 3C) or 10 μg/ml cytochalasin D (Fig. S2A). In both cases the spindles elongated at rates similar to control (0.9±0.3 μm/min, n=5), suggesting that stiff cytoplasmic or cortical actin meshworks are not required for spindle elongation. Given these results, we were forced to the alternative hypothesis that forces generated within the spindle push poles outward following compression.
Kinesin-5 does not power spindle elongation upon compression
Kinesin-5 generates forces that push spindle poles apart [25, 26], and is thus an obvious candidate for powering spindle elongation under compression. To test this, cells were pre-incubated in kinesin-5 inhibitors, 5 μM S-trityl-L-cysteine (STLC) (Fig. 3D) or 500 nM EMD534085 [27] (Fig. S2B), and compressed with drug in the pad. For both drugs, elongation rates of pre-existing bipolar spindle in response to compression (0.8±0.2 μm/min, n=5) were similar to control; newly formed spindles were monopolar, confirming that the drugs were reaching their target. We discuss alternative candidates for outward force production below.
Response to removing compression; k-fibers impede spindle shortening
The morphology of k-fibers during spindle shortening when compression was reversed was also informative (Fig. 3B). When the agarose block was lifted, the cell rounded, and both cell and spindle areas decreased (n=17) (Table 1, Table S1). As observed for expansion, there was an asynchrony in spindle width and length changes: spindle width returned to a value similar to its pre-compression value over ∼3 min, while spindle length changed over ∼10 min, and typically only returned to the pre-compression value for gentler perturbations (Table 1, Fig. 1D-E, Movie S2, supplemental experimental procedures). As with elongation, the cell and spindle responses were highly correlated (Table S1).
When the pad was removed rapidly, we frequently observed strong bending of k-fibers near the poles (Fig. 3B, arrows). These bends slowly disappeared as k-fibers shortened back to their original length. The spindle apparently cannot shorten more rapidly than K-MTs can depolymerize. These bends might reflect individual k-fibers shortening at different rates while being attached to each other at the poles, but their overall appearance was more consistent with a response to pole-pole compression from some mechanical element inside or outside the spindle. The observation that the cell contracts faster than the spindle (Table 1) suggests that the cortex may be one such element. Anecdotally, k-fiber bending was less severe in actin depolymerization drugs, and more pronounced after faster releases, and releases of stronger perturbations. Detailed kinetochore imaging was difficult since the focal plane moved unpredictably when the pad was lifted, but preliminary observations suggest that tension was retained at kinetochores (as judged by the inter-kinetochore distance) even when the polewards ends of k-fibers were strongly bent (Fig. 1E). Just as forces generated by k-fibers drive spindle elongation, the above data suggests that k-fibers impede spindle shortening.
Depolymerization of k-fibers at poles is inhibited by compression
K-fibers in unperturbed Ptk2 cells slide poleward at ∼0.5 μm/min, undergoing net polymerization at kinetochores, and depolymerization at poles [18]. Superimposed on this slow motion, sister kinetochores oscillate around the spindle equator, with individual kinetochore switching every 1-2 min between poleward (P) and away-from-pole (AP) movement at ∼1 μm/min. Electron microscopy suggests that most individual K-MTs span the entire kinetochore-to-pole distance in Ptk cells [28]. To understand how compression of the cell increases net tubulin polymerization in K-MTs (Fig. 2), we imaged kinetochore motion, and photo-marked k-fibers to measure their sliding velocity. It was easy to track kinetochores in compressed cells because they all moved in a single focal plane. Remarkably, inter-kinetochore stretch (s, μm), P- and AP-velocity (vP and vAP, μm/min) were all statistically indistinguishable in control cells (s =2.7±0.8, vP =0.8±0.3, vAP=0.7±0.2, n=8), cells with elongating spindles (s =2.9±0.6, vP =0.8±0.2, vAP=0.8±0.2, n=10), and cells with steady-state elongated spindles (s =2.8±0.8, vP =0.7±0.2, vAP=0.8±0.2, n=5). Thus, forces and polymerization dynamics at kinetochores were very little affected by compression, implying that elongation was not caused by increased pulling forces on kinetochores leading to increased polymerization there. Kinetochore tension and polymerization dynamics were unaffected even in cells like the one shown in Fig. 3A where the poleward end of the k-fibers shows clear signs of compression and buckling. Again, we conclude that the poleward part of the k-fiber can be under compression even when the equatorial part of the k-fiber is under tension, a finding whose significance we discuss below.
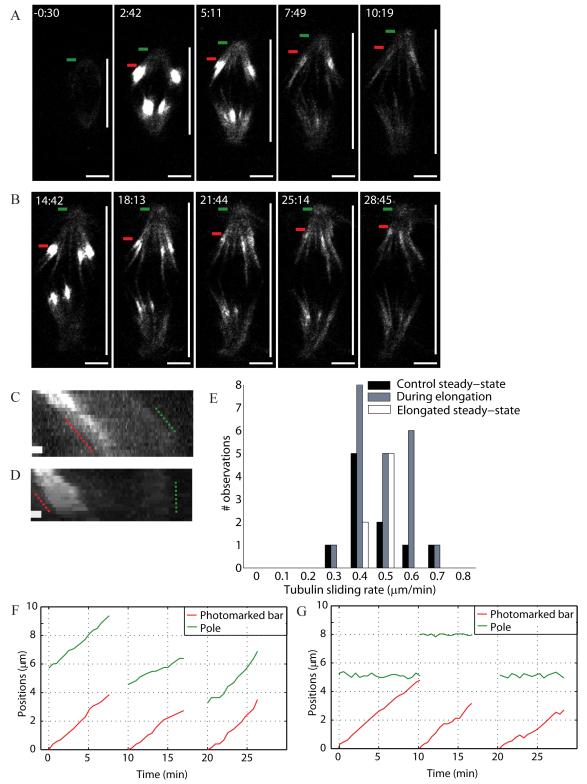
To measure k-fiber sliding rates with respect to the metaphase plate, we locally illuminated (using 405 nm light) cells expressing a photoactivatable GFP (PA-GFP) tubulin. The rate of poleward sliding of the photomark in control was 0.4±0.1 μm/min (n=10), consistent with previous reports for this cell type [18, 29]. The sliding rate was statistically indistinguishable (Fig. 4E) in cells with elongating spindles (0.4±0.1 μm/min, n=21, Fig. 4A, Movie S6), and compressed cells with steady-state elongated spindles (0.4±0.1 μm/min, n=7, Fig. 4B, Movie S6). This sliding rate is roughly half the spindle pole-to-pole elongation rate. In control and steady-state elongated spindles, the photomarked tubulin bar moves towards the stationary pole and disappears as it reaches it (Fig. 4D, G). In elongating spindles, the mark and the pole tend to move outward together (Fig. 4C, F). Thus, spindle elongation correlated with inhibition of k-fiber depolymerization at the poles, with no change in sliding rate, inter-kinetochore stretching or kinetochore dynamics.
Figure 4.
Spindle compression reduces microtubule depolymerization at poles while leaving the tubulin sliding rate unchanged. Tubulin sliding (A) during and (B) after elongation in Ptk1 PA-GFP-α-tubulin spindles being compressed. Green bars mark the poles and red bars the leading edge of photomarked tubulin. Vertical white bar marks spindle length. Tubulin photomarking was performed at 2:25 (min:s) and 14:25. Compression started at 0:00 and scale bar corresponds to 5 μm. Kymographs show the motion of photomarked tubulin with respect to the poles (C) during (over 10 min) and (D) after (over 7.5 min) elongation; scale bar corresponds to 1 μm. (E) Histogram of tubulin sliding rates before (control), during and after spindle elongation. Respective positions of the photomark and pole in three example traces (F) during and (G) after elongation (time translation for clarity).
Discussion
Compression responses reveal spindle mechanics
Figure 5A summarizes our results. Because spindle width and cell width changes were temporally correlated, and widening was insensitive to taxol (Fig. 2B-C), we interpret the spindle width change as a passive mechanical response. Length change, in contrast, appeared to be an active mechanochemical response that required a net increase in tubulin polymer mass in response to compression (Fig. 2), and a decrease in polymer mass in response to decompression. Given the orientation of spindle microtubules, we expect spindles to be much stiffer in the pole-to-pole axis than orthogonal to that axis. Mechanical anisotropy was confirmed by the passive spindle response in the width axis in our experiment (Fig. 1-2, Table 1) and others [12]. It was also previously inferred from chromosome micromanipulation experiments [8, 9]. Our data also reveal regional differences along the spindle axis in the strength of the links between microtubules. Widening occurred by outward pivoting of k-fibers with a pivot-point near the pole, suggesting that fiber-to-fiber linking is tighter at the poles than the equator [9]. Inter-fiber cross-linking at the poles was observed even when centrosomes dissociated (Fig. 3A), consistent with a view of the poles in which factors like NuMA [16], minus-end-directed motors [30] and poly(ADP-ribose) [31] cross-link microtubules there.
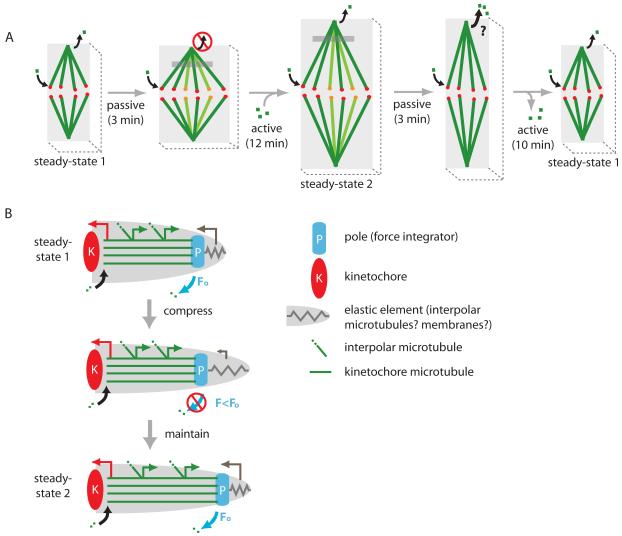
Figure 5.
Compression regulates spindle size. (A) Sketch of the response of a Ptk2 spindle to compression. Curved black arrows depict tubulin polymerization at the kinetochore and depolymerization at the poles (poleward sliding). In panel 2, new k-fibers are in focus (different color). Gray region depicts a photomarked tubulin population. Depolymerization at poles is inhibited during elongation; we predict it to increase during contraction but this has not been shown. For simplicity, active and passive responses are depicted in series, while they actually occur in parallel. (B) Mechanical coupling model for spindle length regulation, where the length of K-MTs is determined by force-dependent effects on microtubule dynamics at poles. Coupling between tension/compression on poles and K-MT dynamics is provided either by force-dependent regulation of the activity of a depolymerase, or by a direct effect of force on depolymerization. In either case, we postulate that the depolymerization rate at poles responds to the sum [43] of all the forces exerted on K-MTs (F), that we divide into pulling on plus-ends by kinetochores (red arrow), an outwards sliding force that is generated along the length of the K-MTs (green arrows), and pushing on minus-ends by other parts of the spindle or cell (gray arrow). K-MTs grow at the sliding rate until the original F (Fo) returns, when depolymerization resumes and a new spindle length steady-state is reached.
Mechanical regulation of spindle length
Compression inhibited k-fiber depolymerization at poles (Fig. 4A-C-F) and this depolymerization resumed, at approximately the pre-compression rate, when the new steady-state in spindle length was reached (Fig. 4B-D-G). We interpret this observation as revealing the existence of a mechanochemical switch at the poles that regulates the depolymerization rate in response to forces acting on spindles, and generated within them, in such a way as to control steady-state length. Below, we discuss two classes of models for how this switch may be controlled.
In our preferred mechanical coupling model (Fig. 5B), depolymerization at poles is controlled mechanically. We propose that compression of the cell and spindle is transmitted into tension on K-MTs at the poles by mechanical coupling at spindle poles. Specifically, we imagine an elastic element (spring in Fig. 5B) that exerts compression or tension on poles, and thus controls the depolymerization rate. Candidate elastic elements include ER membranes surrounding the spindle [17], a putative matrix within the spindle [5, 6] and interpolar microtubules within the spindle (if they are mechanically uncoupled from K-MTs, and have a different elasticity) [12]. Another candidate is the cell cortex; we found no evidence that cortical forces are involved in the response to compression (Fig. 3), but some evidence that it was involved in the response to decompression (Table 1, Fig. 3B). In an enzymatic version of the model, tension at the poles inhibits K-MT depolymerases located there (e.g. [29]). This could occur by repositioning of depolymerases away from minus-ends, or by a direct effect of tension on their enzymatic activity as others have proposed [32]. In a purely physical version of the model, tension on K-MTs directly regulates the affinity of ends for tubulin subunits, independent of any depolymerases, e.g. by a Brownian ratchet mechanism. As required by the equilibrium between chemical potential and mechanical energy, tension on a microtubule favors polymerization, while compression favors depolymerization [33].
In an alternative chemical signaling model (not shown), compression perturbs a morphogen gradient that regulates spindle length. Spatial gradients of Ran-GTP [34] and AuroraB activity [35] have been shown to emanate from chromatin in mitotic cells, presumably generated by reaction-diffusion processes, and proposed to control spindle morphology. Changing the shape of the cell and chromatin mass might perturb the amplitude of these gradients, and thus change the position of biochemical thresholds that affect depolymerase activity (e.g. [36]) and determine spindle length. We also considered a model in which compression drives water out of the cell, increasing the concentration of spindle subunits, and thus driving assembly-disassembly equilibria towards assembly. We think this is unlikely to be the sole explanation of elongation: first, it is unclear that cell compression would have this effect, and osmotic forces would oppose significant water lost; second, while removing water from Ptk cells with strongly hypertonic medium does promote spindle elongation [37], the response is otherwise (in magnitude, morphology and polymer mass) different than that to compression.
Although decisive evidence is lacking, we currently favor the mechanical coupling model for spindle length regulation (Fig. 5B) for several reasons: i) the spindle poles appear to be physically pushed towards the equator at a rate faster than depolymerization during decompression (Fig. 3B), and ii) our preliminary reaction-diffusion modeling suggests that a morphogen gradient model would only work with a very limited set of parameter values (gradient decay length, cell and chromatin shape changes). We take seriously the simplest, purely physical version of the mechanical coupling model (without invoking depolymerases) because the mechanism of depolymerization at poles is far from clear, and microtubule sliding and associated depolymerases are not essential to spindle length determination [38].
Implications for forces on k-fibers
One surprising aspect of our data is that kinetochore tension and dynamics were largely undisturbed at all stages of the compression - new steady-state - decompression cycle (Fig. 1E, Movie S2), despite large changes in spindle shape and size, and, in some cases, dramatic bending of K-MTs near the poles that strongly suggested that K-MTs were under compression there (Fig. 3A-B, arrows). These observations indicate that forces on kinetochores are locally generated and regulated, and that kinetochores are, to a surprising extent, mechanically isolated from poles. Our observations are consistent with laser cutting experiments (e.g. [39]) and suggest that k-fibers experience an outward force generated by a distributed system acting all along their length, similar to the classic traction fiber model [40]. This outward force drives k-fiber poleward sliding at steady-state, and spindle lengthening when depolymerization at poles is transiently inhibited. In agreement with the above, it was recently proposed that artificially high kinetochore tension does not affect depolymerization at poles [41].
What molecular system might generate distributed outward force on k-fibers? In some systems, kinesin-5 activity is required to maintain steady-state spindle length and drive K-MT poleward sliding [25, 26]. However, it is not required for either process in metaphase Ptk spindles [29], and we found that kinesin-5 activity was not required for spindle elongation in compressed cells (Fig. 3D, Fig. S2B) - it is only required for initial bipolarization. Other plus-end directed kinesins might play a role, but they would need to act in a distributed fashion along the k-fiber, which seems to rule out chromokinesins. Given the current lack of evidence for motor involvement, we speculate that outward forces (Fig. 5B, green arrows) are generated by polymerization pressure from growing interpolar microtubules whose minus-ends are anchored in K-MTs [28] (Fig. 5B, diagonal lines). To generate outwards force, the plus-ends of these microtubules would have to push against objects within, or surrounding, the spindle, for example ER membranes. Fast microtubule turnover in spindles was discovered many years ago [4], but its function remains unclear. It probably allows microtubules to rapidly explore different spatial configurations [42], but perhaps its major function is to generate the outward force that keeps spindle poles apart, and balance pulling by kinetochores (Fig. 5B, red arrows).
Experimental Procedures
Cell culture
Ptk2 EGFP-α-tubulin (Alexey Khodjakov, Wadsworth Center) and Ptk2 EYFP-cdc20 (Jagesh Shah, Harvard Medical School) cells were cultured in MEM with Earle’s salts and L-glutamine (Invitrogen 11095), supplemented with sodium pyruvate (Invitrogen 11360), non-essential amino acids (Invitrogen 11140), penicillin/streptomycin and 10% qualified and heat-inactivated fetal bovine serum (Invitrogen 10438). Ptk1 PA-GFP-tub cells (Alexey Khodjakov) were cultured in F-12 (Invitrogen 11765) with non-essential amino acids, antibiotics and serum as above. Imaging was performed in Leibovitz’s L-15 media with L-glutamine without phenol-red (Invitrogen 21083) with antibiotics and serum as above.
Spindle compression and live imaging
A solution of 2 % ultra pure agarose (Invitrogen 15510) in PBS was prepared, brought to boil and 2 ml was put in a 35 mm petri dish to solidify with ∼2 mm thickness. A 1 cm × 1 cm pad area was cut out, soaked in L-15 medium overnight at 4 °C for equilibration, and warmed to 29 °C just before use. Cells were plated on 25 mm HCl-cleaned poly-L-lysine (Sigma P1524) coated coverslips 24-48 h before imaging and placed on a water-heated coverslip holder set for 29 °C at the coverslip. A flat metaphase cell was chosen among 80 % confluent cells and the pad deposited gently, centered on the cell. L-15 was added to cover the pad. Using an oil hydraulic fine manipulator (Narashige MO-202) and a coarse manipulator attached to the Nikon TE-300 microscope, a metal rod was centered on the cell (Bertrand lens) and lowered (z-axis) until weak contact was made with the pad for pre-perturbation imaging (rod diameter ≫ cell diameter).
The rod was lowered slowly (over ∼10 s) for several μm until the cell area expanded, and its position kept constant as the cell and spindle responses were imaged. Ptk2 EGFP-α-tubulin cells were imaged every 30 s and EYFP-cdc20 cells every 15-30 s with a 60X 1.4 NA Plan Apo Ph3 oil objective and cooled CCD Orca-ER camera (Hamamatsu) with 100 W mercury lamp. For EGFP, 480/40 and 535/50 filters (HQ-FITC, Chroma, with ND4) were used; for EYFP, 500/20 and 535/30 filters were used (ET-EFP, Chroma). Both phase contrast (∼0.5 s) and fluorescence (∼200 ms) widefield images were acquired (bin=1, Metamorph 7.5.3.0, MDS Analytical Technologies) before, during and after mechanical perturbation. Continuous phase contrast imaging (80%) and binocular viewing (20%) allowed manual refocusing of the spindle during manipulation. A motorized stage (Prior ProScan II) was used to acquire z-stacks of the spindle. Cell health was monitored through the presence of metaphase oscillations, a microtubule-dense spindle excluding mitochondria, and the ability of the cell to enter anaphase perturbation.
Drug treatments
Drugs were added to the pad during the overnight L-15 pre-incubation. Drug-containing pads and media were added at a fixed time before compression. Taxol (paclitaxel, Sigma T7191) was added 10 min before compression at 10 μM (10 mM DMSO stock). Latrunculin was added 10 min before compression at 5 μM (10 mM DMSO stock). Cytochalasin D was added 10 min before compression at 10 μg/ml (10 mg/ml DMSO stock, Sigma C8273). S-trityl-L-cysteine (STLC, Sigma 164739) was added at 5 μM (10 mM DMSO stock) 30 min before compression. EMD534085 (Merck-Serono)[27] was added at a saturating concentration of 500 nM (100 mM DMSO stock, diluted) 30 min before compression. EMD534085 is a novel allosteric kinesin-5 inhibitor, a hexahydropyranoquinoline that acts at a lower concentration than STLC.
Photomarking
Photomarking experiments were performed using a Nikon TE-2000U microscope, a Perkin Elmer Ultraview Spinning Disk Confocal, a cooled CCD Orca-ER camera (Hamamatsu), a Micropoint Laser System (Photonic Instruments) for photoactivation at 405 nm and a 100X 1.3 NA Plan Fluor Ph3 objective. A tubulin population was photomarked parallel to the metaphase chromosome plate, on either one or two sides of the plate. Photomarking was performed either immediately before or after compression, or as soon as the spindle reached its longer steady-state under compression. Phase and fluorescence imaging were performed every 10-30s.
Supplementary Material
Acknowledgements
We thank Alexey Khodjakov (Wadsworth Center) for the stable Ptk2 EGFP-α-tubulin and Ptk1 PA-GFP-α-tubulin lines, Jagesh Shah (Harvard Medical School) for the stable Ptk2 EYFP-cdc20 line, Xiaohu Wan (UNC Chapel Hill) for sharing his SpeckleTracker Matlab program, Martin Howard (BBSRC, UK) for gradient analysis advice, Fred Lanni (Carnegie Mellon University) for spindle thickness measurement advice, Jennifer Waters for quantitative microscopy advice, and Merck-Serono (Darmstadt, Germany) for EMD534085. Photomarking experiments were performed at the Nikon Imaging Center @ HMS. We also thank Martin Wühr, Kendra Burbank, Lisa Cameron, Guillaume Charras, Jay Gatlin, Aaron Groen, Hao Yuan Kueh, L. Mahadevan, Tom Maresca, Daniel Needleman, Ted Salmon and Jagesh Shah for critical reading of the manuscript and/or discussions. S.D. received support from a Milton Fund (Harvard University) and T.J.M. was supported by NIH grants GM039565 and P50 GM068763.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Inoué S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Molecular Biology of the Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 3.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 4.Inoué S, Sato H. Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. Journal of General Physiology. 1967;50:259–292. [PMC free article] [PubMed] [Google Scholar]
- 5.Pickett-Heaps JD, Forer A, Spurck T. Traction fibre: toward a “tensegral” model of the spindle. Cell Motility and the Cytoskeleton. 1997;37:1–6. doi: 10.1002/(SICI)1097-0169(1997)37:1<1::AID-CM1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 6.Mitchison TJ, Maddox P, Gaetz J, Groen AC, Shirazu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Molecular Biology of the Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 8.Begg DA, Ellis GW. Micromanipulation studies of chromosome movement. I. Chromosome-spindle attachment and the mechanical properties of chromosomal spindle fibers. Journal of Cell Biology. 1979;82:528–541. doi: 10.1083/jcb.82.2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicklas RB, Kubai DF, Hays TS. Spindle microtubules and their mechanical associations after micromanipulation in anaphase. Journal of Cell Biology. 1982;95:91–104. doi: 10.1083/jcb.95.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. Journal of Cell Biology. 1983;97:532–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skibbens RV, Salmon ED. Micromanipulation of chromosomes in mitotic vertebrate tissue cells: tension controls the state of kinetochore movement. Experimental Cell Research. 1997;235:314–324. doi: 10.1006/excr.1997.3691. [DOI] [PubMed] [Google Scholar]
- 12.Itabashi T, Takagi J, Shimamoto Y, Onoe H, Kuwana K, Shimoyama I, Gaetz J, Kapoor TM, Ishiwata S. Probing the mechanical architecture of the vertebrate meiotic spindle. Nature Methods. 2009;6:167–172. doi: 10.1038/nmeth.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gatlin JC, Matov A, Groen AC, Needleman DJ, Maresca TJ, Danuser G, Mitchison TJ, Salmon ED. Spindle fusion requires dynein-mediated sliding of oppositely oriented microtubules. Current Biology. 2009;19:287–296. doi: 10.1016/j.cub.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stout JR, Rizk RS, Kline SL, Walczak CE. Deciphering protein function during mitosis in PtK cells using RNAi. BMC Cell Biology. 2006;7:26. doi: 10.1186/1471-2121-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukui Y, Yumura S, Yumura TK. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and their changes during chemotaxis. Methods in Cell Biology. 1987;28:347–356. doi: 10.1016/s0091-679x(08)61655-6. [DOI] [PubMed] [Google Scholar]
- 16.Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. Journal of Cell Biology. 2003;160:671–683. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. Journal of Cell Biology. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchison TJ. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. Journal of Cell Biology. 1989;109:637–652. doi: 10.1083/jcb.109.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwen BF, Ding Y, Heagle AB. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Research. 1998;6:123–132. doi: 10.1023/a:1009239013215. [DOI] [PubMed] [Google Scholar]
- 20.Hays TS. Volume Ph.D. University of North Carolina at Chapel Hill; Chapel Hill: 1985. The force-balance mechanism of chromosome congression; p. 156. [Google Scholar]
- 21.Inoué S. The effect of colchicine on the microscopic and submicroscopic structure of the mitotic spindle. Experimental Cell Research Supplement. 1952;2:305–318. [Google Scholar]
- 22.Waters JC, Mitchison TJ, Rieder CL, Salmon ED. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Molecular Biology of the Cell. 1996;7:1547–1558. doi: 10.1091/mbc.7.10.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grill SW, Gönczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- 24.Jang CY, Coppinger JA, Seki A, Yates JR, Fang G. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. Journal of Cell Science. 2009;122:1334–1341. doi: 10.1242/jcs.044321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Molecular Biology of the Cell. 2009;20:1749–1762. doi: 10.1091/mbc.E08-10-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Current Biology. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- 27.Shi J, Orth JD, Mitchison TJ. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Research. 2008;68:3269–3276. doi: 10.1158/0008-5472.CAN-07-6699. [DOI] [PubMed] [Google Scholar]
- 28.McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. Kinetochore microtubules in PTK cells. Journal of Cell Biology. 1992;118:369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cameron L, Yang G, Cimini D, Canman JC, Kisurina-Evgenieva O, Khodjakov A, Danuser G, Salmon ED. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. Journal of Cell Biology. 2006;173:173–179. doi: 10.1083/jcb.200601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. Journal of Cell Biology. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature Cell Biology. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 32.Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Current Biology. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 33.Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- 34.Kaláb P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 35.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohi R, Sapra T, Howard J, Mitchison TJ. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Molecular Biology of the Cell. 2004;15:2895–2906. doi: 10.1091/mbc.E04-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder JA. Effect of metabolic inhibitors on sucrose-induced metaphase spindle elongation and spindle recovery. Cell Motility and the Cytoskeleton. 1988;11:291–302. doi: 10.1002/cm.970110407. [DOI] [PubMed] [Google Scholar]
- 38.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Current Biology. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 39.Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. Journal of Cell Biology. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hays TS, Wise D, Salmon ED. Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. Journal of Cell Biology. 1982;93:374–389. doi: 10.1083/jcb.93.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 42.Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- 43.Gordon MB, Howard L, Compton DA. Chromosome movement in mitosis requires microtubule anchorage at spindle poles. Journal of Cell Biology. 2001;152:425–434. doi: 10.1083/jcb.152.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.