Abstract
The tumor suppressors p16INK4a and p53 have been implicated as contributors to age-associated stem cell decline. Key functions of p53 are the induction of cell cycle arrest, senescence, or apoptosis in response to DNA damage. Here, we examine senescence, apoptosis, and DNA damage responses in a mouse accelerated aging model that exhibits increased p53 activity, the p53+/m mouse. Aged tissues of p53+/m mice display higher percentages of senescent cells (as determined by senescence-associated β-galactosidase staining and p16INK4a and p21 accumulation) compared to aged tissues from p53+/+ mice. Surprisingly, despite having enhanced p53 activity, p53+/m lymphoid tissues exhibit reduced apoptotic activity in response to ionizing radiation compared to p53+/+ tissues. Ionizing radiation treatment of p53+/m tissues also induces higher and prolonged levels of senescence markers p16INK4a and p21, suggesting that in p53+/m tissues the p53 stress response is enhanced and is shifted away from apoptosis toward senescence. One potential mechanism for accelerated aging in the p53+/m mouse is a failure to remove damaged or dysfunctional cells (including stem and progenitor cells) through apoptosis. The increased accumulation of dysfunctional and senescent cells may contribute to reduced tissue regeneration, tissue atrophy, and some of the accelerated aging phenotypes in p53+/m mice.
Keywords: p53, Apoptosis, Senescence, p21, p16INK4a, Mutation frequency
1. Introduction
Organismal aging is characterized by a progressive deterioration of physiological function during adult life that leads to an increased vulnerability to challenges and a decreased ability of the organism to survive stress. It has been proposed that age-associated tissue dysfunction is due to the accumulation of molecular and cellular damage, which leads to generalized homeostatic failure either as a direct consequence of this damage or the cellular response to it (Kirkwood, 2005). Of particular interest is the accumulation of DNA mutations of various forms with age (Dolle et al., 2002; Hill et al., 2004; Stuart et al., 2000). A number of mutant mouse models with DNA repair deficiencies do exhibit accelerated aging phenotypes (Hasty et al., 2003). Mice with enhanced stress resistance, including IGF1r+/− mice and p66Shc−/− mice, exhibit increased longevity (Holzenberger et al., 2003; Migliaccio et al., 1999). Conversely, mice deficient in DNA repair, including TTD mutant mice, XPD null mice, Ku80 null mice, and XPF-ERCC1−/− mice, exhibit a broad array of accelerated aging phenotypes (de Boer et al., 2002; Niedernhofer et al., 2006; van der Pluijm et al., 2007; Vogel et al., 1999).
The ability to respond to multiple types of stress and repair the resulting damage is critical for an organism’s survival and longevity. The inability to repair DNA damage may result in cellular responses that contribute to aging (Hasty, 2005). In response to genotoxic stress, the cell will activate tumor suppressors, which can halt cell proliferation through the initiation of cell cycle arrest and induce DNA repair, apoptosis, or cellular senescence (Lowe et al., 2004; Macip et al., 2003). Tumor suppressors may be an example of antagonistic pleiotropy, where natural selection selects for genes that ensure the reproductive success of young organisms, even if that trait or gene has deleterious effects on the organism at post-reproductive stages (Kirkwood and Austad, 2000; Williams, 1957). Hence, while tumor suppression is critical for the prevention of cancers, an unregulated increase in tumor suppression may accelerate the age-related loss of tissue homeostasis by aberrantly increasing apoptosis or senescence, ultimately compromising stem cell proliferation and regenerative capabilities (Rando, 2006; Sharpless and DePinho, 2007).
The ability of tumor suppressors to induce apoptosis and senescence in response to cellular stress may play a role in the development of aging pathologies, as alterations in apoptosis and senescence have been associated with aging (Campisi, 2003; Serrano and Blasco, 2007). Senescent cells have been shown to accumulate with age and have been detected in several types of age-related pathologies (Choi et al., 2000; Fenton et al., 2001; Matthews et al., 2006; Paradis et al., 2001). Recently, p16INK4a, a tumor suppressor which plays a role in the induction and maintenance of senescence, has been identified as a biomarker of cellular senescence and aging, as p16INK4a expression level has been shown to correlate with age in multiple tissues (Krishnamurthy et al., 2004). An accumulation of senescent cells over time may contribute to aging pathologies and may also contribute to tumorigenesis (Campisi, 2005; Michaloglou et al., 2005; Ressler et al., 2006; Rodier et al., 2007). In addition, evidence suggests that aging leads to a general dysregulation of apoptosis, as aging has been shown to both suppress and increase levels of apoptosis (Adams and Horton, 1998; Ikeyama et al., 2002; Suh, 2002). An increase in either senescence or apoptosis may contribute to loss of tissue homeostasis and potential exhaustion of stem and progenitor cells, while a decrease in apoptosis may facilitate the accumulation of non-functional cells leading to an age-related loss of tissue function.
The tumor suppressor p53 has recently been linked to aging (Bauer et al., 2005; Cao et al., 2003; Feng et al., 2007; Gatza et al., 2005; Maier et al., 2004; Rodier et al., 2007; Tyner et al., 2002; Van Heemst et al., 2005; Varela et al., 2005). The ability of p53 to induce DNA repair, apoptosis, or senescence in response to cellular stress may contribute to its role in aging. Multiple mouse models with premature aging phenotypes demonstrate increased p53 function, including the Brca1Δ11/Δ11 mice, Ku80 null mice, Zmpste24 null mice, and telomerase-deficient mice (Cao et al., 2003; Rudolph et al., 1999; Varela et al., 2005; Vogel et al., 1999). Crossing these models to p53 heterozygous or null mice can partially rescue some of the aging phenotypes (Cao et al., 2003; Chin et al., 1999; Lim et al., 2000; Varela et al., 2005). Additionally, two recent models, the p53+/m mouse and the p44 transgenic mouse, directly implicate p53 in aging by demonstrating that increased p53 function leads to accelerated aging (Maier et al., 2004; Tyner et al., 2002). These models reveal that truncated forms of p53 are capable of enhancing the activity of full-length p53, leading to a reduced lifespan and multiple traits associated with accelerated aging (Moore et al., 2007; Tyner et al., 2002).
The p53+/m mice generated by our laboratory express one wildtype p53 allele and a truncated C-terminal p53 mutant allele, referred to as the “m” allele, which consists of exons 7–11 and contains a deletion of p53 exons 1–6 and an upstream region (Tyner et al., 2002). The m allele is missing the transactivation domain, the Mdm2 binding site, and the majority of the DNA binding domain, yet retains the C-terminal oligomerization domain, which allows it to interact with wildtype p53, as demonstrated in in vitro binding assays (Moore et al., 2007; Tyner et al., 2002). p53+/m mice are highly resistant to tumors, yet display a reduction in median lifespan and exhibit a variety of accelerated aging phenotypes, including osteoporosis, sarcopenia, reduced stress resistance, and generalized skin and tissue atrophies (Tyner et al., 2002). The p53+/m mice also exhibit alterations in hematopoietic stem cell number, proliferation, and engraftment capabilities in aged animals, suggesting that altered p53 function affect stem cell functionality (Chambers et al., 2007; Dumble et al., 2007). In response to γ-irradiation, p53+/m tissues exhibit increased levels of wildtype p53 protein, demonstrating that the m protein can both elevate and stabilize wildtype p53 protein levels.
It should be pointed out, however, that a deletion upstream of the p53 m allele renders the p53+/m mice hemizygous for 24 genes, which could also affect the aging genotypes. Moreover, the m allele RNA is expressed at low levels in most tissues and the m protein has not yet been detected in the p53+/m tissues. These caveats have generated hypotheses that non-p53-associated mechanisms could be responsible for the observed p53+/m cancer resistance and early aging phenotypes (Gentry and Venkatachalam, 2005). However, we and others have argued from earlier studies that p53 hyperactivity in at least some of the p53+/m tissues is likely to be the primary driver of the aging and cancer phenotypes (Tyner et al., 2002; Donehower, 2002; Vijg and Hasty, 2005; Moore et al., 2007). Evidence supporting a hyperactive p53 model include (1) in vitro studies showing that the mallele encoded protein binds with high affinity to wildtype p53, and promotes increased wildtype p53 nuclear localization and transcriptional activity on p53 target genes, and (2) in vivo studies of the p53+/m and p53−/m mice showing that m allele associated cancer resistance was wholly dependent on p53 allele status (Tyner et al., 2002). We believe that the data in this paper showing an elevated and prolonged p53 DNA damage response in the p53+/m mice further support our argument that the p53+/m mouse is primarily a model of hyperactive p53.
Here we demonstrate that p53+/m mice exhibit an increase in a variety of senescence markers with age in spleen, liver, and kidney in comparison to age-matched wildtype (p53+/+) mice. The p53+/m mice also display a modestly increased accumulation of mutations in these same tissues with age as compared to p53+/+ mice. We show that with various dosages of ionizing radiation, senescence markers increase in the p53+/m tissues to a much greater extent than in p53+/+ tissues. Surprisingly, both young and old p53+/m lymphoid tissues exhibit decreased apoptosis in response to ionizing radiation compared to p53+/+ tissues. These data indicate that the m allele contributes to the increase in senescence and decrease in apoptosis observed in the p53+/m mice. Moreover, such alterations in apoptosis and senescence may contribute to the aging phenotypes observed in these mice.
2. Materials and methods
2.1. Mice
All mice were bred and maintained in a specific pathogen free animal facility at Baylor College of Medicine, where cages, chow (Irradiated LabDiet 5053 PicoLab Rodent Diet 20), and water were changed weekly. The WT and p53+/m mice were generated by crossing p53+/+ mice to p53+/m mice and were genotyped by PCR as previously described (Dumble et al., 2007).
2.2. Murine embryonic fibroblasts (MEFs)
Mating pairs of p53+/+ and p53+/m mice were housed together and female mice were checked each morning until a vaginal plug was identified. To obtain p53+/− MEFs, p53+/− mice were also mated and monitored in this way. The day of the plug was determined to be day 0.5. Embryos were harvested on day 12.5. The uterus was removed from the mouse and placed in a sterile petri dish containing sterile phosphate buffered saline (PBS). Each embryo was separated from the yolk sac and surrounding uterine tissue and transferred to a 6 well dish with sterile PBS. Genomic DNA was prepared from the yolk sac following proteinase K digestion and phenol-chloroform extraction and was used for genotyping. Each embryo was homogenized by forcing the embryo through an 18 gauge needle two to three times in 3 ml 15% FBS DMEM. The embryo solution was plated in a 10 mm tissue culture dish with 15% FBS (Invitrogen, Carlsbad, CA), DMEM (Invitrogen) and was left undisturbed for 2 days to allow the fibroblasts to adhere to the plate (P0). Once the cells became confluent, MEFs were split twice (P2) before freezing aliquots. MEF aliquots were frozen in 10% DMSO 90% FBS and stored in liquid nitrogen until use.
MEFs were cultured in a 3T3 protocol to prevent transformation while being used in experiments (Harvey et al., 1993b). MEFs were plated in three 10 cm tissue culture dishes at 106 cells per dish with 15% FBS, DMEM (Invitrogen). Every 3 days the cells from the three dishes were pooled, counted by Trypan Blue (Sigma, St Louis, MO) dye exclusion, and replated at 106 cells per dish in three 10 cm tissue culture dishes. Each split was considered one passage.
2.3. Antibodies
Primary antibodies used included cleaved caspase 3 (Cell Signaling Technology [CST], Danvers, MA), caspase 3 (CST), caspase 9 (CST), actin (Neomarkers, Fremont, CA and Santa Cruz Biotechnology [SCBT], Santa Cruz, CA), cleaved PARP (CST), p16INK4a (M-156) (SCBT), p21(F-5) (SCBT) and p53 (R-19) (SCBT). All primary antibodies were incubated in 3% Milk PBS-Tween 0.05% (PBST) at varying antibody dilutions. Secondary antibodies included: anti-mouse-HRP (SCBT), bovine anti-rabbit-HRP (SCBT), and donkey anti-goat-HRP (SCBT). All secondary antibodies were incubated in 3% Milk PBST at 1:10,000 dilution.
2.4. Senescence-associated β-galactosidase assay in MEFs
MEFs were passaged in a 3T3 protocol as above. Prior to staining, p53+/+, p53+/−, and p53+/m MEFs were plated at 2 × 105 cells per well in a 6 well dish and grown overnight. The next day, cells were rinsed with sterile PBS, fixed with 0.5% gluteraldehyde at room temperature for 15 min, and then rinsed with sterile PBS. Cells were stained overnight with fresh senescence-associated β-galactosidase staining solution (1 mg/ml X-gal in 40 mM citric acid/sodium phosphate, pH 6.0, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 150 mM sodium chloride, 2 mM magnesium chloride) at 37 °C. Post staining, the staining solution was removed and cells were stored in PBS. Senescent cells show marked perinuclear blue staining and non-senescent cells do not exhibit this stain. A standard light microscope was used to count the number of senescence-associated β-galactosidase positive cells and the total number of cells over 5 microscope fields per sample. The percent senescence was calculated by dividing the average number of senescence-associated β-galactosidase positive cells by the average number of total cells and multiplying by 100.
2.5. Senescence-associated β-galactosidase assay in tissues
Spleen, liver, and kidney were harvested from young (3–4 months) and old (18–22 months) p53+/+, p53+/−, and p53+/m mice. The tissue was frozen in a cryomold with OCT by flash freezing in liquid nitrogen, allowing the OCT and tissue to gradually freeze in liquid nitrogen vapor. Embedded frozen tissue was stored at −80 °C until use. A cryotome was used to cut 4 µm (spleen and kidney) or 6 µm sections (liver). The tissue was mounted on positively charged glass slides and sections were kept on dry ice until fixed. Immediately after cutting, the sections were fixed in 0.5% glutaraldehyde at room temperature for 15 min and then rinsed with PBS. Sections were incubated in a humidity chamber at 37 °C with fresh senescence-associated β-galactosidase staining solution from 6 h to up to 18 h (10 h for liver, 6 h for kidney, 6 h for spleen). Tissues were counterstained with hematoxylin and then cover slipped with mounting media. A standard light microscope was used to count the average number of senescence-associated β-galactosidase positive cells over 30 microscope fields per sample. The average total cell number was counted on a hematoxylin and eosin-stained slide. The percent senescence was calculated by dividing the average number of senescence-associated β-galactosidase positive cells by the average number of total cells and multiplying by 100. This value is the percent senescent cells in a given tissue.
2.6. TUNEL assay for detection of apoptotic cells
Young (3 months) and old (18–23 months) p53+/+, p53+/−, and p53+/m mice were exposed to 5 Grays whole body γ-irradiation using a cesium-137 source (MDS Nordion GammaCell 40 Exactor). At 0, 4, or 6 h post-irradiation, the spleen was harvested and fixed in 4% paraformaldehyde at room temperature for 2 h followed by 70% ethanol. Tissue was processed, paraffin embedded, and sections (6 µm) were cut at the Baylor College of Medicine Histology Core. Immunostaining for apoptosis was performed using the in situ Apoptosis Detection Kit (Trevigen, Helgerman, CT) per manufacturer’s instructions. For each tissue section, 5 fields were counted and cells were scored positive (dark brown/black nuclear staining) or negative for TUNEL staining. The percent apoptosis was calculated by dividing the average of the percent TUNEL positive cells by the average number of total cells from 3 separate spleens and multiplying by 100.
2.7. Protein lysates
Freshly excised tissue samples were harvested, snap frozen in liquid nitrogen, and stored at −80 °C until use. Tissues lysates were prepared by homogenization of approximately 75 mg tissue in 500 µl NP40 Lysis Buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 0.75% NP-40 + 1 mini EDTA protease inhibitor tablet/10 ml (Roche Applied Science, Mannheim Germany). Tissue lysates were cleared of debris by centrifugation for 10 min at 12,000 × g, and stored at −80 °C until use. MEF lysates were prepared using Tris Lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.1% NP-40 + 1 EDTA protease inhibitor tablet/10 ml). Cultured cells were exposed to 200–500 µl of lysis buffer depending on cell number, passed through a 20 gauge needle 3 times, and then cleared by centrifugation. The supernatant was transferred to a new tube and stored at −80 °C until use.
2.8. Western blots
25–40 µg lysates were loaded per well for SDS-PAGE using 10% NuPAGE Bis-Tris or 4–12% NuPAGE Bis-Tris gels with MES (Invitrogen) running buffer using the Invitrogen Mini-blot apparatus. Gels were transferred to PVDF membrane (PVDF Filter Paper Sandwich, 0.45 µm pore, Invitrogen) with 10–20% Methanol Transfer Buffer (Invitrogen). Blots were blocked with 3% Milk-PBST. Protein was detected using SuperSignal West Pico Chemiluminescent substrate (Pierce, Rockford, IL).
2.9. Big Blue assay for detection of mutation frequencies
Wildtype C57BL/6 Big Blue mice (Stratagene, La Jolla, CA) were crossed with p53+/m mice and aged to 3, 6, 12, 18, 24 months of age. Where possible, tissues of littermates were harvested concurrently such that at least 3 mice from each time point from each genotype were recovered. Tissues were excised, rinsed with PBS, snap frozen in liquid nitrogen and stored at −80 °C until use. High molecular weight genomic DNA was prepared from kidney, liver and spleen using the Stratagene RecoverEase DNA Isolation Kit (Stratagene) or the Big Blue DNA Isolation Kit (Stratagene) for small intestine (due to its resiliency to mechanical disruption) and stored at 4° until use. DNA was packaged into lambda phage using Transpack Packaging Extract (Stratagene) and 1:100 dilutions were made for titering purposes. Mutation frequency was assessed using the λ Select-cII Mutation Detection System (Stratagene) via the manufacturer’s protocols. Top agar was supplemented with 10 mM MgSO4 to aid in plaque visibility at permissive temperatures. Mutation frequency was determined by dividing total mutant plaque counts at 23.5 °C divided by the total plaque titer for each reaction. This number was averaged with that of other mice of the same age and genotype.
3. Results
3.1. Late passage p53+/m MEFs exhibit increased senescence compared to p53+/+ MEFs
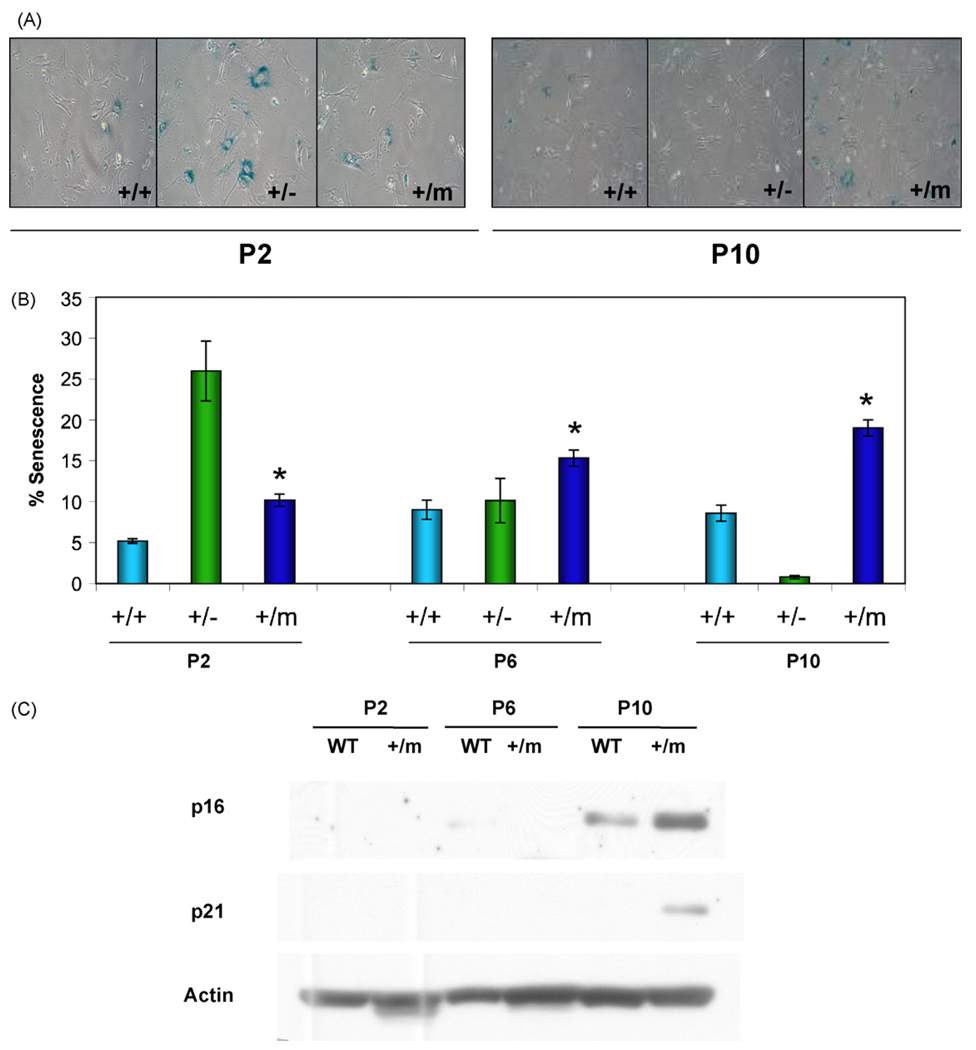
The evidence for augmented p53 activity in the p53+/m mice suggested the possibility that increased p53 induction of senescence could partially account for the aging phenotypes and cancer resistance seen in these mice. To determine if p53+/m MEFs exhibit increased senescence, p53+/+, p53+/−, and p53+/m MEFs were examined by utilizing the senescence-associated β-galactosidase assay (SA-β-gal). At low passage number (P2), p53+/+ MEFs exhibited low levels of senescence, while the p53+/m MEFs exhibited a nearly two-fold increase in senescence (Fig. 1A and B). p53+/m MEFs continue to exhibit significantly higher levels of senescence compared to wildtype at passages 6 and 10. As an additional comparison, we examined MEFs derived from p53+/− mice, which have a single null p53 mutation, and develop early tumors but do not exhibit premature aging phenotypes (Harvey et al., 1993a). Surprisingly, p53+/− MEFs had much higher percentages of senescent cells at passage 2 than either p53+/+ or p53+/m MEFs, but by passage 10, these cells showed virtually no senescence (perhaps due to outgrowth of immortalized clones after the loss of the p53 wildtype allele) (Harvey et al., 1993b).
Fig. 1.
p53+/m MEFs exhibit increased levels of senescence markers compared to p53+/+ MEFs. Mouse embryonic fibroblasts (MEFs) were passaged and evaluated at passages 2 (P2), 6 (P6) and 10 (P10). (A) SA-β-gal staining of MEFs, as indicated by blue perinuclear staining, reveals increased accumulation of positive staining in p53+/− MEFs at early passage (P2) and p53+/m MEFS at late passage (P10) compared to wildtype (p53+/+) MEFs. (B) Quantitation of the mean percent senescence (±s.e.m.) show that p53+/m MEFs exhibit significantly higher levels of staining at each passage than p53+/+ MEFs (*p < 0.02; n = 4 per p53 genotype/passage number). (C) Western blots for cellular senescence initiator p21 and senescence marker p16INK4a verify increased senescence in p53+/m MEFs compared to p53+/+ MEFs with passage.
p53 initiates senescence through the induction of p21, a critical mediator of the G1 arrest typical of senescent cells (Tahara et al., 1995). A subsequent increase in p16INK4a, which can induce G1 arrest by preventing the phosphorylation of Rb, may maintain the senescent growth arrest (Alcorta et al., 1996). Consistent with the results of the SA-β-gal assay, Western blot analysis demonstrated that p53+/m MEFs exhibit an increase in both p21 and p16INK4a protein levels by passage 10 relative to levels of these proteins in P10 p53+/+ MEFs, supporting an increased level of senescence in these cells compared to wildtype cells (Fig. 1C).
3.2. Aged p53+/m tissues exhibit increased numbers of senescent cells
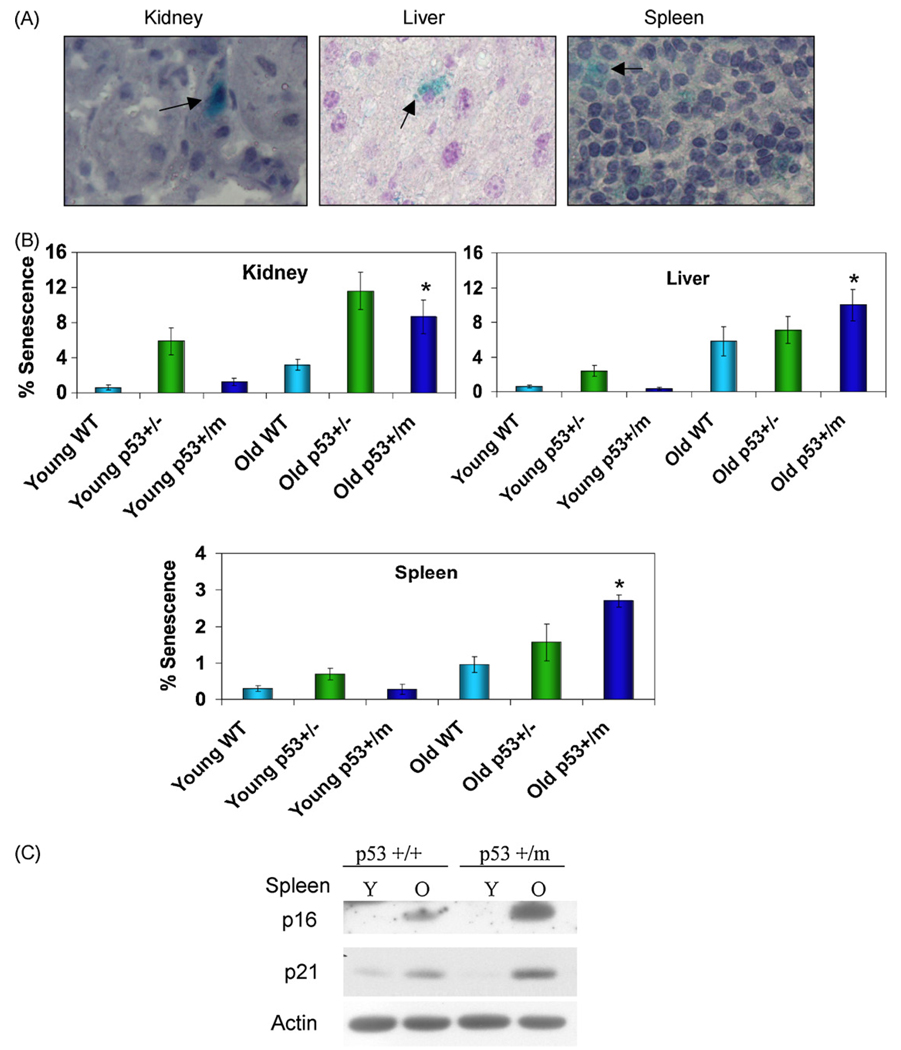
Levels of senescence in young (3 months) and old (18–22 months) p53+/+, p53+/−, and p53+/m mice were examined using an in vivo SA-β-gal assay (Fig. 2A). Young p53+/+ and p53+/m kidney, liver, and spleen exhibit comparable low levels of senescence, while p53+/− tissues show somewhat higher fractions of senescent cells. Old p53+/+ kidney, liver, and spleen demonstrated a two to three-fold increase in levels of senescence with age compared to young p53+/+ tissues. Old p53+/m liver, kidney, and spleen also displayed a dramatic increase in senescence with age compared to young p53+/m tissues. Interestingly, old p53+/m tissues displayed a nearly two-fold increase in levels of senescence compared to age-matched p53+/+ samples (Fig. 2B). The old p53+/− tissues also exhibited an age-associated increase in senescent cells and a higher fraction of senescent cells relative to p53+/+ tissues, but exhibited lower levels of senescence in spleen and liver relative to p53+/m spleen and liver (though not kidney). The age-associated increase in levels of senescence in p53+/m tissues was confirmed by examining p21 and p16INK4a protein levels in young and old spleen from p53+/+ and p53+/m mice. Young p53+/+ and p53+/m spleen exhibited comparable low levels of p16INK4a and p21 (Fig. 2C). Both old p53+/+ and old p53+/m spleens exhibited significant increases in levels of these proteins compared to young spleens, but p53+/m spleens showed higher levels of p21 and p16INK4a than their age-matched p53+/+ counterparts (Fig. 2C). These data support an increase in senescent cell numbers with age in the p53+/m mice, which may be due in part to increased p53 activity.
Fig. 2.
p53+/m tissues exhibit increased levels of senescence markers compared to p53+/+ tissues. Kidney, liver, and spleen from young 3 month (Y) and old 18–22 month (O) p53+/−, p53+/+ and p53+/m mice were analyzed for markers of cellular senescence. (A) Representative SA-β-gal staining of tissues, as indicated by blue perinuclear staining and arrows. All images are shown at 40× magnification. (B) Quantitation of senescent cells in multiple microscope fields for four mice of each age/genotype group reveals increased accumulation of positive staining in p53+/m tissues with age as compared to p53+/+ tissues (*p < 0.05). (C) Western blots for cellular senescence initiator p21 and senescence marker p16INK4a verify increased senescence in p53+/m spleen compared to p53+/+ spleen with age.
3.3. Increased accumulation of point mutations in aged p53+/m tissues
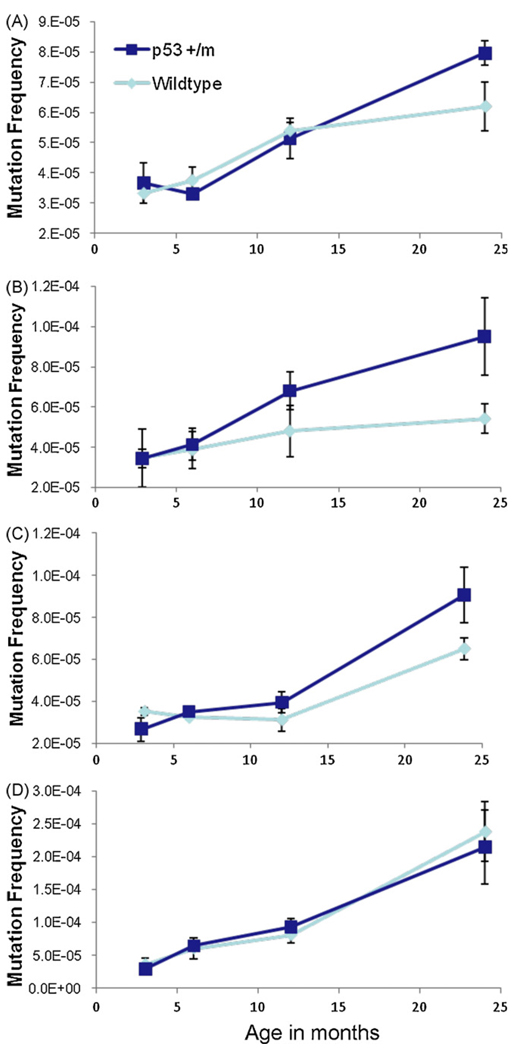
It has been shown in many studies that almost every mouse tissue accumulates mutations with age (Dolle et al., 2000; Hill et al., 2004; Ono et al., 2000; Stuart et al., 2000; Turker et al., 2007). As the p53+/m mouse is a model of increased p53 function, it is likely to have an augmented DNA damage response that could affect mutation frequencies over the mouse lifespan. To assess the effects of them allele on mutation frequency, we crossed it into the Big Blue Mouse, a transgenic model that uses a lambda shuttle vector carrying λ-cII as the mutational target to efficiently detect mutation frequencies in all tissues (Hill et al., 2004). Kidneys, livers, spleens, and small intestine were harvested and analyzed from p53+/+ and p53+/m mice at 3, 6, 12, and 24 months of age. p53+/− and p53−/− tissues were not examined as these were shown not to differ in mutation frequencies from age-matched p53+/+ tissues (Hill et al., 2006; Nishino et al., 1995). As previously reported by other groups, we also observed increases in mutation frequency with age in all tissues examined. In kidney, liver, and spleen (tissues which displayed increased levels of senescent cells with age in p53+/m mice) the p53+/m samples displayed a consistent trend toward higher mutation frequencies at 24 months compared to their p53+/+ counterparts. However, these differences were not quite statistically significant. In the small intestine, a tissue characterized by particularly high mutation frequency, high cellular turnover, and an apoptotic response to stress, there was no difference between p53 genotypes. No significant differences in the types of nucleotide alterations between p53+/+ and p53+/m young and aged tissues were observed in our analyses (data not shown) (Fig. 3).
Fig. 3.
Tissues that exhibit accelerated senescence in p53+/m mice also show increased mutation frequency with age. Kidney, liver, spleen, and small intestine were examined for mutation frequency in genomic DNA of p53+/+ and p53+/m mice at 3, 6, 12, and 24 months of age using the Big Blue Transgenic Mouse system. Wildtype p53 tissue mutation frequencies are represented by light blue lines, p53+/m mutation frequencies are in dark blue (n ≥ 3 at each data point). Increased mutation frequency is observed in all tissues in both genotypes with age. (A) Kidneys of p53+/m mice trend towards increased mutations versus p53+/+ kidneys in the oldest animals (p = 0.051). (B) Livers of 24 month p53+/m mice exhibit increased mutation frequencies compared to 24 month p53+/+ livers (p = 0.056). (C) Spleens of p53+/m mice display increases in mutation frequency with age as compared to p53+/+ spleens (p = 0.076). (D) In small intestine, a tissue where cellular senescence is neither expected nor reported in the literature, p53+/+ and p53+/m mutation frequencies are indistinguishable (p = 0.388).
3.4. p53+/m mice exhibit a reduced apoptotic response to IR stress
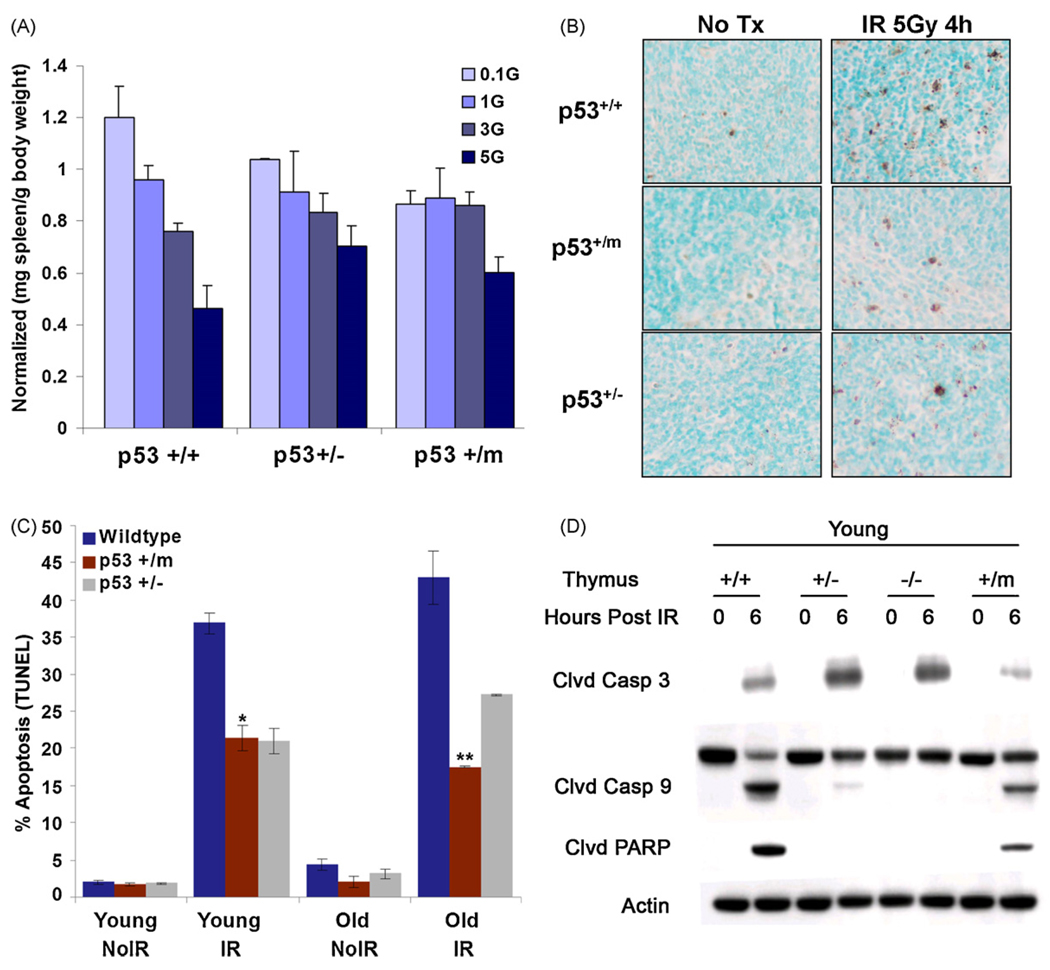
p53 is a potent regulator of apoptosis in response to DNA damage and other stresses (Vousden and Lu, 2002). One effect of altered p53 activity in p53+/m mice could be dysregulation of apoptosis. Because apoptotic cells are infrequently detected in tissues of unstressed mice, mice were treated with differing doses of IR and lymphoid organs were harvested one week post-irradiation. In particular, mouse spleen cells are susceptible to p53-dependent radiation-induced apoptosis (MacCallum et al., 1996). As IR dosage was increased, p53+/+ spleen mass correspondingly decreased one week after treatment. In contrast, p53+/− and p53+/m spleens tended to retain splenic mass with increased IR dosage, suggesting a resistance to apoptotic effects of radiation (Fig. 4A). To more accurately quantitate levels of apoptosis in irradiated spleens, a TdT dUTP nick end labeling (TUNEL) assay was performed to determine if p53+/m mice exhibit altered apoptosis in the presence and absence of ionizing radiation (Fig. 4B and C). In the absence of IR stress, both young (3 months) and old (18–20 months) spleens of all p53 genotypes exhibit comparably low levels of apoptosis. In response to 5 Gy whole body IR, young and old p53+/+ mice exhibited a comparable large induction of apoptosis at 4 h post-irradiation. However, both young and old p53+/m and p53+/− spleens demonstrated a significantly reduced apoptotic index in response to irradiation compared to age-matched p53+/+ mice (Fig. 4B and C). Additionally, both an Annexin V flow cytometry assay and an active caspase 3 flow cytometric assay demonstrated that young p53+/m thymocytes exhibited a decreased apoptotic index in response to whole body irradiation compared to age-matched p53+/+ thymocytes (data not shown).
Fig. 4.
Apoptosis is reduced in p53+/m lymphoid tissues compared to wildtype lymphoid tissues. (A) One week after IR p53+/+ male spleens exhibit a steep decrease in spleen mass (mg spleen per gram body weight) at IR dosage of 3 Gray (Gy) and 5 Gy, whereas p53+/m spleens are refractory until the highest levels of IR (n = 3 per data point). p53+/− spleens exhibit a steady decline in spleen mass with increasing IR dosage. The data are normalized to non-irradiated spleens. (B) Representative TUNEL staining of young and old p53+/+, p53+/−, and p53+/m non-irradiated and 4 h post 5 Gy irradiation spleens. Dark brown-black nuclear staining indicates TUNEL positive, apoptotic cells. All images are shown at 20× magnification. (C) Quantitation of percent average TUNEL positive staining (±s.e.m) in young and old untreated and 5 Gy IR treated spleens from p53+/+, p53+/− and p53+/m animals. In both young and old spleens, p53+/m tissue sections exhibit significantly lower levels of positive TUNEL staining as compared to p53+/+ tissue sections (**p < 4.2 × 10−5 and *p < 0.008, respectively) (n = 4). (D) Western blot showing reduced cleavage of apoptosis-facilitating proteins (caspase 3, caspase 9, and PARP) in young p53+/m thymus relative to young p53+/+ thymus, 6 h post 5 Gy IR.
The decreased level of apoptosis in the p53+/m mice was further confirmed by examining levels of pro-apoptotic proteins in spleen and thymus in response to irradiation. Both young and old p53+/m spleen exhibited decreased levels of cleaved caspases 3 and cleaved PARP, two pro-apoptotic proteins which play an important role in the induction of apoptosis (data not shown). Young p53+/m thymi also exhibited decreased levels of cleaved caspase 3, cleaved caspase 9, and cleaved PARP relative to p53+/+ thymi (Fig. 4D). As expected, p53+/− thymi also showed reduced caspase 9 and PARP cleavage, though caspase 3 cleavage was increased. These data collectively demonstrate that both young and old p53+/m mouse lymphoid tissues exhibit a reduced apoptotic index in response to stress.
3.5. p53+/m tissues show increased induction of senescence markers in response to IR stress
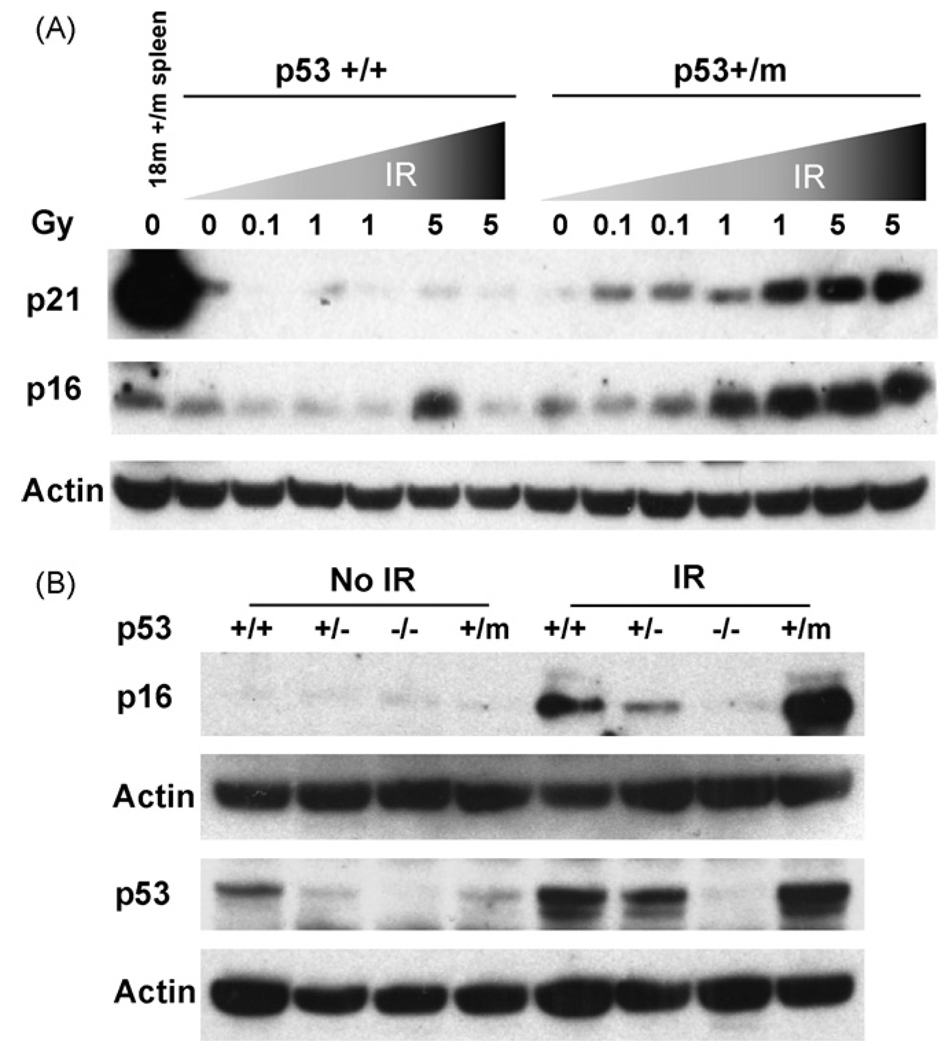
The resistance to IR-induced apoptosis exhibited by p53+/m spleen and thymus (Fig. 4) and the increased level of senescence observed in aged unstressed p53+/m tissues (Fig. 2) is consistent with a higher retention of senescent or dysfunctional cells. Thus, p53 in p53+/m tissues may preferentially induce senescence rather than apoptosis in response to IR stress. To examine this possibility further we measured the accumulation of the senescence markers p16INK4a and p21 in irradiated spleen tissues. Young p53+/+ and p53+/m mice were irradiated with 0, 0.1, 1.0, and 5.0 Gy and spleens were harvested one week post-irradiation. Levels of p16INK4a and p21 in spleen lysates were compared by Western blot analysis. In p53+/+ spleens p21 returned to basal levels within one week post-irradiation regardless of IR dose and there was little to no upregulation of p16INK4a (Fig. 5A). Conversely, in the apoptosis-deficient p53+/m spleens, higher levels of both p21 and p16INK4a were observed with increasing IR dosage. We also examined short term responses to IR by harvesting spleens 6 h post-irradiation. As in spleens harvested at one week post-irradiation, p53+/m spleens harvested 6 h after 5 Gy IR exhibited higher levels of p16INK4a compared to p53+/+ spleens (Fig. 5B). p53+/− spleens showed reduced short term induction of p16INK4a. In addition, IR induction of p53 protein was higher in the p53+/m spleens compared to p53+/+ and p53+/− spleens (Fig. 5B). These data are consistent with the hypothesis that high levels of DNA damage in the p53+/m tissues leads to an enhanced and prolonged p53 response and senescence rather than apoptosis, compared with the p53+/+ tissue response.
Fig. 5.
Increased IR dosage leads to increased and prolonged induction of molecular senescence markers in p53+/m mice. (A) Six-week-old male p53+/+ and p53+/m mice were irradiated with a dosage curve ranging from 0 to 5 Gy IR and spleens were harvested one week post-irradiation. Western blots show little or no increase in p21 or p16INK4a senescence markers in p53+/+ spleens, but dramatic increases in both markers in the p53+/m spleens. (B) Three-month-old p53+/+, p53+/−, p53−/−, and p53+/m male mice were irradiated with 5 Gy IR and spleens were harvested 6 h post-irradiation. Western blot analysis indicates significant increase in the levels of p16INK4a in p53+/m spleens. Levels of p53 protein induced 6 h post IR are also shown. When normalized to loading control, levels of p53 induced in p53+/m spleens are at least three fold higher than in p53+/+ spleens. Representative blots are illustrated.
4. Discussion
The data reported here demonstrate that the p53+/m mouse, a model of accelerated aging, exhibits increased age-associated senescence, decreased apoptosis, and an enhanced and prolonged response to DNA damage relative to p53+/+ mice. Wildtype p53 is a potent effector of both senescence and apoptosis through its response to various cellular stresses, including DNA damage. We propose that these aberrant senescence, apoptosis and damage response phenotypes are a result of altered wildtype p53 regulation and activity in the p53+/m tissues. We have previously shown that the truncated C-terminal form of p53 encoded by the “m” allele interacts with wildtype p53, alters p53 transcriptional activation functions, enhances p53 stability, and promotes nuclear localization of p53 (Moore et al., 2007; Tyner et al., 2002). Nevertheless, despite the evidence that wildtype p53 activities are generally enhanced by them protein, consistent with the enhanced DNA damage responses and increased cellular senescence in aged p53+/m MEFs and tissues, the pro-apoptotic functions of p53 are unexpectedly inhibited in these same tissues.
Multiple molecular and cellular assays on the spleens and thymi of young and old irradiated p53+/m mice indicate a significant reduction of apoptosis compared to that observed in lymphoid organs of similarly treated p53+/+ mice. A reduction in p53+/m apoptotic response relative to the p53+/+ apoptotic response seems counterintuitive given the enhanced p53-associated DNA damage and senescence responses in the p53+/m mice. One possible explanation may derive from our earlier finding that them protein interacts with the wildtype p53 protein and drives it into the nucleus, even in the absence of stress (Moore et al., 2007). Because a significant component of the p53 apoptotic response is non-transcriptional and dependent on direct p53 migration to mitochondrial membranes (Moll et al., 2005), the p53+/m apoptosis response may be reduced because of an absence of cytoplasmic wildtype p53 and consequently a deficiency in p53 mitochondrial-induced apoptosis.
The decrease in apoptosis in the p53+/m mice may contribute to the observed aging phenotypes, although the mechanism for how it would do so is unclear. A consensus does not appear to have formed on the role of apoptosis in organismal aging (Higami and Shimokawa, 2000; Kujoth et al., 2006; Suh and Vijg, 2006; Zhang et al., 2003). A reduction in apoptosis, as observed in the p53+/m model, may contribute to aging by failing to remove damaged and dysfunctional cells, leading to an accelerated loss of tissue function with age. Some of these retained dysfunctional cells could include stem and progenitor cells, which could inhibit tissue regeneration. However, p53+/− tissues also exhibit reduced apoptotic phenotypes similar to those in p53+/m mice, yet p53+/− mice do not show early aging phenotypes and are cancer susceptible.
One possibility is that wildtype p53 activity in p53+/m and p53+/− mice preferentially induces cellular senescence rather than apoptosis in response to stress. p53+/m and p53+/− mice exhibit an age-related increase in levels of senescence in kidney, liver, and spleen in comparison to age-matched p53+/+ control mice. Supporting these data, p53+/m tissues also exhibit an age-related increase in p16INK4a protein levels, which is known to play an important role in the induction and maintenance of senescence (Janzen et al., 2006; Krishnamurthy et al., 2006, 2004;Molofsky et al., 2006). In addition, MEFs derived from p53+/m and p53+/− mice exhibit a significant increase in levels of senescence as examined by the SA-β-gal assay, even at an early passage (P2). Late passage p53+/m MEFs exhibit an increase in p21 and p16INK4a protein levels as well. One possible mechanism for the increased senescence in early passage p53+/− cells are studies showing that reduced dosage of p53 in some contexts results in a shift away from apoptosis and towards cell cycle arrest or senescence (Macip et al., 2003). However, despite high senescence levels, the older p53+/− mice are profoundly susceptible to tumors (Harvey et al., 1993a), unlike the highly cancer resistant p53+/m mice (Tyner et al., 2002). We hypothesize that these differences are due to the reduced upregulation of p53 (and p21 and p16INK4a) after stress in p53+/− tissues, while p53+/m tissues show an abnormally high and prolonged response to stress compared to p53+/+ and p53+/− tissues (Fig. 5A and B).
In this paper we attempted to determine which major p53 function most closely correlated with the cancer resistance and early aging phenotypes observed in the p53+/m mice. By examining p53-mediated apoptosis, senescence, mutation frequencies, and DNA damage responses in three mouse strains with differing levels of p53 activity, we hoped to better understand how p53 may influence longevity and aging. Spontaneous mutation frequencies were only modestly increased in some tissues in older p53+/m mice in comparison to their p53+/+ counterparts (Fig. 3), so we do not believe increased mutation rates are a major factor in p53+/m premature aging phenotypes. It is possible that the increased p53+/m mutation frequencies are related to increased retention of senescent or dysfunctional cells.
Aged cancer resistant p53+/m mice exhibit significantly increased cellular senescence in multiple tissues relative to age-matched p53+/+ mice, but only marginally increased senescence relative to cancer-susceptible p53+/− mice (Fig. 6A). Likewise, p53+/m and p53+/− mice both display attenuated apoptosis relative to p53+/+ mice, indicating a failure to mount a proper p53 response. In this context age-associated senescent cells and attenuated apoptotic responses do not necessarily correlate with aging, given their similar appearance in a cancer-susceptible normally aging mouse (p53+/−) and a cancer resistant premature aging mouse (p53+/m). But the p53+/m mouse does display a robust and prolonged DNA damage response relative to both p53+/+ and p53+/− mice, as measured by high p53 protein levels, and high levels of p21 and p16INK4a. Such a prolonged anti-proliferative response to damage could be the critical p53-associated property that is most relevant to the early aging phenotypes observed in the p53+/m mice. The robust and extended upregulation of p21 and p16INK4a in stem and progenitor cells could produce senescence or dysfunction in these cells that over time may contribute to the p53+/m aging phenotypes and pathologies through the loss of proliferative and tissue regenerative capacities (Fig. 6B). Consistent with this model, we have shown that hematopoietic and mammary gland stem cells in aged p53+/m mice display reduced proliferative and regenerative function compared to their age-matched p53+/+ counterparts (Dumble et al., 2007; Gatza et al., 2008).
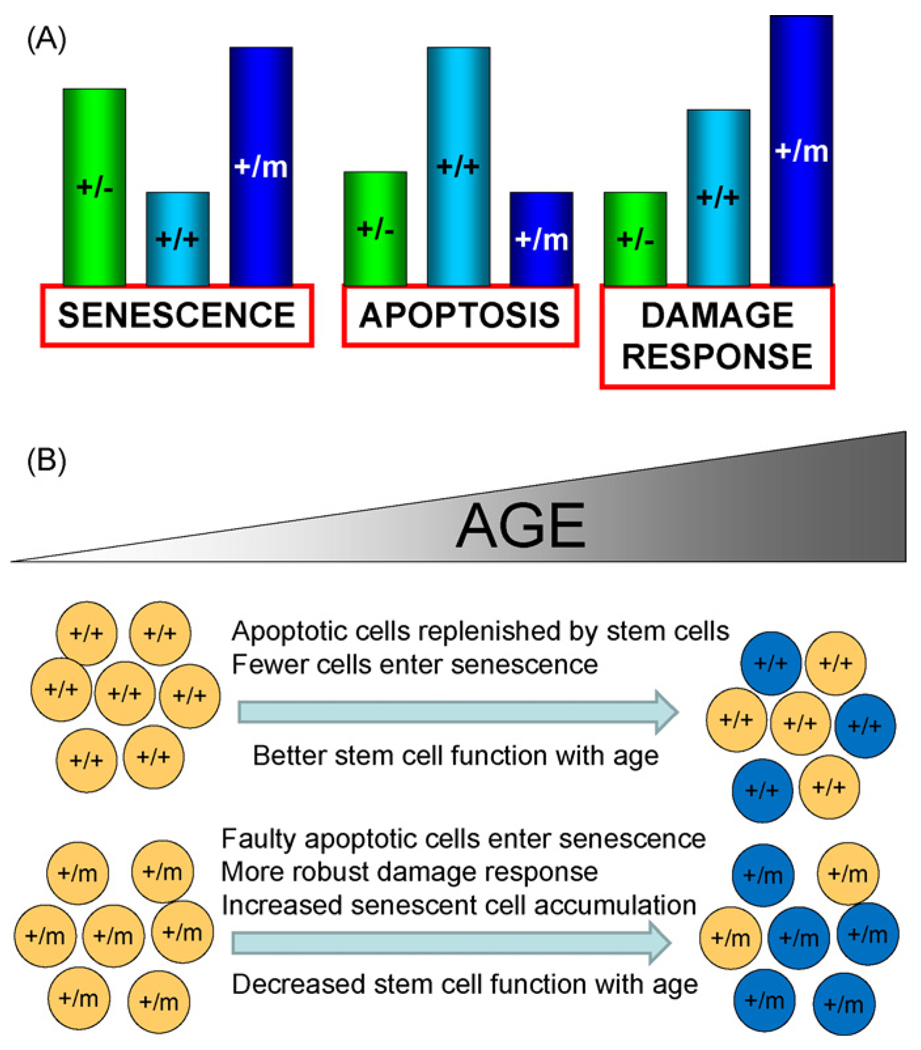
Fig. 6.
Models showing relative p53 responses in p53+/−, p53+/+, and p53+/m mice (A) and p53+/m accelerated organismal and cellular aging resulting from altered stress responses (B). (A) In response to DNA damage and other stresses, p53 induces differential senescence, apoptosis, and damage responses dependent on p53 dosage and activity. Relative responses are indicated by bar heights for each effect for p53+/−, p53+/+, and p53+/m mice. p53+/m mice exhibit enhanced senescence and DNA damage responses and reduced apoptosis relative to p53+/+ mice. (B) Enhanced damage responses in p53+/m tissues are associated with inadequate apoptotic removal of damaged cells and retention of senescent cells in p53+/m tissues and may contribute to the accelerated aging phenotypes observed in p53+/m mice. As p53+/+ mice age, damaged cells are efficiently removed by p53-mediated apoptosis and replaced by functional adult tissue stem cells (top part of panel). Senescent cells accumulate at a modest rate and aging phenotypes occur late in life. In contrast, p53+/m tissues exhibit enhanced anti-proliferative damage responses and are defective in apoptotic elimination of damaged or dysfunctional cells and these accumulate with age, perhaps as senescent cells. Such cells may include stem and progenitor cells that more rapidly lose the ability to replenish cells in the tissues as the organism ages. The result is tissue atrophy and some of the accelerated aging phenotypes observed in the p53+/m mouse.
In summary, the data presented here demonstrate that tissues in aging p53+/m mice exhibit an increase in senescent cell numbers, a reduced apoptotic index and an enhanced and prolonged DNA damage response in comparison to their wildtype counterparts. Because only the DNA damage response is consistently enhanced in p53+/m mice relative to both p53+/+ and p53+/− mice, we hypothesize that this p53 property is the best correlate for the p53+/m premature aging phenotypes. We postulate that the truncated p53 m protein could disrupt normal wildtype p53 regulation and even in the absence of exogenous stress provide a chronic low level stimulation of wildtype p53 that might gradually impact on stem and progenitor cell functionality and cause the p53+/m early aging effects. This work strengthens the link between p53 and aging, suggesting that p53 activity must be maintained under tight regulation, as alterations in activity may contribute to aberrant aging phenotypes. Increasing p53 dosage while retaining normal p53 regulation can sometimes be beneficial by promoting cancer resistance without early aging (Garcia-Cao et al., 2002), but if normal p53 regulation is disrupted, deleterious effects such as increased tumor incidence or even accelerated aging can result (Donehower et al., 1992; Maier et al., 2004; Tyner et al., 2002). Further studies will be necessary to precisely identify the mechanisms by which p53 impacts on aging and longevity.
Acknowledgments
We are grateful to Michelle Weiss for technical assistance. We thank Sharra Hammond for analysis of mutational spectra in the Big Blue assay. This work was supported by funds from the National Institute for Aging and the Ellison Medical Foundation to L.A.D. G.H. was also supported by training grant funds from the National Institute for Aging.
References
- Adams CS, Horton WE., Jr Chondrocyte apoptosis increases with age in the articular cartilage of adult animals. Anat. Rec. 1998;250(4):418–425. doi: 10.1002/(SICI)1097-0185(199804)250:4<418::AID-AR4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, Barrett JC. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 1996;93(24):13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr. Biol. 2005;15(22):2063–2068. doi: 10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cancer and ageing: rival demons? Nat. Rev. Cancer. 2003;3(5):339–349. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120(4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Cao L, Li W, Kim S, Brodie SG, Deng CX. Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev. 2003;17(2):201–213. doi: 10.1101/gad.1050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5(8):e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97(4):527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Choi J, Shendrik I, Peacocke M, Peehl D, Buttyan R, Ikeguchi EF, Katz AE, Benson MC. Expression of senescence-associated beta-galactosidase in enlarged prostates from men with benign prostatic hyperplasia. Urology. 2000;56(1):160–166. doi: 10.1016/s0090-4295(00)00538-0. [DOI] [PubMed] [Google Scholar]
- de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296(5571):1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- Dolle ME, Snyder WK, Dunson DB, Vijg J. Mutational fingerprints of aging. Nucleic Acids Res. 2002;30(2):545–549. doi: 10.1093/nar/30.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc. Natl. Acad. Sci. U.S.A. 2000;97(15):8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA. Does p53 affect organismal aging? J. Cell. Physiol. 2002;192(1):23–33. doi: 10.1002/jcp.10104. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA, Donehower LA. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109(4):1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, Teresky AK, Hernando E, Cordon-Cardo C, Levine AJ. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proc. Natl. Acad. Sci. U.S.A. 2007;104(42):16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton M, Barker S, Kurz DJ, Erusalimsky JD. Cellular senescence after single and repeated balloon catheter denudations of rabbit carotid arteries. Arterioscler. Thromb. Vasc. Biol. 2001;21(2):220–226. doi: 10.1161/01.atv.21.2.220. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21(22):6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatza C, Hinkal G, Moore L, Dumble M, Donehower LA. p53 and mouse aging models. In: Masoro Edward J, Austad SN., editors. Handbook of the Biology of Aging. sixth ed. San Diego, CA: Academic Press; 2005. pp. 147–178. [Google Scholar]
- Gatza CE, Dumble M, Kittrell F, Edwards DG, Dearth RK, Lee AV, Xu J, Medina D, Donehower LA. Altered mammary gland development in the p53+/m mouse, a model of accelerated aging. Dev. Biol. 2008;313(1):130–141. doi: 10.1016/j.ydbio.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry A, Venkatachalam S. Complicating the role of p53 in aging. Aging Cell. 2005;4(3):157–160. doi: 10.1111/j.1474-9726.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- Harvey M, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat. Genet. 1993a;5(3):225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- Harvey M, Sands AT, Weiss RS, Hegi ME, Wiseman RW, Pantazis P, Giovanella BC, Tainsky MA, Bradley A, Donehower LA. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993b;8(9):2457–2467. [PubMed] [Google Scholar]
- Hasty P. The impact of DNA damage, genetic mutation and cellular responses on cancer prevention, longevity and aging: observations in humans and mice. Mech. Ageing Dev. 2005;126(1):71–77. doi: 10.1016/j.mad.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299(5611):1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- Higami Y, Shimokawa I. Apoptosis in the aging process. Cell Tissue Res. 2000;301(1):125–132. doi: 10.1007/s004419900156. [DOI] [PubMed] [Google Scholar]
- Hill KA, Buettner VL, Halangoda A, Kunishige M, Moore SR, Longmate J, Scaringe WA, Sommer SS. Spontaneous mutation in Big Blue mice from fetus to old age: tissue-specific time courses of mutation frequency but similar mutation types. Environ. Mol. Mutagen. 2004;43(2):110–120. doi: 10.1002/em.20004. [DOI] [PubMed] [Google Scholar]
- Hill KA, Buettner VL, Heidt A, Chen LL, Li W, Gonzalez KD, Wang JC, Scaringe WA, Sommer SS. Most spontaneous tumors in a mouse model of Li-Fraumeni syndrome do not have a mutator phenotype. Carcinogenesis. 2006;27(9):1860–1866. doi: 10.1093/carcin/bgl029. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Ikeyama S, Kokkonen G, Shack S, Wang XT, Holbrook NJ. Loss in oxidative stress tolerance with aging linked to reduced extracellular signal-regulated kinase and Akt kinase activities. FASEB J. 2002;16(1):114–116. doi: 10.1096/fj.01-0409fje. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, Depinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16(INK4a) Nature. 2006 doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120(4):437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Austad SN. Why do we age? Nature. 2000;408(6809):233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16(INK4a) induces an age-dependent decline in islet regenerative potential. Nature. 2006 doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 2004;114(9):1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations and apoptosis in mammalian aging. Cancer Res. 2006;66(15):7386–7389. doi: 10.1158/0008-5472.CAN-05-4670. [DOI] [PubMed] [Google Scholar]
- Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA, Hasty P. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 2000;20(11):3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Hupp TR, Midgley CA, Stuart D, Campbell SJ, Harper A, Walsh FS, Wright EG, Balmain A, Lane DP, Hall PA. The p53 response to ionising radiation in adult and developing murine tissues. Oncogene. 1996;13(12):2575–2587. [PubMed] [Google Scholar]
- Macip S, Igarashi M, Berggren P, Yu J, Lee SW, Aaronson SA. Influence of induced reactive oxygen species in p53-mediated cell fate decisions. Mol. Cell. Biol. 2003;23(23):8576–8585. doi: 10.1128/MCB.23.23.8576-8585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18(3):306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews C, Gorenne I, Scott S, Figg N, Kirkpatrick P, Ritchie A, Goddard M, Bennett M. Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ. Res. 2006;99(2):156–164. doi: 10.1161/01.RES.0000233315.38086.bc. [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402(6759):309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Moll U, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr. Opin. Cell. Biol. 2005;17(6):631–636. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16(INK4a) expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006 doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L, Lu X, Ghebranious N, Tyner S, Donehower LA. Aging-associated truncated form of p53 interacts with wild-type p53 and alters p53 stability, localization, and activity. Mech. Ageing Dev. 2007;128(11–12):717–730. doi: 10.1016/j.mad.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeij-makers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444(7122):1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Nishino H, Knoll A, Buettner VL, Frisk CS, Maruta Y, Haavik J, Sommer SS. p53 wild-type and p53 nullizygous Big Blue transgenic mice have similar frequencies and patterns of observed mutation in liver, spleen and brain. Oncogene. 1995;11(2):263–270. [PubMed] [Google Scholar]
- Ono T, Ikehata H, Nakamura S, Saito Y, Hosoi Y, Takai Y, Yamada S, Onodera J, Yamamoto K. Age-associated increase of spontaneous mutant frequency and molecular nature of mutation in newborn and old lacZ-transgenic mouse. Mutat. Res. 2000;447(2):165–177. doi: 10.1016/s0027-5107(99)00200-6. [DOI] [PubMed] [Google Scholar]
- Paradis V, Youssef N, Dargere D, Ba N, Bonvoust F, Deschatrette J, Bedossa P. Replicative senescence in normal liver, chronic hepatitis C, and hepatocellular carcinomas. Hum. Pathol. 2001;32(3):327–332. doi: 10.1053/hupa.2001.22747. [DOI] [PubMed] [Google Scholar]
- Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441(7097):1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- Ressler S, Bartkova J, Niederegger H, Bartek J, Scharffetter-Kochanek K, Jansen-Durr P, Wlaschek M. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5(5):379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- Rodier F, Campisi J, Bhaumik D. Two faces of p53: aging and tumor suppression. Nucleic Acids Res. 2007;35(22):7475–7484. doi: 10.1093/nar/gkm744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96(5):701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Serrano M, Blasco MA. Cancer and ageing: convergent and divergent mechanisms. Nat. Rev. Mol. Cell. Biol. 2007;8(9):715–722. doi: 10.1038/nrm2242. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell. Biol. 2007;8(9):703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Stuart GR, Oda Y, de Boer JG, Glickman BW. Mutation frequency and specificity with age in liver, bladder and brain of lacI transgenic mice. Genetics. 2000;154(3):1291–1300. doi: 10.1093/genetics/154.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y. Cell signaling in aging and apoptosis. Mech. Ageing Dev. 2002;123(8):881–890. doi: 10.1016/s0047-6374(02)00025-8. [DOI] [PubMed] [Google Scholar]
- Suh Y, Vijg J. Maintaining genetic integrity in aging: a zero sum game. Antioxid. Redox Signal. 2006;8(3–4):559–571. doi: 10.1089/ars.2006.8.559. [DOI] [PubMed] [Google Scholar]
- Tahara H, Sato E, Noda A, Ide T. Increase in expression level of p21sdi1/cip1/waf1 with increasing division age in both normal and SV40-transformed human fibroblasts. Oncogene. 1995;10(5):835–840. [PubMed] [Google Scholar]
- Turker MS, Lasarev M, Connolly L, Kasameyer E, Roessler D. Age-related accumulation of autosomal mutations in solid tissues of the mouse is gender and cell type specific. Aging Cell. 2007;6(1):73–86. doi: 10.1111/j.1474-9726.2006.00264.x. [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee Park S, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415(6867):45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, de Wit J, Mitchell JR, van Oostrom C, Beems R, Niedernhofer LJ, Velasco S, Friedberg EC, Tanaka K, van Steeg H, Hoeijmakers JH, van der Horst GT. Impaired genome maintenance suppresses the growth hormone—insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5(1):e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heemst D, Mooijaart SP, Beekman M, Schreuder J, de Craen AJM, Brandt BW, Slagboom PE, Westendorp RGJ Long Life Study Group. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp. Gerontol. 2005;40(1–2):11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437(7058):564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Vijg J, Hasty P. Aging and p53: getting it straight. A commentary on a recent paper by Gentry and Venkatachalam. Aging Cell. 2005;4(6):335–338. doi: 10.1111/j.1474-9726.2005.00179.x. [DOI] [PubMed] [Google Scholar]
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl. Acad. Sci. U.S.A. 1999;96(19):10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat. Rev. Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- Zhang JH, Zhang Y, Herman B. Caspases, apoptosis and aging. Ageing Res. Rev. 2003;2(4):357–366. doi: 10.1016/s1568-1637(03)00026-6. [DOI] [PubMed] [Google Scholar]