Abstract
MALAT1 is a long non-coding RNA whose expression level was originally identified as a predictor of metastasis of non-small cell lung tumors and was subsequently shown to be over-expressed in many human cancers. Here, we have identified a highly conserved tRNA-like small RNA of 61 nucleotides that originates from the MALAT1 locus and is broadly expressed in human tissues. In contrast to the long MALAT1 transcript that localizes to nuclear speckles, the small RNA exclusively localizes to the cytoplasm. We show that RNase P cleaves downstream of a genomically encoded poly(A)-rich tract to simultaneously generate the 3’ end of the abundant MALAT1 transcript and the 5’ end of the small RNA. Our findings reveal a novel 3’ end processing mechanism by which a single locus can yield both a stable nuclear retained non-coding RNA with a short poly(A) tail-like moiety and a small tRNA-like cytoplasmic RNA.
INTRODUCTION
Although it has been generally assumed that most genetic information is expressed as and transacted by proteins, the majority of the mammalian genome is transcribed yielding a complex network of overlapping transcripts that includes tens of thousands of long non-coding RNAs (ncRNAs) with little or no protein-coding capacity (Carninci et al., 2005; Birney et al., 2007; Kapranov et al., 2007). Few long ncRNAs have been assigned a function; however, a significant number of these transcripts are unlikely to represent transcriptional “noise” as they have been shown to exhibit cell-type specific expression, localization to specific subcellular compartments, and association with human diseases (reviewed in Prasanth and Spector, 2007; Szymanski et al., 2005; Costa, 2005; Mercer et al., 2008). Among those shown to be functional, Xist ncRNA is involved in mammalian X-inactivation (Masui and Heard, 2006), the roX genes mediate X-chromosome hyperactivation in Drosophila (reviewed in Deng and Meller, 2006), and several long ncRNAs, including Air, have been implicated in genomic imprinting (Braidotti et al., 2004).
A recent study suggested that the function of a significant fraction (~40%) of long unannotated transcripts may be to serve as precursors for small RNAs less than 200-nt in length (Kapranov et al., 2007). Consistent with this hypothesis, the H19 imprinted gene was originally suggested to function as a ~2.3-kb ncRNA, but has recently been shown to serve as a microRNA precursor (Cai and Cullen, 2007). In addition to microRNAs that regulate RNA degradation and translation (Filipowicz et al., 2008; Vasudevan et al., 2007) and piRNAs that silence transposons in the germline (Brennecke et al., 2007; Carmell et al., 2007), additional classes of small RNAs have been recently discovered. These include small RNAs that are present near the 5’ and 3’ ends of genes (PASRs and TASRs, respectively), although the mechanisms of their biogenesis and their functions remain to be determined (Kapranov et al., 2007).
MALAT1 (Metastasis associated lung adenocarcinoma transcript 1) was originally identified in a screen for transcripts whose expression was altered in early-stage non-small cell lung cancers that had a propensity for metastasis (Ji et al., 2003). Expression of the previously uncharacterized RNA was increased in tumors that later metastasized compared to those that did not, indicating the prognostic value of MALAT1 for metastasis. Additionally, high expression of MALAT1 was found to be associated with significantly decreased patient survival. Curiously, MALAT1 lacks open reading frames of significant length and in vitro translation of MALAT1 yielded no peptides, suggesting that MALAT1 functions as a long (~7-kb) non-coding RNA (Ji et al., 2003; Lin et al., 2006).
MALAT1 shows broad expression in normal human and mouse tissues (Ji et al., 2003; Hutchinson et al., 2007) and is over-expressed in many human carcinomas, including those of the breast, pancreas, lung, colon, prostate, and liver (Lin et al., 2006), implying that MALAT1 misregulation may play a role in the development of numerous cancers. Chromosomal translocation breakpoints associated with cancer have also been identified within the MALAT1 locus (Davis et al., 2003; Kuiper et al., 2003; Rajaram et al., 2007). Recently, it has been shown that MALAT1 is specifically retained in the nucleus in nuclear speckles (Hutchinson et al., 2007), domains that are thought to be involved in the assembly/modification and/or storage of the pre-mRNA processing machinery (reviewed in Lamond and Spector, 2003). Such localization is indicative of MALAT1 playing a potential role in the organization or regulation of gene expression.
In the present study, we investigated whether MALAT1 may also function as a precursor for the production of small RNAs. We have identified a highly conserved, tRNA-like non-coding RNA that maps near the 3’ end of the MALAT1 locus. Northern blot analysis showed that the 61-nt RNA is broadly expressed in normal human tissues. In contrast to the stable nuclear-retained MALAT1 transcript, the 61-nt RNA is exclusively localized to the cytoplasm and has a relatively short half-life (and thus we have named it mascRNA, MALAT1-associated small cytoplasmic RNA). We show that 3’ end processing of MALAT1 yields the mascRNA transcript. RNase P cleaves downstream of a genomically encoded poly(A)-rich tract to simultaneously generate the 3’ end of MALAT1 and the 5’ end of mascRNA. This 3’ end processing mechanism results in a short poly(A) tail-like moiety being present on the 3’ end of MALAT1. These results reveal a novel 3’ end processing mechanism by which a single locus can yield both a stable nuclear retained ncRNA and a small tRNA-like cytoplasmic RNA.
RESULTS
Identification of a highly conserved small non-coding RNA originating from the MALAT1 locus
MALAT1 is a long non-coding transcript that specifically localizes to nuclear speckles, shows broad expression in mouse and human normal tissues, and is over-expressed in numerous cancers (Hutchinson et al., 2007; Lin et al., 2006; Ji et al., 2003). Within the MALAT1 transcript, there are short blocks of high conservation, especially in the 3’ half of the RNA, which may represent functional elements such as binding sites for proteins, nuclear localization signals, or sequences for the generation of small RNAs (Figure 1A). To determine if any small RNAs map to the mouse MALAT1 (mMALAT1) locus, an enriched fraction of small RNAs under 200 nucleotides was isolated from mouse EpH4 mammary epithelial cells. These cells had been stably transfected with an empty vector (EpH4-EV cells) or a vector that over-expresses the ErbB2 oncogene (EpH4-A6 cells) to induce cellular transformation (Fantin et al., 2002). Normal and transformed EpH4 cells were used because expression of mMALAT1 increases 2.5-fold in response to ErbB2 over-expression (Supplementary Figure 1) and over-expression of MALAT1 has been previously linked to various cancers.
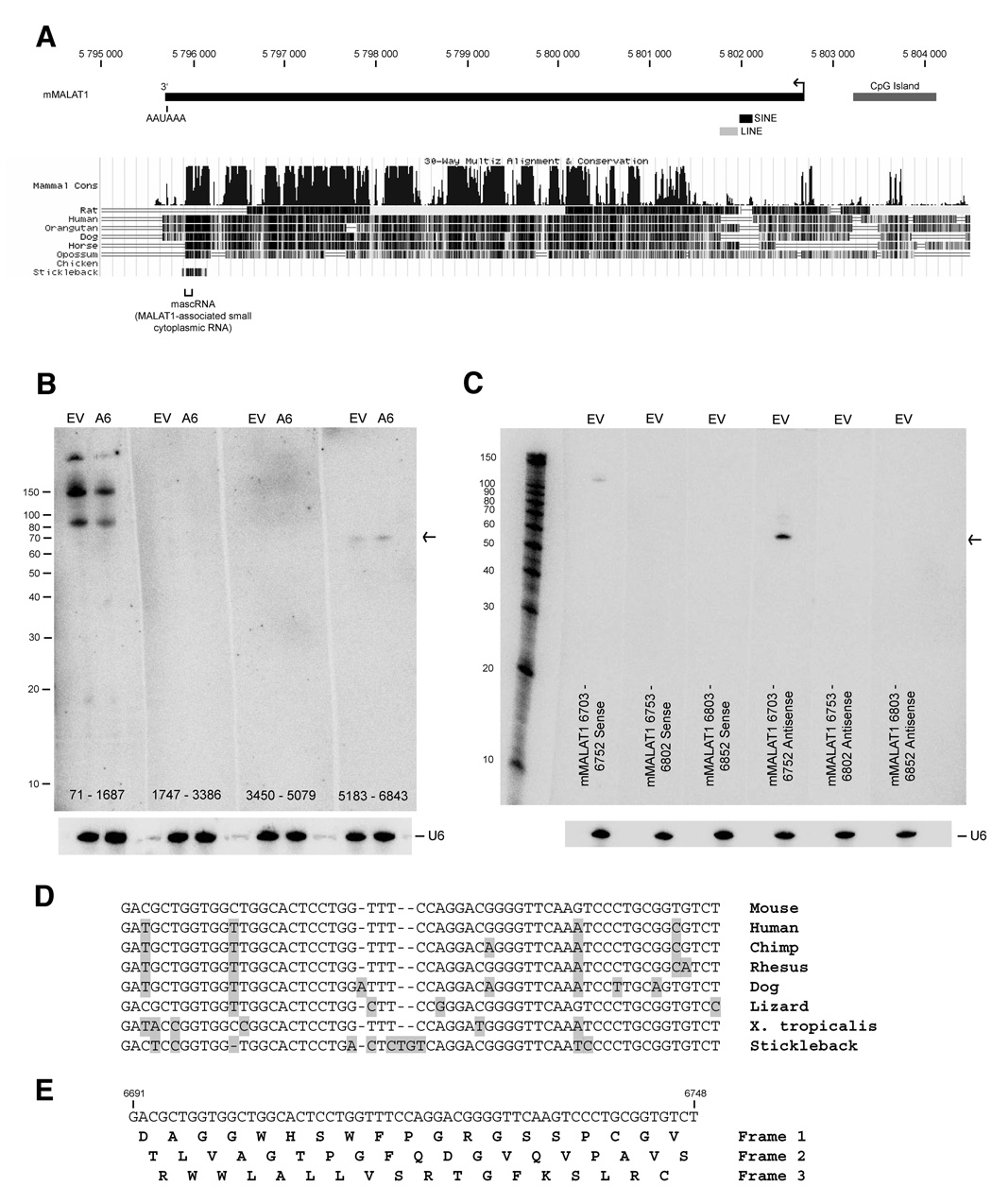
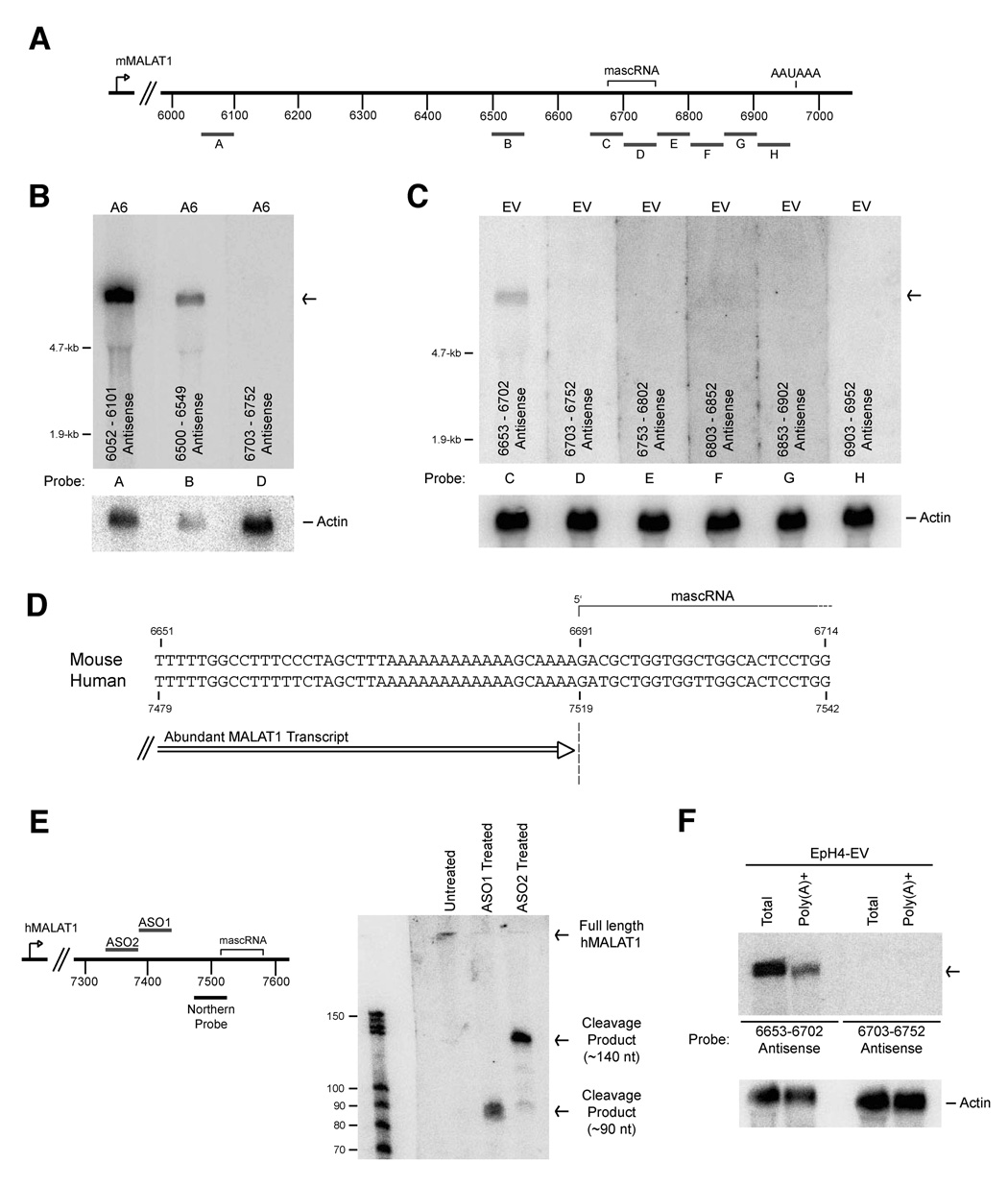
Figure 1. A 61-nt small RNA maps near the 3’ end of mMALAT1.
(A) The MALAT1 locus, located on mouse chromosome 19, has been reported to yield a ~7-kb non-coding transcript (transcription start site denoted by arrow) that contains regions of high conservation. MALAT1 lacks repetitive elements except for a SINE and LINE element near its 5’ end. (B) Small RNAs were isolated from EpH4-EV (denoted EV) and EpH4-A6 (denoted A6) cells. Four different probes that span the mMALAT1 locus were used for Northern blot analysis as designated at the bottom of each blot. U6 snRNA was used as a loading control. (C) Strand-specific oligonucleotide probes were used to further map the 61-nt small RNA. Sense probes detect transcripts originating from the opposite strand of mMALAT1, while antisense probes detect transcripts originating from the same strand as mMALAT1. (D) The 61-nt mascRNA transcript is highly conserved (Sequence shown as DNA and does not include CCA at the 3’ end, which is added during processing). (E) The mascRNA transcript lacks translation initiation and stop codons.
Small RNA Northern blots were then performed using four different probes, each approximately 1.6-kb in length, to span the ~7-kb sequence of mMALAT1 (Figure 1B). Probes to the middle of mMALAT1 did not hybridize to any small RNA, whereas a probe to nt 71 to 1687 hybridized to three RNA species of approximately 90, 150, and >200-nt in length and a probe to nt 5183 to 6843 of mMALAT1 hybridized to a single RNA species of 61-nt in length (Note that all mMALAT1 nucleotide numbering is based on GenBank accession number AY722410). Upon further mapping the small RNAs hybridizing to the 5’ end of mMALAT1, it was determined that all three RNAs map to the SINE element and thus probably do not originate from mMALAT1 (Supplementary Figure 2).
To further map the small RNA originating from the 3’ end of mMALAT1, the original 1.6-kb probe was divided into several shorter probes that were used to show that the 61-nt RNA originated from within nt 6703 to 6843 (Supplementary Figures 3A and 3B). Oligonucleotide probes were then used to show that a probe complementary to nt 6703 to 6752 of mMALAT1 specifically hybridizes to the 61-nt RNA, indicating that the small RNA originates from the same strand as mMALAT1 (Figure 1C). An antisense probe to nt 6653 to 6702 of mMALAT1 was also able to weakly detect the small RNA, suggesting that the small RNA spanned the two probes (data not shown). By performing Northern blots with multiple 20-nt oligonucleotide probes that tiled this region, we concluded that the small RNA maps to approximately nt 6690 to 6750 of mMALAT1 (Supplementary Figure 3C). In contrast to mMALAT1, small RNA expression did not significantly change upon transformation of EpH4 cells. When we examined small RNA transcriptome maps that were generated using genomic tiling arrays (Kapranov et al., 2007), we noted that the 61-nt RNA was not detected by this methodology, possibly due to RNA secondary structure or the threshold employed (data not shown).
We have named this 61-nt RNA mascRNA (MALAT1-associated small cytoplasmic RNA) for the reasons stated below. The mascRNA transcript is highly conserved and present in multiple species, including mouse, human, dog, lizard, X. tropicalis, and stickleback (Figure 1D). There are only 4 mismatches between the mouse and human orthologs. Interestingly, mascRNA is at the 3’ end of a ~160-nt region that is highly conserved among these species, suggesting that the upstream region may be important for the biogenesis of the mascRNA transcript (Supplementary Figure 4). Immediately upstream of mascRNA is a conserved poly(A)-rich tract (16/18 adenines in mouse, 17/19 adenines in human). Further upstream, there are two nearly perfectly conserved 10-nt U-rich motifs separated by a conserved predicted stem loop. No translation initiation codons or stop codons are present within the mascRNA transcript (Figure 1E), indicating that it likely functions as a small non-coding RNA.
The mascRNA transcript is broadly expressed in human tissues
Because the mascRNA transcript is highly conserved, we examined whether the 61-nt RNA is also expressed in human cell lines and tissues. Using an antisense probe to the syntenic region of human MALAT1 (hMALAT1), we were able to detect expression of the 61-nt human mascRNA ortholog in multiple cell lines, including HeLa (Figure 2A) and MCF10A (data not shown) (Note that all hMALAT1 nucleotide numbering is based on GenBank accession number BK001418). Next, we examined the expression of the human mascRNA transcript in twenty normal human tissues and found that the small RNA is broadly expressed (Figure 2B), which is not surprising given that the long MALAT1 transcript is also broadly expressed (Ji et al., 2003; Hutchinson et al., 2007).
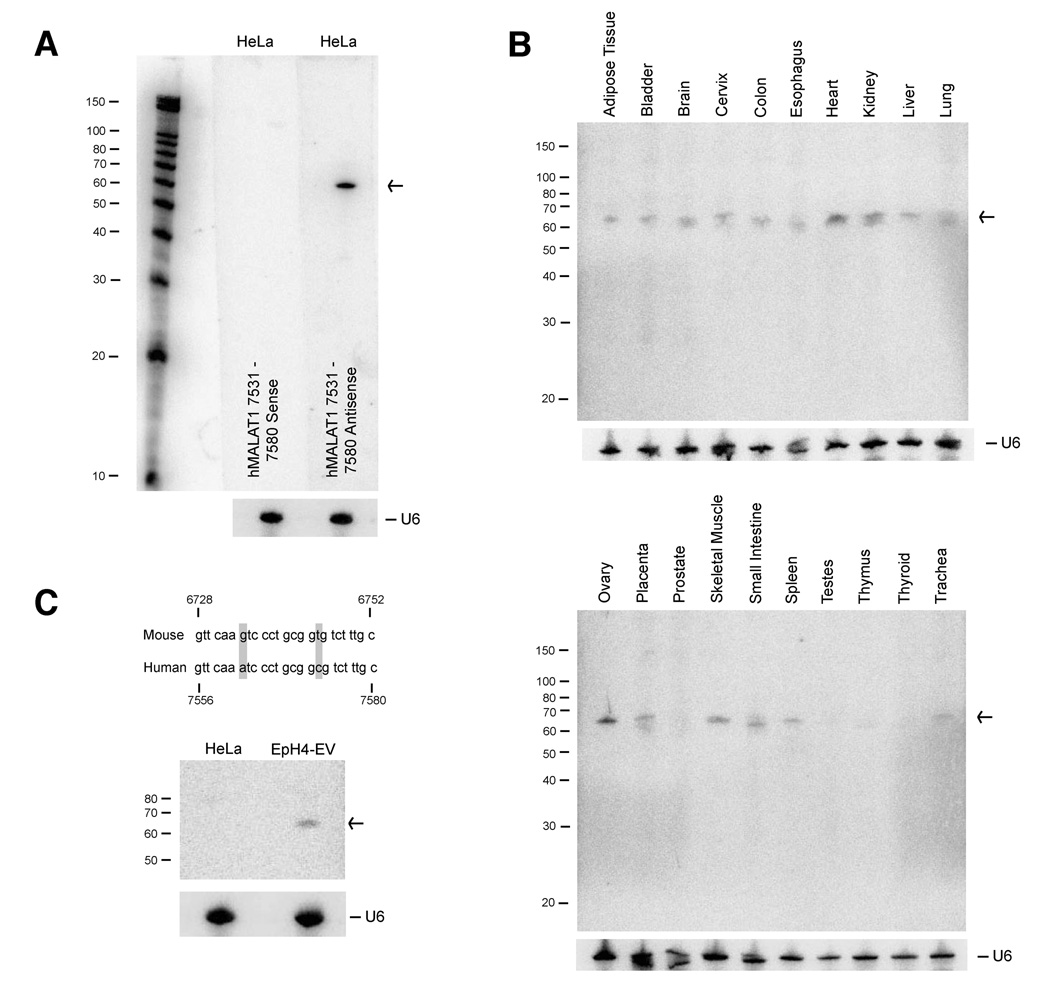
Figure 2. mascRNA is broadly expressed in normal human tissues.
(A) Northern blot analysis using strand-specific oligonucleotide probes showed that the human mascRNA ortholog is expressed in HeLa cells. U6 snRNA was used as a loading control. (B) 10 µg of total RNA from twenty normal human tissues were probed for mascRNA expression. (C) A 25-nt probe complementary to nt 6728–6752 of mMALAT1 was designed. The 25-nt probe specifically distinguished expression of the mouse isoform in EpH4-EV cells from the human isoform in HeLa cells.
To further verify that the 61-nt RNA we detected on the Northern blots originates from the MALAT1 locus, we took advantage of several point mutations between the mouse and human mascRNA sequences. Based upon these differences, we designed a 25-nt probe complementary to nt 6728 to 6752 of mMALAT1 that should be able to distinguish between the mouse and human mascRNA orthologs when hybridized at 42 degrees by only detecting mouse mascRNA (Figure 2C). As shown, the 25-nt probe only detected the mouse isoform, confirming that the small RNA detected on the Northern blots originates from the MALAT1 locus.
mascRNA is a tRNA-like RNA polymerase II transcript that localizes to the cytoplasm
Upon treating total RNA with a 5’ phosphate-dependent exonuclease, we found that mascRNA is not susceptible to degradation, suggesting that it lacks a 5’ monophosphate (Figure 3A, Lanes 1 and 2). However, this exonuclease fails to digest double-stranded RNA or RNA with extensive secondary structure such as tRNAs. We thus reasoned that the mascRNA secondary structure may make the transcript insensitive to digestion, and mascRNA may actually have a 5’ monophosphate. To disrupt the mascRNA secondary structure, total RNA was hybridized to DNA oligonucleotides complementary to mascRNA and then subjected to digestion by RNase H (Figure 3A, Lanes 3–6). The 5’ phosphate-dependent exonuclease was able to digest the shorter mascRNA 5’ fragments, indicating that there is a 5’ monophosphate on mascRNA. Using a ligation-based small RNA cloning procedure, we then defined the 5’ end of mascRNA to be at exactly nt 6691 of mMALAT1 (which is equivalent to nt 7519 of hMALAT1) (Supplementary Figure 5).
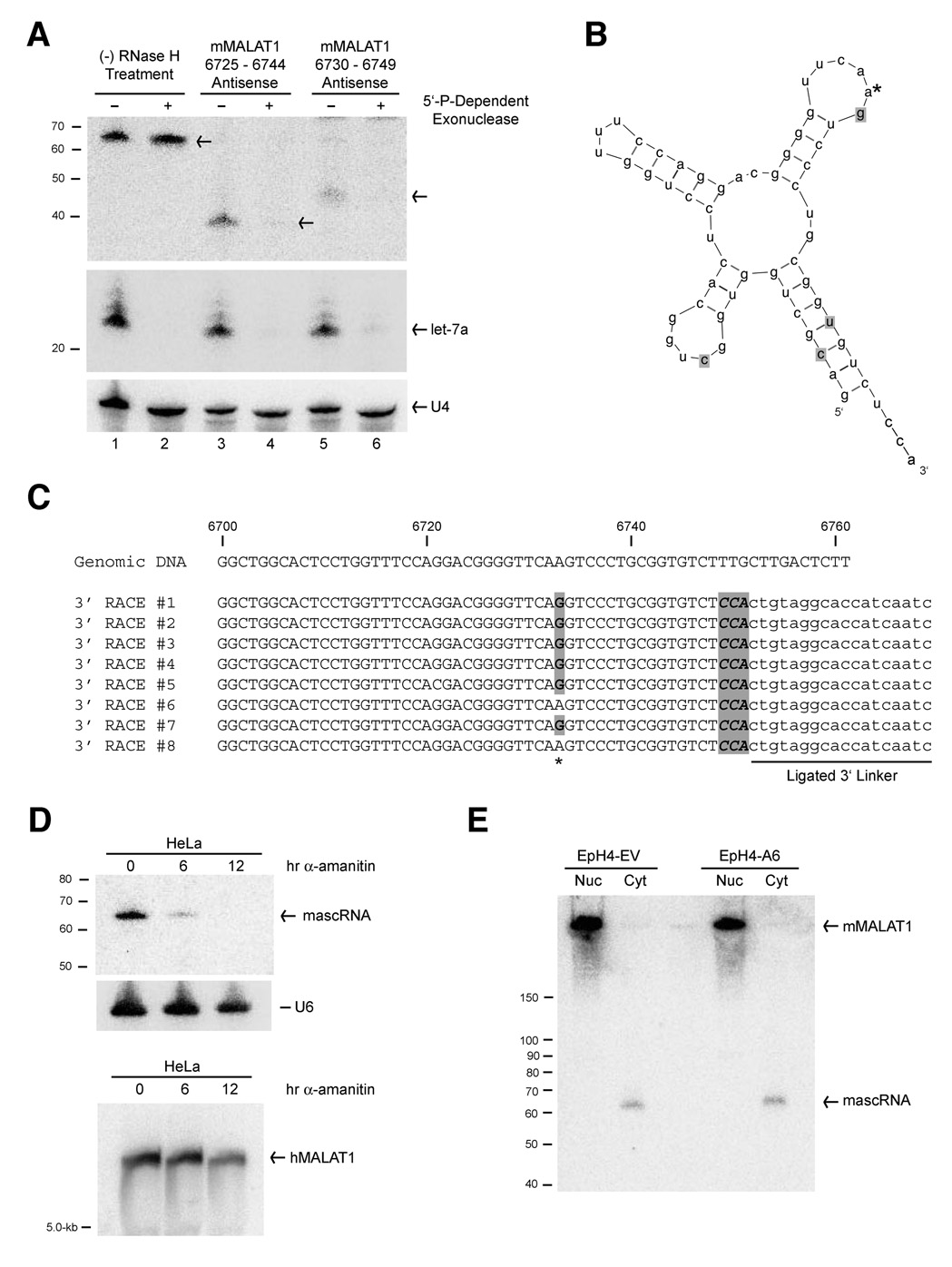
Figure 3. mascRNA is a RNA polymerase II tRNA-like transcript that localizes to the cytoplasm.
(A) A 5’-phosphate-dependent exonuclease was used to show that mascRNA has a 5’-monophosphate group. In lanes 3–6, EpH4-A6 total RNA was first treated with antisense oligonucleotides (as designated at top) complementary to mascRNA and subjected to RNase H treatment. A probe complementary to nt 6690–6709 of mMALAT1 was used for the Northern blot. The microRNA let-7a was used as a positive control for exonuclease activity, whereas U4 snRNA is capped and serves as a loading control. (B) The mature mouse mascRNA transcript is predicted to form a tRNA-like cloverleaf secondary structure. The four point mutations between the mouse and human orthologs are shaded. The * designates the nucleotide that is commonly modified. (C) The 3’ end of mouse mascRNA was cloned using a ligation-based approach (3’ linker oligo is designated). All sequenced mascRNA cDNA clones end in CCA and six of eight clones show a common nucleotide modification. (D) Total RNA from HeLa cells treated with 50 µg/mL of α-amanitin, an RNA polymerase II inhibitor, for 6 or 12 hours was subjected to Northern blot analysis. (E) EpH4-EV and EpH4-A6 cells were fractionated to isolate nuclear and cytoplasmic total RNA, which was then subjected to Northern blot analysis using a probe to nt 6563–6843 of mMALAT1. The long mMALAT1 transcript was exclusively nuclear, whereas the mascRNA transcript was exclusively cytoplasmic.
We similarly cloned the 3’ end of mascRNA using a ligation-based approach and found that mascRNA has a 3’ hydroxyl group (Figure 3C). To our surprise, all sequenced mouse (Figure 3C) and human (data not shown) mascRNA cDNA clones ended in 3’-terminal CCA, which is not encoded in the genome and is a hallmark of the 3’ ends of tRNAs and tRNA-like structures (reviewed in Hopper and Phizicky, 2003). Upon folding the 61-nt mascRNA transcript using Mfold, we found that mascRNA is predicted to fold similar to a tRNA cloverleaf secondary structure (Zuker, 2003; Matthews et al., 1999) (Figure 3B). This tRNA-like structure of mascRNA has been preserved through evolution, as the four mismatches between the mouse and human orthologs maintain the cloverleaf secondary structure. Although similar in structure to a tRNA and containing a well-conserved B-box (Supplementary Figure 4), the 61-nt mascRNA transcript is smaller than most tRNAs (~76-nt) and has a small, relatively poorly conserved anticodon loop. This suggests that mascRNA does not function in transferring a specific amino acid to a growing polypeptide chain during protein synthesis. By performing polyacrylamide gel electrophoresis under acidic conditions to look at aminoacylation levels of tRNAs in vivo (Varshney et al., 1991), we found no evidence that mascRNA is aminoacylated in HeLa cells (Supplementary Figure 6). mascRNA cloning also revealed that at least one nucleotide of mascRNA is commonly modified because the genomic A at mMALAT1 nt 6733 was sequenced as a G in 6 of 8 mouse clones (Figure 3C), suggestive of post-transcriptional modification of adenine to inosine (A-to-I editing) (reviewed in Bass, 2002).
MALAT1 is known to localize to nuclear speckles (Hutchinson et al., 2007), and thus we were interested in determining to which subcellular compartment the 61-nt mascRNA transcript localizes. Using biochemical fractionation to separate nuclear and cytoplasmic total RNA from EpH4-EV and EpH4-A6 cells, we were able to confirm that the long mMALAT1 transcript is exclusively nuclear (Figure 3E and Figure 6A). In contrast, mascRNA is exclusively cytoplasmic in EpH4 (Figure 3E) and HeLa cells (data not shown). To examine if mascRNA is associated with polysomes, exponentially growing HeLa cells were harvested and cytoplasmic extracts were prepared and displayed on sucrose gradients (Supplementary Figure 8). Unlike microRNAs, mascRNA does not associate with polysomes, but instead associates with lighter fractions where tRNAs are present. In summary, two non-coding transcripts are produced from the MALAT1 locus; interestingly, each localizes to a different subcellular compartment, implying that they may have distinct functions.
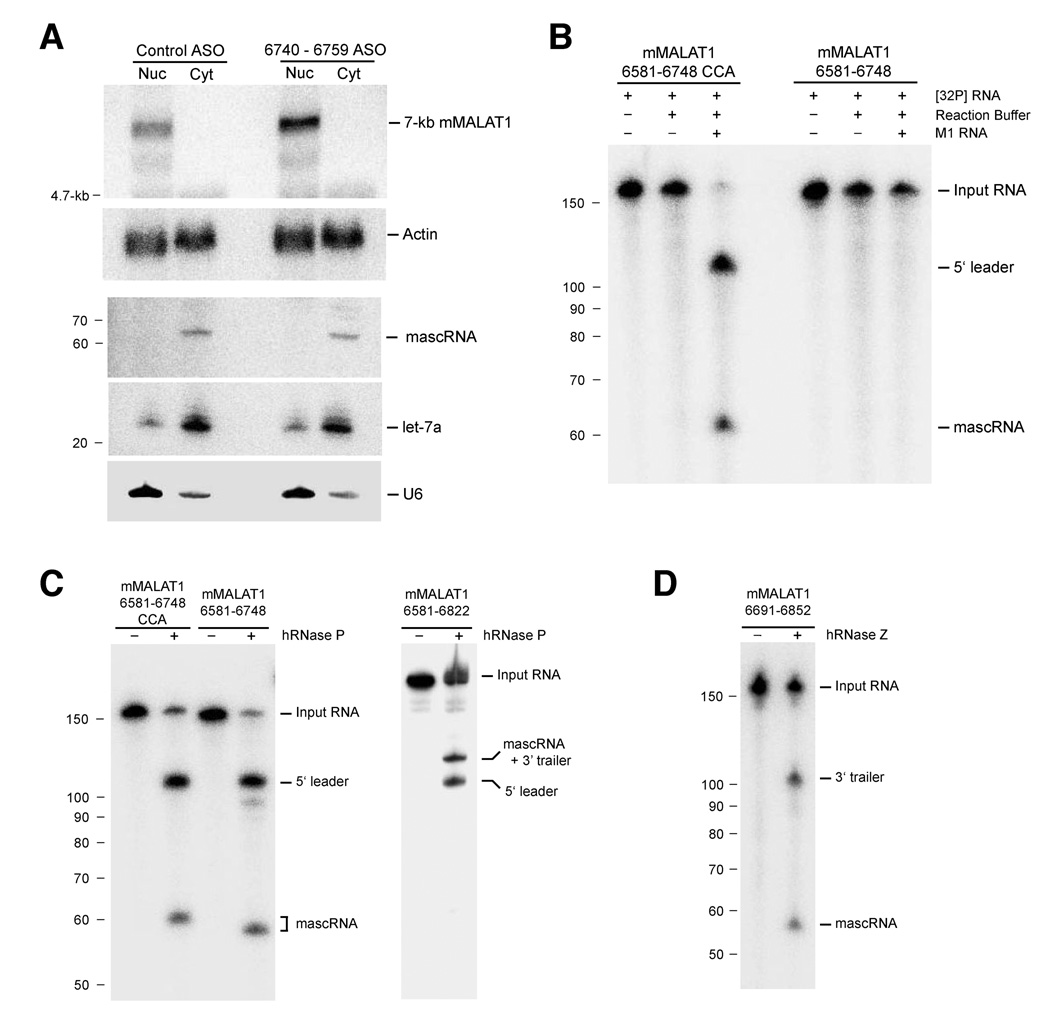
Figure 6. RNase P and RNase Z cleave MALAT1 in the nucleus to yield the mascRNA transcript.
(A) After ASO transfection, EpH4-EV cells were fractionated to isolate nuclear and cytoplasmic total RNA. The ~7-kb mMALAT1 transcript was nuclear-retained and ASO treatment did not affect the cytoplasmic localization of mascRNA. U6 and let-7a were used as controls for fractionation efficiency. (B) MALAT1 can be cleaved in vitro by E. coli RNase P. M1 RNA from E. coli was incubated for 1 hr at 37 degrees with uniformly labeled MALAT1 substrates. Samples were then electrophoresed in a 8% polyacrylamide/8 M urea gel. (C) MALAT1 can be cleaved in vitro by partially purified human RNase P. (D) Recombinant His-tagged human RNase Z cleaves MALAT1 in vitro at the 3’ end of mascRNA.
We were next interested in determining if mascRNA is made via processing of MALAT1 or if the small RNA is derived from a separate, overlapping transcriptional unit. First, we addressed whether mascRNA is an RNA polymerase II transcript by incubating HeLa cells with α-amanitin, an RNA polymerase II inhibitor (irreversible in tissue culture cells at 50 µg/ml). mascRNA expression decreased greater than 70% after 6 hr of α-amanitin treatment and was undetectable after 12 hr, consistent with the small RNA being an RNA polymerase II transcript with a relatively short half-life (Figure 3D, top panel; Supplementary Figure 7A). Interestingly, MALAT1 was present at significant levels in HeLa cells even after prolonged transcriptional inhibition, indicating low turnover and a RNA half-life of greater than 12 hr (Figure 3D, bottom panel). As further support that mascRNA is a RNA polymerase II transcript, mascRNA expression rapidly decreased when HeLa cells were incubated with a lower concentration of α-amanitin (Supplementary Figure 7B) or with DRB, another specific RNA polymerase II inhibitor (Supplementary Figure 7C). These results are consistent with a model in which mascRNA is derived from an RNA polymerase II transcript, but do not definitively prove that MALAT1 is the precursor.
The abundant MALAT1 transcript ends immediately upstream of mascRNA
Mouse MALAT1 has been reported to be an abundant ~7-kb polyadenylated RNA (Lin et al., 2007; Hutchinson et al., 2007). However, the exact size of long transcripts can be difficult to precisely determine using Northern blot analysis. Therefore, we used multiple oligonucleotide probes to map what regions are and are not part of the abundant mMALAT1 transcript (Figure 4A). Consistent with previous reports, multiple oligo probes complementary to mMALAT1 were able to detect a single long transcript (Figure 4B, Probes A and B). Unexpectedly, a probe complementary to the mascRNA region was unable to detect the long mMALAT1 transcript (Figure 4B, Probe D), indicating that this region may be spliced out of mMALAT1 or that the 3’ end of mMALAT1 is further upstream than has been reported. We were unable to detect any mMALAT1 transcripts that were spliced in this region by RT-PCR (data not shown), suggesting that RNA splicing can not explain our observation.
Figure 4. The abundant MALAT1 transcript ends immediately upstream of mascRNA.
(A) Antisense probes complementary to mMALAT1 were designed and used in Parts (B) and (C). (B) 10 µg of total RNA from EpH4-A6 cells was probed by Northern blot analysis using the designated oligonucleotide probes. β-actin was used as a loading control. (C) Northern blots revealed that the 3’ end of the abundant mMALAT1 transcript is further upstream than previously reported and, therefore, the abundant transcript is only ~6.7-kb in length. (D) A conserved poly(A)-rich tract is present immediately upstream of mascRNA. Numbers at the top denote the mouse nucleotide position, whereas numbers at the bottom denote the corresponding human nucleotide position. (E) RNase H digestion followed by Northern blot analysis was used to map the 3’ end of the abundant human MALAT1 transcript in HeLa cells. ASO1 is complementary to nt 7381–7430 of hMALAT1. ASO2 is complementary to nt 7331–7380 of hMALAT1. Both antisense oligonucleotides result in RNase H cleavage products that suggest the 3’ end of the long hMALAT1 transcript is at ~nt 7520. (F) Total and poly(A)+ RNA were isolated from EpH4-EV cells and probed by Northern blot analysis to show that the ~6.7-kb mMALAT1 transcript is present in the poly(A)+ fraction.
To roughly determine the location of the 3’ end of mMALAT1, we used multiple probes complementary to the region the mascRNA transcript originates from and flanking sequences (Figure 4A, Probes C–H). Using an antisense probe to the region immediately upstream of mascRNA, we were able to detect the long mMALAT1 transcript (Figure 4C, Probe C). In contrast, an antisense probe to the mascRNA region or any other probe after nt 6700 was unable to detect mMALAT1 (Figure 4C, Probes D–H). These Northern blots thus show that the abundant mMALAT1 transcript is only ~6.7-kb in length and ends immediately upstream of the mascRNA transcript. 3’ RACE PCR showed that the previously reported ~7-kb polyadenylated mMALAT1 transcript is present in cells (data not shown), but it is at a low level that is not detectable by Northern blot analysis using 10 µg of total RNA. Therefore, we conclude that mMALAT1 can generate two long transcripts (~6.7-kb and ~7-kb in mouse) that differ in the location of their 3’ ends. Using 5’ RACE PCR, we were only able to detect mMALAT1 transcripts starting at the previously identified start site (Hutchinson et al., 2007), suggesting that there are no other mMALAT1 promoters (data not shown).
Human MALAT1 has been reported to be a ~8.7-kb polyadenylated transcript that utilizes a promoter ~1.3-kb upstream of the promoter that the mouse ortholog utilizes (Hutchinson et al., 2007) (Supplementary Figure 9A). We determined that the hMALAT1 transcript observed on a Northern blot utilizes the same promoter as the mouse ortholog and is only ~7-kb in length (Supplemental Figure 9B). We then addressed whether the abundant hMALAT1 transcript ends immediately upstream of the mascRNA region similar to mMALAT1. Indeed, any probe complementary to regions 5’ of the small RNA was able to detect the long hMALAT1 transcript, whereas probes to the mascRNA region or sequences further downstream were unable to detect the long transcript by Northern blot analysis (Supplemental Figure 10).
The abundant MALAT1 transcript has a short poly(A) tail-like moiety
There are no canonical cleavage/polyadenylation signals located in the immediate vicinity of where the abundant MALAT1 transcript ends (Figure 4D and Supplementary Figure 4). Instead, we identified a conserved poly(A)-rich tract (16/18 adenines in mouse, 17/19 in human) encoded in the genome immediately upstream of the mascRNA transcript (Figure 4D). To better define the location of the 3’ end of the abundant MALAT1 transcript and determine if it has a canonical poly(A) tail, we carried out RNase H digestion followed by Northern analysis. Total RNA from HeLa cells was hybridized to specific DNA oligos, subjected to digestion by RNase H, and run on a small RNA Northern blot to allow high resolution mapping of the 3’ end of the hMALAT1 transcript (Figure 4E). In contrast to a normal polyadenylated RNA which would give a smear due to variable poly(A) tail lengths, the abundant hMALAT1 transcript yielded defined bands. These results indicated that MALAT1 3’ end cleavage occurs at a defined position and that there is not subsequent addition of nucleotides as occurs during polyadenylation.
To further confirm the RNase H-based mapping studies, we performed a modified form of 3’ RACE PCR in which an RNA adaptor was ligated to the 3’ ends of HeLa and EpH4-EV total RNA followed by reverse transcription using a primer complementary to the adaptor. Upon sequencing multiple clones, we determined that the RNA adaptor is able to be ligated to the 3’ end of the abundant MALAT1 transcript, confirming that there is a 3’ hydroxyl group, and that 3’ end cleavage occurs exactly after nt 7518 of hMALAT1 (nt 6690 of mMALAT1) (Figure 4D and Supplementary Figure 11). The 3’ cleavage site is immediately after the genomically encoded poly(A)-rich tract, indicating that 3’ end processing of the abundant MALAT1 transcript results in a short poly(A) tail-like moiety. When poly(A)+ RNA from mouse EpH4-EV cells was isolated, the abundant ~6.7-kb mMALAT1 transcript is indeed present in the poly(A)+ fraction (Figure 4F). No signal was detected when using a probe (6703–6752 Antisense) that can detect only the ~7-kb transcript, indicating that the observed poly(A)+ signal with the 6653–6702 Antisense probe is not due to the low level of the ~7-kb polyadenylated mMALAT1 transcript present in cells.
Despite having a short poly(A) tail-like moiety, MALAT1 is a stable non-coding RNA (Figure 3D, bottom panel). It has previously been shown that the poly(A) tail of a viral nuclear-retained RNA interacts with an upstream U-rich element to stabilize the RNA by inhibiting deadenylation (Conrad et al., 2006). We reasoned that the conserved U-rich motifs present upstream of mascRNA (Supplementary Figure 4) may play a similar role in stabilizing the poly(A) tail-like moiety of MALAT1. Indeed, upon mutating one of the U-rich motifs, MALAT1 was rapidly deadenylated in vitro when incubated with HeLa extracts under decay conditions (Ford et al., 1999), implicating the U-rich motifs in stabilizing the abundant MALAT1 transcript (Supplementary Figure 12).
3’ end processing of MALAT1 in the nucleus yields the mascRNA transcript
Although the mapping of the 3’ end of MALAT1 and the 5’ end of mascRNA to the same nucleotide position (Figure 4D) suggests that mascRNA may be generated via a novel form of a 3’ end processing reaction, it does not formally prove the precursor-product relationship between the MALAT1 primary transcript and mascRNA. For example, it is possible that although not detectable in our analysis, mascRNA may use a different RNA polymerase II promoter than that of the ~6.7-kb mMALAT1 transcript. To address this issue, we reasoned that transfection of antisense oligonucleotides (ASOs) complementary to mascRNA and/or flanking sequences may inhibit the RNA processing reaction by altering the local RNA secondary structure or by sterically blocking the binding of proteins involved in the cleavage reaction. If mascRNA is indeed a product of 3’ end processing of the ~6.7-kb mMALAT1 transcript, active ASOs must cause both (i) an increase in the expression of the ~7-kb polyadenylated mMALAT1 transcript (because cleavage upstream at ~6.7-kb will be blocked) and (ii) a corresponding decrease in mascRNA expression.
We designed ten 20-mer antisense oligonucleotides with a 2’-O-methoxy-ethyl ribose (MOE) phosphorothioate backbone that are complementary to mMALAT1 as well as an unrelated oligonucleotide which was used as a negative control (Figure 5A). This backbone modification imparts a very high affinity for targeted RNA, resistance to both exo- and endonucleases, and does not support cleavage of hybridized RNA by RNase H (Monia et al., 1993; McKay et al., 1999). The ASOs were transfected into EpH4-EV cells and total RNA was isolated 24 hours post transfection. To first determine if any ASOs caused an increase in the expression of the ~7-kb mMALAT1 transcript, a probe that can only detect the ~7-kb transcript was used (Figure 5A, Probe B). Little to no ~7-kb transcript was observed in EpH4-EV cells that were mock transfected or treated with the control oligonucleotide (Figure 5B, Lanes 1 and 2), consistent with our previous observation that the abundant mMALAT1 transcript is only ~6.7-kb in length.
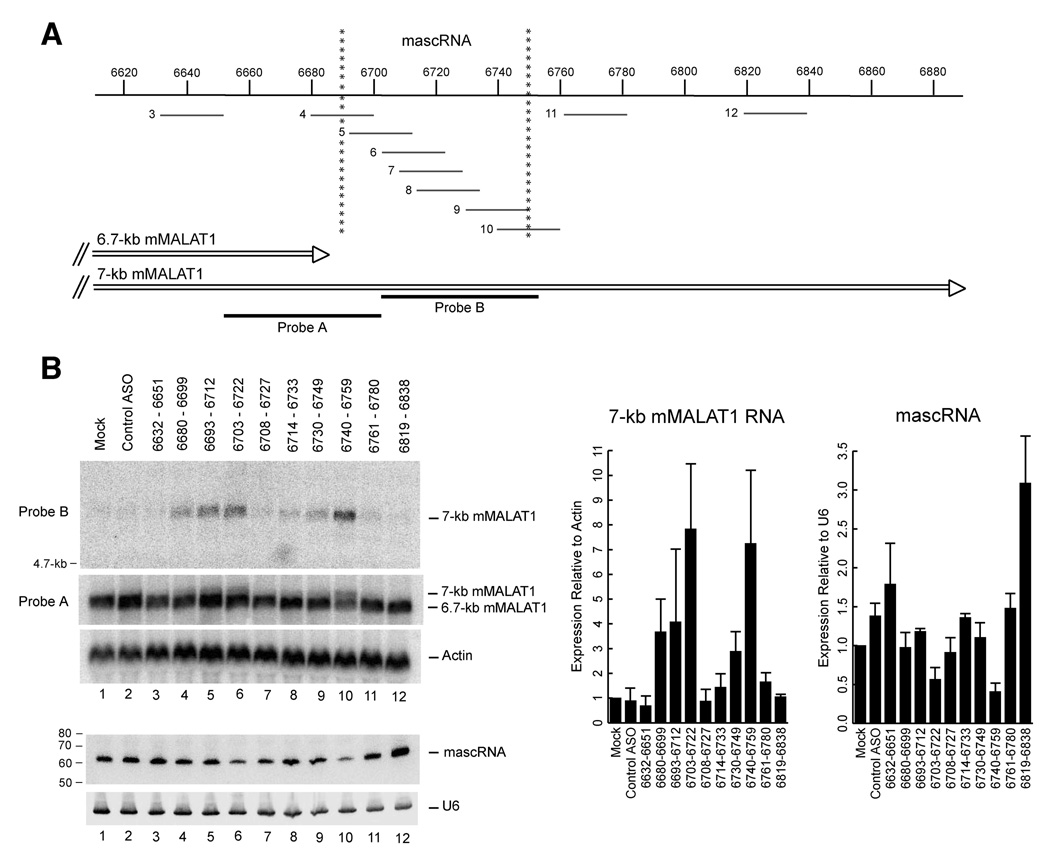
Figure 5. Antisense oligonucleotides inhibit mMALAT1 upstream 3’ end processing and knock-down mascRNA expression.
(A) Antisense oligonucleotides (ASOs) complementary to the mascRNA transcript and flanking regions were designed. Numbers to the left of each ASO represent lane number in Panel B. Probe A recognizes both the ~6.7-kb and ~7-kb mMALAT1 isoforms, whereas Probe B only recognizes the ~7-kb mMALAT1 isoform. (B) Northern blots were performed to determine the effect of ASO treatment on expression of the ~7-kb mMALAT1, ~6.7-kb mMALAT1, and mascRNA transcripts. The data in the bar graphs are shown as mean and standard deviation values of three independent transfections. Representative Northern blots are shown.
Numerous ASOs complementary to mMALAT1 caused an accumulation of the ~7-kb transcript, indicating that they were active in inhibiting 3’ end processing upstream at ~6.7-kb (Figure 5B). Two ASOs consistently caused expression of the ~7-kb transcript to increase greater than 7-fold over Mock (Figure 5B, Lanes 6 and 10). Perhaps not surprisingly, one of these most active ASOs (6640–6659, Lane 10) is complementary to the 3’ end of the mascRNA transcript and thus directly interfered with one of the cleavage sites (Figure 5A). The ASO complementary to the mascRNA 5’ cleavage site (6680–6699, Lane 4) was also weakly active, but this ASO is complementary to the poly(A)-rich tract, which has low sequence complexity and thus reduces efficacy. The other most active ASO (6703–6722, Lane 6) targets within mascRNA and thus probably disrupts its secondary structure. We verified that the observed band on the Northern blots was the ~7-kb mMALAT1 polyadenylated transcript by using additional probes complementary to the regions upstream and downstream of the cleavage/polyadenylation site at nt 6982 (Supplementary Figure 13). After stripping the blot and using a probe that can detect both the ~6.7 and ~7-kb transcripts (Figure 5A, Probe A), a doublet was observed (Figure 5B, Lanes 4, 5, 6, and 10) showing that the ASOs do not affect the total amount of mMALAT1 but change the ratio between the ~6.7 and ~7-kb isoforms.
Next, we tested if the ASOs caused a corresponding decrease in mascRNA expression. Indeed, the two ASOs that were most active in causing an accumulation of the ~7-kb mMALAT1 transcript also caused the largest decreases (50–60% knock-down) in mascRNA expression (Figure 5B, Lanes 6 and 10). Interestingly, one ASO (6819–6838, Lane 12) that was targeted greater than 60-nt downstream of the mascRNA region consistently caused an increase in mascRNA expression (Figure 5B). We propose that this ASO inhibited cleavage/polyadenylation downstream, resulting in a signal that caused upstream 3’ end processing to occur more often. The region this ASO targets may play a role in regulating the choice between downstream polyadenylation and upstream 3’ cleavage. In summary, we were able to identify several ASOs that caused an accumulation of the ~7-kb mMALAT1 transcript and a corresponding decrease in mascRNA expression, supporting a precursor-product relationship in which mascRNA is generated from 3’ end processing of the ~6.7-kb mMALAT1 transcript.
Because the ~6.7-kb mMALAT1 transcript localizes to nuclear speckles yet mascRNA localizes to the cytoplasm, two models are possible for where the 3’ end processing event occurs in the cell. First, processing could occur in the nucleus (either co-transcriptionally or post-transcriptionally at nuclear speckles) with the mascRNA transcript subsequently exported to the cytoplasm. Alternatively, MALAT1 could be transcribed in the nucleus as a ~7-kb polyadenylated transcript and exported to the cytoplasm where it is further processed to yield the small RNA. Following processing, the ~6.7-kb MALAT1 transcript would be imported back into the nucleus while mascRNA remained in the cytoplasm. To distinguish between these two possibilities, the control ASO or the ASO complementary to nt 6740–6759 were transfected into EpH4-EV cells. 24 hours post transfection, cells were fractionated to determine where the ~7-kb mMALAT1 transcript localized. If the processing reaction occurs in the nucleus, we expected the ~7-kb transcript to be in the nuclear fraction. In contrast, if the processing reaction occurs in the cytoplasm, we expected the ~7-kb transcript to accumulate in the cytoplasmic fraction upon transfection of the ASO complementary to nt 6740–6759. As shown in Figure 6A, the ~7-kb mMALAT1 transcript localizes specifically to the nucleus, suggesting that the processing reaction occurs in the nucleus followed by small RNA export to the cytoplasm. The ASOs had no effect on the localization of mascRNA, which remained exclusively cytoplasmic (Figure 6A).
MALAT1 is a substrate for RNase P and RNase Z
Because the processing event occurs in the nucleus and mascRNA resembles a tRNA, we reasoned that RNase P, which generates the 5’ termini of mature tRNAs and is known to be nuclear (Jacobson et al., 1997), may catalyze the endonucleolytic cleavage event that forms the 5’ end of mascRNA and the 3’ end of the abundant MALAT1 transcript. A ~170-nt region of mMALAT1 that includes mascRNA at the 3’ end (with or without CCA added) was cloned, transcribed in vitro, and employed for RNase P in vitro cleavage assays. The catalytic RNA component of E. coli RNase P (M1 RNA) was able to cleave mMALAT1 in vitro, but only when CCA was already present on the 3’ end (Figure 6B). Partially purified human RNase P from HeLa cells was also able to cleave mMALAT1 in vitro, regardless if CCA was already present on the 3’ end (Figure 6C, left panel). Using ligation-based small RNA cloning procedures, the in vitro RNase P cleavage site was mapped to the 5’ end of mascRNA, confirming that both in vitro systems accurately recapitulate in vivo MALAT1 processing (data not shown).
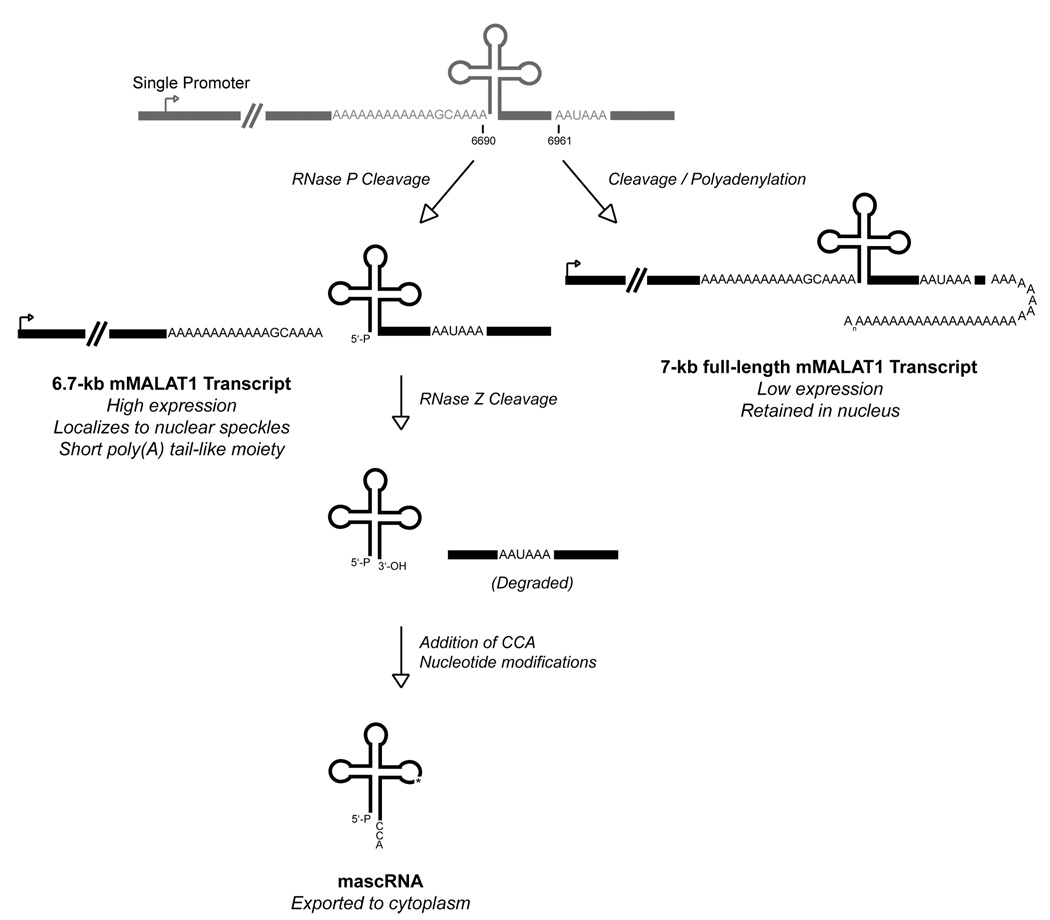
RNase Z is an endoribonuclease that generates the 3’ ends of eukaryotic tRNAs prior to addition of the CCA motif (reviewed in Vogel et al., 2005). Recombinant human RNase Z was able to accurately cleave mMALAT1 in vitro to generate the 3’ end of mascRNA (Figure 6D). To then determine whether RNase P or RNase Z cleavage occurs first, a mMALAT1 RNA substrate that includes both 5’ leader and 3’ trailer sequences was used for in vitro analysis. Human RNase P was able to accurately cleave the mMALAT1 6581–6822 substrate at the 5’ end of mascRNA, indicating that RNase P cleavage can precede RNase Z cleavage (Figure 6C, right panel). In contrast, RNase Z failed to accurately cut the mMALAT1 6581–6822 substrate at the 3’ end of mascRNA in vitro (data not shown), consistent with previous reports that RNase Z activity is inhibited by long 5’ extensions (de la Sierra-Gallay et al., 2005). We thus conclude that RNase P first cleaves to simultaneously generate the 3’ end of MALAT1 and the 5’ end of mascRNA, followed by RNase Z cleavage to generate the 3’ end of mascRNA, followed by CCA addition to the 3’ end of mascRNA (Figure 7).
Figure 7. MALAT1 is processed at its 3’ end to yield mascRNA.
Cleavage/polyadenylation can occur to yield a ~7-kb mMALAT1 transcript, but this represents a minor product that is expressed at a very low level in cells. MALAT1 is primarily processed via an upstream cleavage mechanism, which yields a mature ~6.7-kb mMALAT1 transcript with a short poly(A) tail-like moiety at its 3’ end. Endonucleolytic cleavage by RNase P simultaneously generates the 3’ end of the ~6.7-kb MALAT1 transcript and the 5’ end of mascRNA. mascRNA is then cleaved by RNase Z and subjected to CCA addition to generate the mature 61-nt mascRNA transcript, which is subsequently exported to the cytoplasm.
DISCUSSION
MALAT1 has been suggested to function as a long non-coding RNA that localizes to nuclear speckles and is misregulated in numerous cancers (Ji et al., 2003; Lin et al., 2006; Hutchinson et al., 2007). In the present study, we demonstrated that the nascent MALAT1 transcript is processed to yield two non-coding RNAs via a newly identified 3’ end processing mechanism. A highly conserved tRNA-like structure present near the 3’ end of the MALAT1 locus is recognized and cleaved by the tRNA processing machinery to yield a 61-nt small RNA. Unlike the stable nuclear-retained long MALAT1 transcript, the 61-nt mascRNA transcript localizes to the cytoplasm and has a relatively short half-life.
A single locus yields both a stable nuclear retained ncRNA and a small tRNA-like cytoplasmic RNA
Long RNA polymerase II transcripts have been previously shown to be precursors for multiple types of small RNAs, including snoRNAs (Kiss et al., 2006), microRNAs (Lee et al., 2004; Cai et al., 2004), and possibly piRNAs (Brennecke et al., 2007; Gunawardane et al., 2007). Here, we have shown that RNA polymerase II transcripts can also be precursors for tRNA-like small RNAs. Via a single cleavage event by RNase P, the 3’ end of the abundant MALAT1 transcript and the 5’ end of the mascRNA transcript are simultaneously generated (Figure 7). Interestingly, mascRNA is at the 3’ terminus of MALAT1, and tRNA-like structures at the 3’ ends of RNAs have been proposed to have served as genomic tags in the RNA world (Weiner and Maizels, 1987). After processing, the long MALAT1 transcript remains in the nucleus, while mascRNA is exported to the cytoplasm. This regulatory mechanism thus allows a single locus to produce two non-coding RNAs that localize to different subcellular compartments and have separate (possibly unrelated) functions.
mascRNA folds similar to a tRNA cloverleaf secondary structure, allowing it to be recognized by several members of the canonical tRNA processing machinery, such as RNase P, RNase Z, and the CCA-adding enzyme. Unlike tRNAs, however, mascRNA probably does not participate in transferring a specific amino acid to a growing polypeptide chain during protein synthesis because of its lack of a conserved anticodon loop and failure to be aminoacylated. Other tRNA mimics have been shown to be involved in viral replication (by being present at the 3’ end of the viral genomic RNA), translational regulation of gene expression, group I intron splicing, and tagging of abnormal proteins for proteolysis (reviewed in Giegé et al., 1998; Fechter et al., 2001). mascRNA does not likely participate in any of these processes as MALAT1 is not translated nor spliced, and mascRNA fails to resemble bacterial tmRNAs that tag abnormal proteins.
Although unclear what the exact function of the mascRNA transcript is, its biogenesis is a direct consequence of transcription of MALAT1. By being exported to the cytoplasm, mascRNA may function as a transmittable signal to allow components of the cytoplasm to sense that MALAT1 has been produced in the nucleus without having to export MALAT1. Consistent with this model, we determined that mascRNA has a relatively short half-life, thus allowing it to potentially serve a sensitive readout of MALAT1 promoter activity. In addition, there is emerging evidence that RNAs may function as signaling molecules, both within and between cells (Dinger et al., 2008). Alternatively, mascRNA may have a unique function limited to the cytoplasm. For example, because of its similarity to tRNAs, mascRNA may be involved in translational regulation by serving as a tRNA mimic.
The abundant MALAT1 transcript has a short poly(A) tail-like moiety
Practically all eukaryotic mRNAs, with the exception of histone mRNAs, are thought to terminate in a poly(A) tail that is approximately 200–250 residues in length in vertebrates (reviewed in Proudfoot, 2004; Colgan and Manley, 1997; Zhao et al., 1999). These poly(A) tracts are not encoded in the genome but are added to nascent transcripts in a two-step reaction that involves endonucleolytic cleavage followed by polyadenylation. It is, therefore, not surprising that there are conserved polyadenylation signals at the 3’ end of the MALAT1 locus that can be used to generate a long MALAT1 isoform (~7-kb in mouse). Unexpectedly, we found that this polyadenylated isoform is present at a very low level in cells. Instead, the 3’ end of MALAT1 is almost always generated several hundred nucleotides upstream of the polyadenylation signals (to yield a ~6.7-kb isoform in mouse) via a mechanism that is in stark contrast to classical cleavage/polyadenylation (Figure 7).
Within the nascent MALAT1 transcript, we identified a conserved genomically encoded short poly(A)-rich tract. Our data demonstrate that RNase P cleaves downstream of the poly(A)-rich tract to yield a mature MALAT1 transcript with a poly(A) tail-like moiety at its 3’ end. In contrast to classical cleavage/polyadenylation, no adenine residues are added subsequent to cleavage. Because MALAT1 is nuclear-retained and not translated, a long poly(A) tail may be unnecessary. Upon searching the mouse and human genomes for sequences similar to the 3’ end of MALAT1, we found one other instance. Perhaps not surprisingly, it is at the 3’ end of another long (>20-kb) non-coding transcript that is retained in the nucleus (Sunwoo et al., In preparation). All of the motifs we identified in MALAT1 (Supplementary Figure 4) are present at the 3’ end of the >20-kb transcript: the two upstream U-rich motifs, the poly(A)-rich tract, and a predicted tRNA-like structure. Similar to MALAT1, the nascent >20-kb transcript is cleaved immediately downstream of the poly(A)-rich motif by RNase P, such that the mature transcript has a short poly(A) tail-like moiety. These results suggest that short poly(A) tail-like moieties may be a common feature at the 3’ ends of at least some long nuclear-retained transcripts.
Short poly(A) tails (<20-nt) have previously been shown to be present at the 3’ ends of numerous mRNAs, especially in developing oocytes (Richter, 1999; Gu et al., 1999; Choi and Hagedorn, 2003). However, these short poly(A) tails are added by a classical cleavage/polyadenylation mechanism. In some cases, short poly(A) tails are just as effective as long poly(A) tails at ensuring RNA stability (Peng et al., 2005). It is unclear whether the poly(A)-rich tract at the 3’ end of the mature MALAT1 transcript functions in the same manner as these previously identified short poly(A) tails. The abundant MALAT1 transcript does, however, have a long half-life, suggesting that its poly(A) tail-like moiety and nearby U-rich motifs are sufficient to ensure RNA stability and resistance to exonucleases.
It is also unclear why the shorter MALAT1 isoform is expressed at such a higher level than the longer polyadenylated isoform. Regardless if the cleavage event occurs co-transcriptionally or post-transcriptionally, processing must be efficient to explain the drastic differences in steady-state levels between MALAT1 isoforms. If cleavage occurs co-transcriptionally, it may occur prior to the transcribing RNA polymerase reaching the AAUAAA and other polyadenylation signals several hundred nucleotides downstream. Rapid cleavage could be accomplished, for example, by having the cleavage factor(s) be present in the transcribing polymerase complex. On rare occasions when upstream cleavage is delayed (e.g., because of improper RNA folding), we propose that the downstream cleavage/polyadenylation signals ensure transcriptional termination and thus production of the longer polyadenylated MALAT1 transcript.
The present study has elucidated a novel mechanism by which a long nuclear-retained non-coding RNA is processed at its 3’ end to yield a short poly(A) tail-like moiety. RNase P cleavage not only generates the 3’ end of the abundant MALAT1 transcript, but also simultaneously generates the 5’ end of a 61-nt tRNA-like small RNA that is subsequently exported to the cytoplasm. Although cleavage/polyadenylation is thought to take place at the 3’ ends of almost all long eukaryotic RNA polymerase II transcripts, the 3’ end of the abundant MALAT1 transcript is not defined in this manner. Instead, cleavage several hundred nucleotides upstream of the polyadenylation signals is the preferred way by which the 3’ end of MALAT1 is generated. Our findings reveal a new paradigm for how the 3’ ends of certain RNA polymerase II transcripts are produced. In addition, our results suggest a general mechanism by which genetic loci are able to generate multiple non-coding RNA transcripts, each of which localize to different subcellular compartments.
EXPERIMENTAL PROCEDURES
Cell Culture
EpH4-EV, EpH4-A6, and HeLa cells were grown at 37 degrees, 5% CO2 in DMEM containing high glucose (Invitrogen, Carlsbad, California) supplemented with penicillin-streptomycin and 10% fetal bovine serum (Hyclone Laboratories, Logan, Utah). To inhibit RNA polymerase II transcription, HeLa cells were incubated with α-amanitin (20 or 50 µg/ml; Sigma, St. Louis, Missouri) for 2 to 12 hours at 37 degrees.
RNA Isolation
To isolate small RNAs under 200 nt, the mirVana miRNA Isolation Kit was used (Ambion, Austin, Texas). Trizol was used for all total RNA isolations as per the manufacturer’s instructions (Invitrogen, Carlsbad, California). To isolate poly(A)+ RNA, the Oligotex mRNA Mini Kit was used (Qiagen, Germantown, Maryland). Nuclear and cytoplasmic fractionation and RNA isolation were performed as described previously (Topisirovic et al., 2003). The FirstChoice Human Total RNA Survey Panel (Ambion, Austin, Texas) was used to examine expression of mascRNA in normal human tissues.
Northern blots
Small RNAs were separated by 15% denaturing polyacrylamide gel electrophoresis and electroblotted to Hybond N+ membrane (GE Healthcare, Piscataway, New Jersey). For all Northern blots using oligonucleotide probes, ULTRAhyb-Oligo Hybridization Buffer was used as per the manufacturer’s instructions (Ambion, Austin, Texas). Oligo probes were labeled with [γ-32P] ATP using T4 polynucleotide kinase (New England Biolabs, Ipswich, Massachusetts). For all Northern blots using random-labeled probes, NorthernMax Prehybridization/Hybridization Buffer was used (Ambion, Austin, Texas). Labeling of random-labeled probes was performed using the Prime-It RmT Random Primer Labeling Kit (Stratagene, La Jolla, California). Ambion Decade Markers were used as a ladder on small RNA Northern blots. Terminator 5’-Phosphate-Dependent Exonuclease was used as per the manufacturer’s instructions (Epicentre Biotechnologies, Madison, Wisconsin). Blots were visualized and quantified using the Fujifilm Life Science FLA-5100 imaging system. All oligonucleotide probe sequences and PCR primers are included in the Supplemental Experimental Procedures.
For RNase H treatments, 10 µg of total RNA was first mixed with 20 pmol of antisense oligo and heated to 65 degrees for 10 min. After allowing the antisense oligos to anneal by slow cooling, the RNA was treated with RNase H (New England Biolabs, Ipswich, Massachusetts) at 37 degrees for 30 min and then subjected to Northern blot analysis.
Ligation-based cloning
The 5’ end of mascRNA was cloned using the GeneRacer kit (Invitrogen, Carlsbad, California) according to the manufacturer’s instructions, except that CIP and TAP treatments were omitted. To clone the 3’ ends of the mascRNA and MALAT1 transcripts, the Modban oligo (IDT DNA Technologies, Coralville, Iowa) was ligated to the 3’ ends of total RNA similar to a previously described procedure (Lau et al., 2001).
Antisense Oligonucleotide Treatment
Uniform 2’-O-methoxy ethyl substituted oligonucleotides with a full phopshorothioate backbone and 5-methyl U and C as the pyrimidine heterocycle were prepared on solid support using standard phosphoramidite chemistry. All ASO sequences are included in the Supplemental Experimental Procedures. ASOs were administered at a final concentration of 100 nM to EpH4-EV cells using Lipofectamine 2000 as per the manufacturer’s instructions (Invitrogen, Carlsbad, California). Cells were incubated with a mixture of Lipofectamine 2000 and oligonucleotide in OptiMEM medium (Invitrogen) at 37 degrees, 5% CO2. After 5 hr, the transfection mixture was removed from the cells and replaced with fresh DMEM supplemented with 10% fetal bovine serum and incubated at 37 degrees, 5% CO2 for 18–20 hr.
In vitro Cleavage Assays
mMALAT1 RNA substrates were internally labeled using [α-32P] UTP, gel purified, and used at 10,000 cpm per cleavage reaction. Reactions with E. coli M1 RNA were incubated at 37 degrees for 1 hr in 50 mM Tris-HCl, pH 7.5, 100 mM MgCl2, and 100 mM NH4Cl. Partially purified HeLa RNase P was generously provided by Sidney Altman (Yale University). Reactions with human RNase P were incubated at 37 degrees for 1 hr in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 100 mM NH4Cl. Recombinant His-tagged tRNase ZL (delta30) was generously provided by Masayuki Nashimoto (Niigata University) and reactions were incubated at 37 degrees for 30 min in 10 mM Tris-HCl, pH 7.5, 1.5 mM DTT, and 10 mM MgCl2. All reactions were stopped by adding gel loading dye and samples were electrophoresed on 8% polyacrylamide/8 M urea gels.
Accession Numbers
Human and mouse mascRNA sequences have been deposited in GenBank under accession numbers FJ209302 and FJ209303, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Tom Gingeras, Greg Hannon, Adrian Krainer, Kannanganattu Prasanth, Hongjae Sunwoo, Oliver Tam, Jeff Wilusz, Carol Wilusz, Zhenyu Xuan, and members of the Spector laboratory for many helpful discussions. We would also like to thank Chris Black and Frank Bennett for help and discussions regarding the antisense oligonucleotides, Josh Nichols and Walt Lima for performing the sucrose gradients, Sidney Altman and Masayuki Nashimoto for providing tRNA processing reagents, and Philip Leder for providing the EpH4-EV and EpH4-A6 cell lines. We thank Delphine Bernard and Alain Bessis for bringing MALAT1 to our attention. J.E.W. is supported by a Beckman Graduate Studentship at the Watson School of Biological Sciences. Supported by a grant to D.L.S. from NIH/NIGMS (42694).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bass BL. RNA editing by adenosine deaminaes that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identifcation and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidotti G, Baubec T, Pauler F, Seidl C, Smrzka O, Stricker S, Yotova I, Barlow DP. The Air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb. Symp. Quant. Biol. 2004;69:55–66. doi: 10.1101/sqb.2004.69.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as a mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Choi YH, Hagedorn CH. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3’ poly(A) end phenotype. Proc. Natl. Acad. Sci. 2003;100:7033–7038. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Davis IJ, His B, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, et al. Cloning of an alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc. Nat. Acad. Sci. USA. 2003;100:6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Sierra-Gallay IL, Pellegrini O, Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. doi: 10.1038/nature03284. [DOI] [PubMed] [Google Scholar]
- Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem. Sci. 2006;31:526–532. doi: 10.1016/j.tibs.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Mercer TR, Mattick JS. RNA as extracellular signaling molecules. J. Mol. Endocrinol. 2008;40:151–159. doi: 10.1677/JME-07-0160. [DOI] [PubMed] [Google Scholar]
- Fantin VR, Berardi MJ, Scorrano L, Korsmeyer SJ, Leder P. A novel mitochondriotoxic small molecule that selectively inhibits tumor cell growth. Cancer Res. 2002;2:29–42. doi: 10.1016/s1535-6108(02)00082-x. [DOI] [PubMed] [Google Scholar]
- Fechter P, Rudinger-Thirion J, Florentz C, Giegé R. Novel features in the tRNA-like world of plant viral RNAs. Cell. Mol. Life Sci. 2001;58:1547–1561. doi: 10.1007/PL00000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R, Frugier M, Rudinger J. tRNA mimics. Curr. Opin. Struct. Biol. 1998;8:286–293. doi: 10.1016/s0959-440x(98)80060-2. [DOI] [PubMed] [Google Scholar]
- Gu H, Das Gupta J, Schoenberg DR. The poly(A)-limiting element is a conserved cis-acting sequence that regulates the length of poly(A) on nuclear pre-mRNAs. Proc. Natl. Acad. Sci. 1999;96:8943–8948. doi: 10.1073/pnas.96.16.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JD, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. Nuclear domains of the RNA subunit of RNase P. J. Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Ji P, Diedreichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF, van Kessel AG. Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum. Mol. Genet. 2003;12:1661–1669. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2006;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Masui O, Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harb. Symp. Quant. Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- Matthews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McKay RA, Miraglia LJ, Cummins LL, Owens SR, Sasmor H, Dean NM. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of protein kinase C-alpha expression. J. Biol. Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM. Evaluation of 2’-modified oligonucleotides containing 2’-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- Peng J, Murray EL, Schoenberg DR. The poly(A)-limiting element enhances mRNA accumulation by increasing the efficiency of pre-mRNA 3’ processing. RNA. 2005;11:958–965. doi: 10.1261/rna.2020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: An answer to the “genome” complexity conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3’ end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Rajaram V, Knezevich S, Bove KE, Perry A, Pfeifer JD. DNA sequence of the translocation breakpoints in undifferentiated embryonal sarcoma arising in mesenchymal hamartoma of the liver harboring the t(11;19)(q11;q13.4) translocation. Genes Chromosomes Cancer. 2007;46:508–513. doi: 10.1002/gcc.20437. [DOI] [PubMed] [Google Scholar]
- Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syzmanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: Noncoding RNAs. Biochim. Biophys. Acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Culjkovic B, Cohen N, Perez JM, Skrabanek L, Borden KL. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 2003;22:689–703. doi: 10.1093/emboj/cdg069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U, Lee C, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vogel A, Schilling O, Spath B, Marchfelder A. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol. Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- Weiner AM, Maizels N. tRNA-like structures tag the 3’ ends of genomic RNA molecules for replication: Implications for the origin of protein synthesis. Proc. Natl. Acad. Sci. 1987;84:7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3’ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.