Abstract
Arsenic (As) is a well-characterized human carcinogen but is generally not mutagenic. The evidence that As induces both loss of global DNA methylation and gene promoter DNA hypermethylation has suggested that epigenetic mechanisms may play an important role in As-induced carcinogenesis. In the present study, we examined the change in histone methylation by As exposure. In human lung carcinoma A549 cells, exposure to inorganic trivalent As (arsenite) increased H3K9 dimethylation (H3K9me2) and decreased H3K27 trimethylation (H3K27me3), both of which represent gene silencing marks, while increasing the global levels of the H3K4 trimethylation (H3K4me3), a gene-activating mark. The increase in H3K9me2 was mediated by an increase in the histone methyltransferase G9a protein and messenger RNA levels. We also observed strikingly significant altered histone modifications induced by very low-dose (0.1 μM) arsenite. Taken together, these results suggest a potential mechanism by which As induces carcinogenesis through the alteration of specific histone methylations that represent both gene silencing and activating marks. Furthermore, these marks are known to affect DNA methylation, and it is likely that arsenic's effect is not limited to histone modifications alone, but extends, perhaps by them, to DNA methylations as well. Future studies in our laboratory will address the genomic location of these silencing and activating marks using ChIP-on-chip technology.
Introduction
Arsenite is ubiquitously present in the environment from both natural and anthropogenic sources. Epidemiological studies have shown that chronic, low-dose exposure to arsenic (As) is associated with skin, bladder, lung, kidney and liver cancer (1–4). Moreover, occupational As exposures increase lung cancer risk in smokers and uranium miners (5,6). Inorganic As exists in the environment in two predominant forms, arsenite [As(III)] or arsenate [As(V)], the former being more bioreactive and responsible for cancer risk increases (7). The metabolism of arsenite requires the methyl donor, S-adenosyl-methionine, and converts arsenite into mono- and dimethylated metabolites (8).
Since As is not classified as a mutagen, and historically there has been a lack of animal models to study the As-induced cancers observed in humans, the mechanisms of As-mediated carcinogenesis remain unclear. Several hypotheses have been proposed to explain the mechanisms by which As causes cancer, including oxidative stress, inhibition of DNA repair, perturbation of signal transduction pathways and chromosomal aberrations (9–11). The disrupted transcriptional activity and aberrant gene expression caused by As exposure (12) have led to investigations of the possible epigenetic effects of As exposure. In particular, chronic exposure to low levels of As in a rat liver epithelial cell line increased global DNA hypomethylation by depleting S-adenosyl-methionine and subsequently induced the overexpression of a metallothionein gene (13). Consistently, As was found to be able to deplete S-adenosyl-methionine in human HaCaT keratinocytes, repress DNA methyltransferase genes' DNMT1 and DNMT3A transcription and cause DNA hypomethylation (14). As can also induce DNA hypermethylation in the promoter of tumor suppression genes and is associated with transcriptional silencing of genes. For example, in human bladder cancer, As exposure was associated with hypermethylation at the promoter of tumor suppressor genes, RASSF1A and RPSS3 (15). The As-exposed A/J mice had higher rates of methylation at CpG islands of tumor suppressor genes p16INK4a and RASSF1A and decreased expression of p16INK4a and RASSF1A compared with unexposed control mice (16). Similarly, hypermethylation and lower protein expression of death-associated protein kinase (DAPK) were detected in As-treated SV40-immortalized human uroepithelial cells (17). Clearly, these studies suggest that As disrupts DNA methylation. It is believed to have caused cell transformation and increased cancer risks.
In addition to DNA methylation, histone modifications at N-terminal tails that protrude from the nucleosomes are now recognized as crucial epigenetic marks that modulate gene expression and genomic function. At least eight histone modifications have been identified. Among them, acetylation, methylation and phosphorylation are the most commonly studied. These modifications act in combination to modulate chromatin structure and regulate gene expression (18). The ‘histone code’ theory proposes that the nature and extent of histone modifications can be read by different effector proteins, which subsequently translate the marks into transcriptionally silent or active states of gene expression (19). Lysine methylation can be mono-, di- or trimethylated. Generally, H3K9 di- and trimethylation and H3K27 di- and trimethylation are found in transcriptionally silent gene regulatory regions, whereas H3K4 di- and trimethylation, H3K36 di- and trimethylation and H3K79 dimethylation are associated with active transcription (20). However, the function of histone methylation continues to be poorly understood.
In an effort to understand the involvement of epigenetic mechanisms in metal-induced carcinogenesis, our group has reported that water-insoluble nickel compounds such as nickel sulfide (NiS) and nickel subsulfide (Ni3S2) cause transgene silencing by increasing DNA methylation and chromatin condensation in G12 cells in which the gpt transgene is located specifically near a heterochromatin region, but not in the G10 cells where the gpt transgene is inserted into a euchromatic region (21). The gpt gene silencing induced by nickel involved a loss of histone H3 and H4 acetylation and an increase of histone H3 lysine 9 methylation (22). A more recent study has shown that nickel ion increases global dimethylation of H3 lysine 9 and induces the gpt transgene silencing by inhibiting an Fe(II)- and 2-oxoglutarate-dependent H3K9 demethylase (23).
The goal of the present study was to investigate the alterations of histone methylations by arsenite. We used human lung carcinoma A549 cells in the study because it is relevant to the route of exposure. It is also a well-established model to study arsenite's carcinogenic effects (24) as well as the epigenetic effects of metals (23). We found that arsenite increases both the levels of the repressive mark dimethylated H3K9 and the activating mark trimethylated H3K4 and decreases the repressive mark trimethylated H3K27. The increase in dimethylated H3K9 was attributed to an increase in the histone methyltransferase G9a protein and messenger RNA (mRNA) levels. Low-dose arsenite at 0.1 μM was shown to effectively and unambiguously alter global histone methylation levels in A549 cells. Our study provides a plausible explanation of how altered histone modifications may contribute to arsenite-induced aberrant gene expression and carcinogenesis.
Materials and methods
Chemicals
NiCl2•6H2O and NaAsO2 were purchased from Sigma (St Louis, MO).
Cell culture
Cells were grown at 37°C in an incubator with a humidified atmosphere containing 5% CO2. Human lung carcinoma A549 cells were cultured in F-12K medium (Mediatech, Herndon, VA) and normal human bronchial epithelial BEAS-2B cells were grown in Dulbecco's modified Eagle's medium. All medium was supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin.
Histone extraction
Histones were extracted from the cells according to Chen et al. (23). Briefly, cells grown in 10 cm dishes were washed with ice-cold phosphate-buffered saline and lysed in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris–HCl, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM Na3VO4 and 1 mM NaF) supplemented with a protease inhibitor mixture (Roche Applied Sciences, Indianapolis, IN) for 10 min. The pellet was collected by centrifugation at 10 000g for 10 min. The pellet was washed once in 10 mM Tris–Cl and 37 mM ethylenediaminetetraacetic acid (pH 7.4) and resuspended in 200 μl 0.4 N H2SO4. After 1.5 h of incubation on ice, the supernatant was collected by centrifugation at 14 000g for 15 min and mixed with 1 ml cold acetone and kept at −20°C overnight. The histones were collected by centrifugation at 14 000g for 15 min. After one wash with acetone, the histones were air dried and suspended in 4 M urea.
Whole-cell protein extraction
Whole-cell lysates were extracted by incubating with ice-cold radioimmunoprecipitation assay buffer (50 mM Tris–HCl, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl and 1 mM ethylenediaminetetraacetic acid) supplemented with a protease inhibitor mixture (Roche Applied Sciences) for 20 min on ice, followed by centrifugation at 14 000g for 15 min. The supernatant was collected.
Western blot
The protein concentration was determined using Bio-Rad DC protein assay (Bio-Rad, Hercules, CA), and 5 μg histones were separated by 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel and transferred to polyvinylidene difluoride membranes (Bio-Rad). Immunoblotting was performed using monomethyl H3K9 (1:1000; Upstate Biotechnology, Lake Placid, NY), dimethyl H3K9 (1:2000; Upstate Biotechnology), trimethyl H3K9 (1:1000; Upstate Biotechnology), monomethyl H3K4 (1:5000; Upstate Biotechnology), dimethyl H3K4 (1:5000; Upstate Biotechnology), trimethyl H3K4 (1:5000; Abcam, Cambridge, MA), trimethyl H3K27 (1:1000; Upstate Biotechnology), dimethyl H3K36 (1:2000; Upstate Biotechnology), trimethyl H3K36 (1:2000; Upstate Biotechnology), G9a (1:500; Upstate Biotechnology) and JHDM2A (1:500; Aviva Systems Biology, San Diego, CA) primary antibodies and horseradish peroxidase-conjugated anti-rabbit secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The detection was accomplished by chemical fluorescence following an ECL Western blotting protocol (Amersham, Piscataway, NJ). After transfer to polyvinylidene difluoride membranes, the gels were stained with Bio-safe Coomassie stain (Bio-Rad) to assess the loading of histones. The immunoblots were scanned and analyzed using ImageJ software, and values were normalized to those obtained in the control samples.
Northern blot
Total RNA was extracted from cells immediately after exposure using Trizol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. Twenty micrograms of total RNA was separated on formaldehyde–agarose gels and transferred onto BrightStar-Plus positively charged nylon membranes (Ambion, Austin, TX). Polymerase chain reaction-amplified G9a complementary DNA fragment was used as probe. Probe labeling and subsequent hybridization were performed as described previously (22).
Immunofluorescence staining
The cells were cultured in Cultureslides (BD Falcon™, Bedford, MA) and exposed to various chemicals individually. At selected time intervals, cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.2% Triton X-100 for 5 min. Cells were then quenched with fresh 0.1% sodium borohydride for 5 min and blocked in blocking buffer (10% goat serum, 1% bovine serum albumin in phosphate-buffered saline). Monomethyl H3K9 (1:250; Upstate Biotechnology), dimethyl H3K9 (1:250; Upstate Biotechnology), trimethyl H3K9 (1:250; Upstate Biotechnology), trimethyl H3K27 (1:250; Upstate Biotechnology) and G9a (1:100; Upstate Biotechnology) primary antibodies diluted in blocking buffer were incubated overnight at 4°C. The cells were then incubated with Alexa Fluor 488-conjugated secondary antibody (Molecular Probes, Eugene, OR) for 1 h and mounted with ProLong Gold Anti-fade Reagent with 4′, 6-amidino-2-phenyl-indole (DAPI; Molecular Probes, Eugene, OR). The signals were visualized and captured using a fluorescent microscope (Model AX 70; Olympus, Melville, NY).
Dual-color immunofluorescence was performed by staining H3K9 dimethylation and H3K4 trimethylation sequentially. H3K9 dimethylation was stained with 1:1000 dimethylated H3K9 antibody (Abcam) and visualized by Alexa Fluor 594 secondary antibody (red; Molecular Probes). H3K4 trimethylation was stained with 1:5000 trimethylated H3K4 antibody (Upstate Biotechnology) and visualized with Alexa Fluor 488-conjugated secondary antibody (green; Molecular Probes). The image was scanned by a Leica TSC SP5 confocal microscope.
Results
Arsenite alters H3K9 methylation at the global level
H3K9 can be mono-, di- and trimethylated. Mono- and dimethylated H3K9 appear in euchromatin at the promoter regions of silenced genes, whereas trimethylated H3K9 is enriched at pericentric heterochromatin (25). To investigate the effects of arsenite on H3K9 methylation, A549 cells were treated with arsenite (2.5 and 5 μM) for 24 h. The doses of arsenite used in the study (1–10 μM) correspond to the exposure level (250–1000 μg/l) of As that causes diseases, and the time point was based on other studies (26–28). After arsenite exposure, histones were extracted from the cells, and the levels of H3K9 methylation were measured by western blot analysis with antibodies against mono-, di- or trimethylated H3K9. Twenty-four hours exposure to arsenite (2.5 and 5 μM) could increase the global levels of di- (Figure 1B) and trimethylated H3K9 (Figure 1C) significantly but could not increase the levels of monomethylated H3K9 (Figure 1A). Similar results were seen in immunofluorescent staining (Figure 2). Arsenite-induced increase of dimethylated H3K9 was also observed in normal human bronchial epithelial BEAS-2B cells (Figure 1D) much as we saw in A549 lung cancer cells. Thus, these phenomena are not a property only of a cancer cell line.
Fig. 1.
Arsenite exposure for 24 h increased di- (B) and tri- (C) but not monomethylated (A) H3K9 in A549 cells. Histones were extracted and immunoblotted with mono-, di- and trimethyl H3K9 antibodies. (D) As-induced dimethylated H3K9 level in BEAS-2B cells. BEAS-2B cells are more sensitive to arsenite treatment, and thus, the cells were only exposed to 1 or 2 μM arsenite for 24 h. H3K9me2 was detected by western blot analysis. Three independent experiments were performed, and shown are representative results that have been quantitated using imaging software described in the Materials and Methods. All gels were stained with Coomassie blue to assess histone loading in each lane. The numbers below the figure represent the relative intensity of the bands.
Fig. 2.
Low-dose arsenite at 0.1 μM increased mono- (A) and dimethylated (B) H3K9 but had no effect on trimethylated (C) H3K9 in A549 cells. Arsenite at higher doses (1, 5 or 10 μM) increased di- (B) and tri- (C) but not monomethylated (A) H3K9 in A549 cells. A549 cells were treated with different concentrations of arsenite for 24 h, followed by immunofluorescent staining with mono-, di- and trimethylated H3K9 antibodies as described in Materials and Methods.
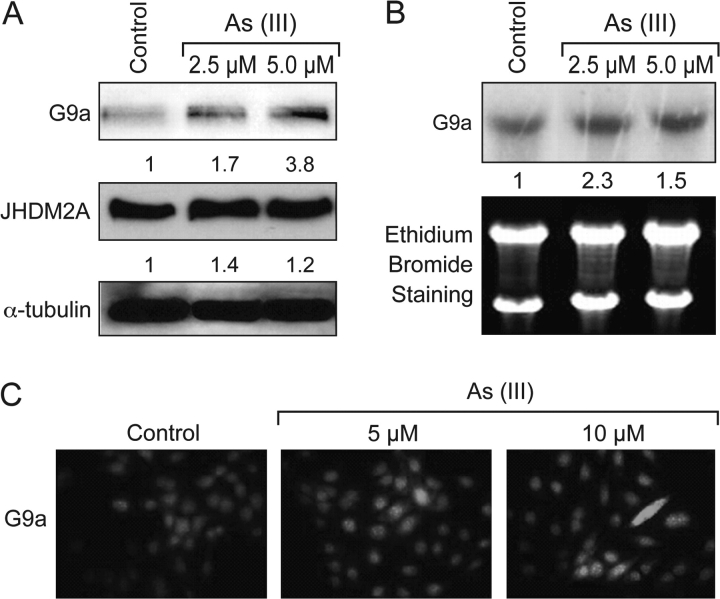
Arsenite increases histone methyltransferase G9a level in A549 cells
Dimethylated H3K9 is a hot spot for gene silencing and plays a pivotal role in carcinogenesis. Therefore, the possible mechanisms by which arsenite increased H3K9 dimethylation were investigated. Since G9a is a major histone methyltransferase responsible for H3K9 dimethylation, we first examined whether G9a played any role in arsenite-induced H3K9 dimethylation. A549 cells were treated with arsenite (2.5 and 5 μM), and the G9a protein levels were analyzed using a specific antibody against human G9a. As shown in Figure 3A, arsenite-treated cells exhibit significantly increased levels of G9a protein as compared with control, which could be responsible for the increased H3K9 dimethylation. In agreement with the western blotting results, an increase of G9a by arsenite treatment was also seen with immunofluorescent staining (Figure 3C).
Fig. 3.
Arsenite-induced histone methyltransferase G9a expression. A549 cells were treated with arsenite (2.5 and 5 μM) for 24 h. A set of representative results is shown from three independent experiments. (A) G9a and JHDM2A protein levels were analyzed by western blotting with antibodies as described in the Materials and Methods. The same membrane was reblotted with α-tubulin to assess protein loading. (B) Arsenite exposure increases G9a mRNA in A549 cells. Total RNA was extracted and subjected to northern blotting. The bottom panel shows the total RNA in the formaldehyde–agarose gels detected by ethidium bromide staining. The numbers below the figure represent the relative intensity of the bands. (C) Increased level of G9a in As-treated A549 cells. A549 cells were treated with arsenite at different concentrations for 24 h, followed by immunofluorescent staining with G9a antibody as described in Materials and Methods.
To address whether the increased G9a protein level was due to transcriptional regulation of the G9a gene by arsenite, total RNA was extracted from arsenite-treated A549 cells and analyzed for G9a mRNA by northern blotting. As shown in Figure 3B, 24 h exposure to arsenite (2.5 and 5 μM) induced a moderate increase in the G9a mRNA level in A549 cells, indicating As regulation of G9a activity at the transcriptional level.
The role of JHDM2A in arsenite-induced H3K9 dimethylation
The removal of H3K9 methyl groups is regulated by a family of JmjC domain-containing demethylases that require iron, oxygen, ascorbate and 2-oxoglutarate for enzymatic activity. JHDM2A (also named JMJD1A) has been identified as a demethylase specific for mono- and dimethylation of H3K9. To examine the role of JHDM2A in arsenite-induced H3K9 dimethylation, total protein lysates were isolated from A549 cells following arsenite exposure (2.5 and 5 μM) and subjected to western blotting using an antibody against human JHDM2A. In contrast to the increase of H3K9 dimethylation, JHDM2A protein levels did not appear to change after arsenite exposure (2.5 and 5 μM) (Figure 3A).
Arsenite alters H3K4 methylation at the global level
H3K4 methylation is also known to impact upon the transcriptional machinery and have the opposite effect on chromatin structure and gene regulation as that of H3K9 methylation. Similar to H3K9, the methylation of H3K4 can exist in three states: mono-, di- or trimethylated. Di- or trimethylated H3K4 occurs at euchromatic regions and is strongly correlated with transcriptional activation when found at promoter sites (29,30). We examined if arsenite affected H3K4 methylation. The different levels of H3K4 methylation were measured using antibodies specific for mono-, di- or trimethylated H3K4. Arsenite exposure (2.5 and 5 μM) in A549 cells resulted in an increase of di- and trimethylated H3K4, but a decrease of monomethylated H3K4 (Figure 4).
Fig. 4.
Arsenite increased di- (B) and tri- (C) but decreased monomethylated (A) H3K4 in A549 cells. A549 cells were treated with arsenite at different concentrations for 24 h. Histones were extracted and immunoblotted with mono-, di- and trimethyl H3K4 antibodies. Two independent experiments were performed; shown are representative results that have been quantitated using the imaging software described in the Materials and Methods. All gels were stained with Coomassie blue to assess the loading of histones in each lane. The numbers below the figure represent the relative intensity of the bands.
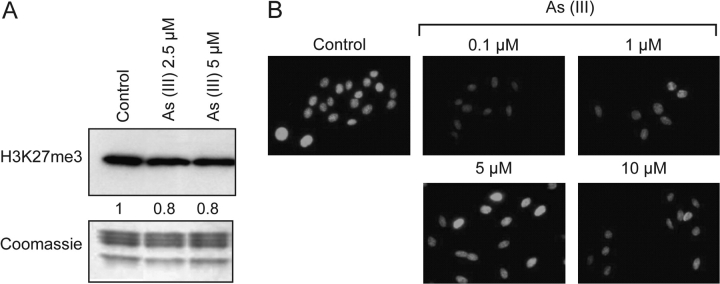
Arsenite alters H3K27 trimethylation at the global level
H3K27 trimethylation is an important mark in genomic silencing. To investigate the effects of arsenite on H3K27 trimethylation, A549 cells were treated with arsenite for 24 h. Histones were then extracted from the cells, and the level of H3K27 trimethylation was measured by western blot analysis with antibodies against trimethyl H3K27. Twenty-four hours exposure to arsenite (2.5 and 5 μM) decreased H3K27 trimethylation (Figure 5A). Consistent with these western blot results, immunofluorescent analysis showed that there was a significant decrease of H3K27 trimethylation levels in cells exposed to arsenite (0.1, 1, 5 and 10 μM) except at the 5 μM concentration (Figure 5B). This effect seemed to be biphasic with a strong reduction seen at 0.1 μM followed by a recovery at 5 μM and then a loss again at 10 μM. It should be noted that 0.1 μM arsenite is a low dose that does not result in much cytoxicity. Thus, the results of our experiments at these low levels are of much current interest, since higher doses are both considered toxic, and believed to render results whose interpretation is of doubtful relevance.
Fig. 5.
Decreased level of H3K27 trimethylation in arsenite-treated A549 cells. A549 cells were treated with arsenite at different concentrations for 24 h, and trimethylated H3K27 was detected by western blotting (A) and immunofluorescent staining (B). The western blot result was repeated three times and representative results are shown and quantitated. Coomassie blue was used to assess equal histone loading. The numbers below the figure represent the relative intensity of the bands.
Low doses of arsenite on other histone modifications
We next looked at the effect of arsenite at 0.1 μM on other histone modifications. Intriguingly, 0.1 μM arsenite increased di- and trimethylated H3K4 and H3K9, results already seen at higher doses. Arsenite at 0.1 μM also increased trimethylated H3K36 and decreased dimethylated H3K36 (Figure 6). Immonofluorescent staining shows that 0.1 μM arsenite increased mono- and dimethylated (Figure 2A and B, respectively) H3K9 but had no effect on trimethylated H3K9 (Figure 2C).
Fig. 6.
Arsenite (0.1 μM) alters various histone modifications. A549 cells were treated with 0.1 μM arsenite for 24 h. Histones were extracted and immunoblotted with di- and trimethyl H3K4, H3K9 and H3K36 antibodies. Two or three independent experiments were performed for each data set, and the bands were quantified using the imaging software described in the Materials and Methods. A composite graph was generated from the quantification of the bands.
Dual immunofluorescent staining of H3K9 dimethylation and H3K4 trimethylation
Since it is unlikely that the transcriptional silencing and activating signal appear simultaneously at the same gene, the fact that arsenite increased both gene repression and gene expression histone marks raised the possibility that these were located at different positions in chromatin. We therefore performed immunofluorescent staining of dimethylated H3K9 and trimethylated H3K4 and analyzed the distribution of these silencing and activating marks by confocal microscopy. As shown in Figure 7, though there is some overlapping of the dimethylated H3K9 (red) and trimethylated H3K4 (green) signal, after 24 h of 5 μM arsenite treatment, the dimethylated H3K9 was located primarily at the periphery of the nucleus, which represents the heterochromatin, whereas trimethylated H3K4 appeared mainly in the euchromatin region.
Fig. 7.
H3K9 dimethylation and H3K4 trimethylation were distributed differently in A549 cells. After 24 h of 5 μM arsenite exposure, cells were costained with dimethylated H3K9 (red) and trimethylated H3K4 (green) antibodies, and DNA was stained with DAPI (blue). The pictures were taken using a confocal microscope. The experiment was repeated two times.
Discussion
Previous findings that arsenite induces global DNA hypomethylation and DNA hypermethylation at the promoter region of tumor suppressor genes suggest that epigenetic mechanisms are involved in As-induced carcinogenesis. Our study is the first to report that arsenite disrupts the epigenetic machinery that regulates histone modifications. Exposure of cells to arsenite results in an increase of both di- and trimethylation of H3K9 and H3K4 as well as a decrease in H3K27 trimethylation.
The posttranslational modifications of histones play an important role in the transcriptional process. H3K9 dimethylation is a critical mark for cytosine methylation of DNA and is correlated with a loss of gene expression (31). A persistent increase of H3K9 dimethylation during extended arsenite exposure is likely to lead to the establishment of DNA methylation, which could affect the normal expression of genes and contribute to arsenite-induced silencing of tumor suppressor genes. This notion is in agreement with the finding that the exposure of human lung adenocarcinoma A549 cells to arsenite results in increased cytosine methylation in the p53 gene promoter (32). Recent epidemiological studies have confirmed that As significantly increases DNA hypermethylation in the promoter region of the p53 gene in a dose-responsive manner in humans (33).
It has now been realized that histone methylations are tightly controlled by both methylation and demethylation processes. Our results show that arsenite exposure increases histone methyltransferase G9a mRNA and protein levels in A549 cells. However, it is still not clear how arsenite increases G9a protein and mRNA. We also investigated whether arsenite may increase H3K9 dimethylation by interfering with the histone lysine demethylation process. Methyl groups on H3K9 can be removed by two families of demethylases, LSD1 and JmjC domain-containing demethylases. Although it was first identified as a demethylase specific for mono- and dimethylated H3K4, LSD1 demethylates H3K9 and stimulates androgen receptor-dependent gene transcription when it interacts with androgen receptor (34). H3K9 can also be demethylated by a family of JmjC-containing dioxygenases that require iron, oxygen, ascorbate and 2-oxoglutarate for their enzymatic activity. The family of demethylases includes JHDM2A (35), JMJD2A/JHDM3A (36,37), JMJD2B (38), JMJD2C/GASC1 (39) and JMJD2D (40). As a major dimethylated H3K9 demethylase, the role of JHDM2A in arsenite-induced H3K9 dimethylation was investigated. We had expected that JHDM2A protein level might decrease after arsenite treatment because H3K9 dimethylation was increased. Since JHDM2A protein did not appear to change during arsenite exposure, it suggested that JHDM2A protein levels were not likely to be responsible for the increase in H3K9 dimethylation.
Other than H3K9 methylation, H3K4, H3K27 and H3K36 methylations were also modified by arsenite. In this study, H3K4 di- and trimethylation were increased by arsenite, whereas H3K27 trimethylation was decreased. Interestingly, H3K36 dimethylation was significantly decreased, whereas H3K36 trimethylation was quite noticeably increased. These changes suggest that arsenite exposure can also activate gene transcription. The repression of H3K27 trimethylation by arsenite could be caused by the inhibition of the enzymatic methylation process. The enhancer of zeste homolog 2 (EZH2) is a methyltransferase known to trimethylate H3K27. It has been reported that arsenite is able to activate the phosphoinositide-3 kinase (PI3K/Akt) signaling pathway (41), and activated Akt phosphorylates EZH2 to suppress its enzymatic activity, resulting in a decrease of H3K27 trimethylation (42). Ubiquitously transcribed tetratricopeptide repeat, X chromosome, or UTX, which is a member of the JmjC-family of proteins, has recently been discovered to exhibit H3K27 di- and tridemethylase activity. Finally, when UTX was complexed to mixed-lineage leukemia and recruited to the HOX gene, H3K27 demethylation and an ordered cycle of H3K4 trimethylation were observed (43). This is consistent with our results that arsenite-treated A549 cells exhibit higher H3K4 trimethylation but lower H3K27 trimethylation.
Our dual staining confocal experiments demonstrated that H3K9 dimethylation and H3K4 trimethylation induced by arsenite were distributed at different regions in the nucleus, although there were some overlaps. These results showed that arsenite had different effects on histone modifications at different regions of chromatin, and through these epigenetic changes, the gene expression pattern on the whole genome may be altered when the repressing or activating chromatin marks are acquired.
In summary, this study provides the first evidence that arsenite disrupts epigenetic events by altering histone modifications, providing a novel view of As's potential mechanism of carcinogenesis. Further analysis on global and gene-specific changes by arsenite should give us more insights into the mechanisms of arsenite carcinogenesis.
Funding
National Institutes of Environmental Health Sciences (ES00260, ES014454, ES005512, ES010344 and T32-ES07324 to T.P.E.); National Cancer Institute (CA16087).
Acknowledgments
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- As
arsenic
- mRNA
messenger RNA
References
- 1.National research council. National Research Council Report: Arsenic in the Drinking Water. Washington, DC: National Academy Press; 2000. [Google Scholar]
- 2.Simeonova PP, et al. Mechanisms of arsenic carcinogenicity: genetic or epigenetic mechanisms? J. Environ. Pathol. Toxicol. Oncol. 2000;19:281–286. [PubMed] [Google Scholar]
- 3.Morales KH, et al. Risk of internal cancers from arsenic in drinking water. Environ. Health Perspect. 2000;108:655–661. doi: 10.1289/ehp.00108655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinmaus C, et al. Arsenic in drinking water and bladder cancer. Cancer Invest. 2000;18:174–182. doi: 10.3109/07357900009038249. [DOI] [PubMed] [Google Scholar]
- 5.Hertz-Picciotto I, et al. Synergism between occupational arsenic exposure and smoking in the induction of lung cancer. Epidemiology. 1992;3:23–31. doi: 10.1097/00001648-199201000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Kusiak RA, et al. Mortality from lung cancer in Ontario uranium miners. Br. J. Ind. Med. 1993;50:920–928. doi: 10.1136/oem.50.10.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barret JC, et al. Mechanisms of arsenic induced cell transformation. Biol. Trace Elem. Res. 1989;21:421–429. doi: 10.1007/BF02917284. [DOI] [PubMed] [Google Scholar]
- 8.Zakharyan RA, et al. Enzymatic methylation of arsenic compounds III. The marmoset and tamarin, but not the rhesus, monkeys are deficient in methyltransferases that methylate inorganic arsenic. Toxicol. Appl. Pharmacol. 1996;140:77–84. doi: 10.1006/taap.1996.0199. [DOI] [PubMed] [Google Scholar]
- 9.Hei TK, et al. Mutagenicity of arsenic in mammalian cells: role of reactive oxygen species. Proc. Natl Acad. Sci. USA. 1998;95:8103–8107. doi: 10.1073/pnas.95.14.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn S, et al. Arsenite retards DNA break rejoining by inhibiting DNA ligation. Mutagenesis. 1997;12:353–358. doi: 10.1093/mutage/12.5.353. [DOI] [PubMed] [Google Scholar]
- 11.Huang C, et al. Molecular mechanisms of arsenic carcinogenesis. Mol. Cell. Biochem. 2004;255:57–66. doi: 10.1023/b:mcbi.0000007261.04684.78. [DOI] [PubMed] [Google Scholar]
- 12.Kann S, et al. Butylhydroquinone protects cells genetically deficient in glutathione biosynthesis from arsenite-induced apoptosis without significantly changing their prooxidant status. Toxicol. Sci. 2005;87:365–384. doi: 10.1093/toxsci/kfi253. [DOI] [PubMed] [Google Scholar]
- 13.Zhao CQ, et al. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc. Natl Acad. Sci. USA. 1997;94:10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reichard JF, et al. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem. Biophys. Res. Commun. 2007;352:188–192. doi: 10.1016/j.bbrc.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsit CJ, et al. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–116. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 16.Cui X, et al. Chronic oral exposure to inorganic arsenate interferes with methylation status of p16INK4a and RASSF1A and induces lung cancer in A/J mice. Toxicol. Sci. 2006;91:372–381. doi: 10.1093/toxsci/kfj159. [DOI] [PubMed] [Google Scholar]
- 17.Chai CY, et al. Arsenic salts induced autophagic cell death and hypermethylation of DAPK promoter in SV-40 immortalized human uroepithelial cells. Toxicol. Lett. 2007;173:48–56. doi: 10.1016/j.toxlet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Strahl BD, et al. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein T, et al. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 20.Peterson CL, et al. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Lee YW, et al. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol. Cell. Biol. 1995;15:2547–2557. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Y, et al. Analysis of specific lysine histone H3 and H4 acetylation and methylation status in clones of cells with a gene silenced by nickel exposure. Toxicol. Appl. Pharmacol. 2003;190:272–277. doi: 10.1016/s0041-008x(03)00169-8. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, et al. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol. Cell. Biol. 2006;26:3728–3737. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mei N, et al. Acute arsenite-induced 8-hydroxyguanine is associated with inhibition of repair activity in cultured human cells. Biochem. Biophys. Res. Commun. 2002;297:924–930. doi: 10.1016/s0006-291x(02)02309-4. [DOI] [PubMed] [Google Scholar]
- 25.Rice JC, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 26.Kessel M, et al. Arsenic induces oxidative DNA damage in mammalian cells. Mol. Cell. Biochem. 2002;234:301–308. [PubMed] [Google Scholar]
- 27.Mei N, et al. Genetic predisposition to the cytotoxicity of arsenic: the role of DNA damage and ATM. FASEB J. 2003;17:2310–2312. doi: 10.1096/fj.02-0093fje. [DOI] [PubMed] [Google Scholar]
- 28.Sun X, et al. Effects of sodium arsenite on catalase activity, gene and protein expression in HaCaT cells. Toxicol. In Vitro. 2006;20:1139–1144. doi: 10.1016/j.tiv.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 30.Sims RJ, 3rd, et al. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 31.Jackson JP, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 32.Mass MJ, et al. Arsenic alters cytosine methylation patterns of the promoter of the tumor suppressor gene p53 in human lung cells: a model for a mechanism of carcinogenesis. Mutat. Res. 1997;386:263–277. doi: 10.1016/s1383-5742(97)00008-2. [DOI] [PubMed] [Google Scholar]
- 33.Chanda S, et al. DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol. Sci. 2006;89:431–437. doi: 10.1093/toxsci/kfj030. [DOI] [PubMed] [Google Scholar]
- 34.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 35.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 36.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 37.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 38.Fodor BD, et al. JMJD2B antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cloos PA, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 40.Shin S, et al. Diversity within the JMJD2 histone demethylase family. Biochem. Biophys. Res. Commun. 2006;353:973–977. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 41.Ouyang W, et al. Essential roles of PI-3K/Akt/IKKß/NFkappaB pathway in cyclin D1 induction by arsenite in JB6 Cl41 cells. Carcinogenesis. 2006;27:864–873. doi: 10.1093/carcin/bgi321. [DOI] [PubMed] [Google Scholar]
- 42.Cha TL, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 43.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]