Abstract
Peroxisome proliferator-activated receptor-alpha (PPARA) has been shown to increase fatty acid oxidation and decrease cytokine levels and has been implicated in insulin production. Genetic variants of PPARA have been associated with cardiovascular disease, obesity and type II diabetes mellitus. Although no research to date has investigated the possible link between PPARA and breast cancer, the function of this gene suggests that it could play a role in breast cancer development. Six PPARA polymorphisms were evaluated in association with incident breast cancer in a population-based case–control study (n = 1073 cases and n = 1112 controls) using unconditional logistic and multilevel regression and haplotype-based analyses. The odds of breast cancer were doubled among women with PPARA polymorphism rs4253760 (odds ratio = 1.97 for rare versus common homozygote alleles; 95% confidence interval: 1.14, 3.43). This association remained constant with the inclusion of all interrogated polymorphisms studied in hierarchical models. No additive interactions with body mass index or weight gain were present, but there was some evidence of interaction between PPARA variants and aspirin use, defined as use at least once per week for 6 months or longer. Fourteen haplotypes were imputed with frequencies >1% among postmenopausal women, but no statistically significant differences in haplotype frequencies between cases and controls were evident. Our results are the first to evaluate the relationship between PPARA and breast cancer incidence and suggest that replication in an independent cohort is warranted.
Introduction
The peroxisome proliferator-activated receptor (PPAR) family is composed of three nuclear hormone receptor genes: PPAR-gamma, peroxisome proliferator-activated receptor-alpha (PPARA) and PPAR-delta. In general, nuclear hormone receptors encode proteins that induce gene transcription by binding to the promoter region of a target gene. PPARs are activated when small lipophilic hormones (ligands) bind to a ligand-specific nuclear hormone receptor (1). PPARA ligands include palmitic acid, arachidonic acid and stearic acid in addition to compounds such as fenofibrate, bezafibrate and non-steroidal anti-inflammatory drugs (2,3).
PPARA is primarily expressed in organs with high fatty acid oxidation rates (3,4), such as the liver, kidney, heart, brown adipose tissue and, in small quantities, white adipose tissue (2). PPARA has also been found in human breast cancer cell lines, where its activation has been associated with increased proliferation (5). PPARA has been shown to regulate lipid metabolism by controlling the uptake and oxidation of fatty acids (3). This regulation can lead to an excess of free fatty acids, which may contribute to insulin resistance (6). Fibrates, which are PPARA agonists, have also been shown to reduce the expression of multiple cytokines, including interleukin-6, fibrinogen and C-reactive protein in humans (3).
Two PPARA isoforms have been characterized (PPARA1 and PPARA2). Both isoforms are expressed in human tissue (7,8). PPARA1 encodes the entire gene, whereas PPARA2 is truncated at exon 6. This truncation results in the absence of the ligand-binding domain in the gene’s protein and, consequently, prevents activation by the ligand. Therefore, all study inferences pertain to PPARA1 because of its protein’s known activity.
Given its role in energy homeostasis, it seems feasible that genetic variation in PPARA could influence disease incidence. The epidemiologic literature has focused on a functional polymorphism that results in a leucine to valine substitution at codon 162 of exon 5 (L162V, rs1800206) and a subsequent cytosine to guanine base change in the DNA-binding domain region of PPARA protein. This missense polymorphism has been shown to have a functional impact based on cotransfection assays; specifically, the V162 allele showed elevated ligand-dependent transcription activity compared with the L162 allele (9).
Although no research to date has investigated the possible link between PPARA and breast cancer, the biology and epidemiology of the gene suggest that it could play a role in breast cancer incidence. Genetic variants of PPARA have been linked to lipoprotein levels (10–12), cardiovascular disease (13–15), obesity (16,17) and type II diabetes (18–20). These conditions arise through etiologic mechanisms that may also be relevant to breast carcinogenesis, including inflammation and insulin resistance. Therefore, the goals of this study were to examine the association between PPARA genetic polymorphisms and breast cancer development using single polymorphism and haplotype-based approaches and semi-Bayesian techniques. Interactions with body mass index (BMI), weight gain, aspirin use and menopausal status were also explored.
Materials and methods
To evaluate the study aims, we utilized data and samples from the Long Island Breast Cancer Study Project (LIBCSP), a large population-based case–control study; details of the parent study population and data collection methods have been published previously (21). The study protocol was approved by the institutional review boards of participating institutions.
Study population
English-speaking women newly diagnosed with a primary in situ or invasive breast cancer between 1 August 1996 and 31 July 1997 were eligible to be study cases, if they were 20 years of age or older at diagnosis and were residents of Nassau or Suffolk counties on Long Island, New York. Cases were identified through daily contact with the 33 Long Island and New York City hospitals that served women with breast cancer in these two counties. Physician permission was obtained prior to case contact.
Controls were randomly selected from among English-speaking female residents of the same two Long Island counties and were frequency matched to the expected age distribution of case subjects by 5-year age group. Potentially eligible controls were identified by Waksberg’s method of random digit dialing (22) for women <65 years of age and by Health Care Finance Administration rosters for women ≥65 years of age.
Participants in the LIBCSP included 1508 (82.1%) eligible case women and 1556 (62.7%) eligible control women. Study subjects ranged in age from 24 to 98 years, and 93.8% of cases and 91.8% of controls were Caucasian, whereas 4.6% of cases and 5.5% of controls were African-American. Approximately 68% of cases and 67% of controls were postmenopausal (n = 1010 and 993, respectively).
Exposure assessment
Questionnaire.
Case–control interviews were administered by trained interviewers in respondents’ homes. Interviews took an average of 101 min to complete. In previous analyses, weight gain, particularly after age 50, was positively associated with postmenopausal breast cancer (23), and aspirin use was inversely associated with breast cancer among women of all ages (24). Other factors found to be associated with breast cancer in the LIBCSP have also been described previously (21).
Biologic specimens.
Among respondents who completed the interview, 73.0% of cases and 73.3% of controls donated a blood sample. DNA was isolated using methods described previously (25). Of the 1102 cases and 1141 controls who donated blood, 19 (1.7%) and 22 (1.9%), respectively, were later found to have insufficient DNA. Thus, there were a total of 1083 case and 1119 control samples available for genotyping. For analyses restricted to postmenopausal women, genotyping was available for 708 cases and 692 controls.
Genetic polymorphisms
Selection of tagging single-nucleotide polymorphisms.
Tag single-nucleotide polymorphisms (SNPs) were selected to represent comprehensive coverage of the PPARA gene by binning SNPs with a minor allele frequency >0.10 and an estimated minimum pairwise correlation of 0.80. For PPARA, sequence data were available for 23 European American and 24 African-American Coriell samples on the University of Washington-Fred Hutchinson Cancer Research Center Variation Discovery Resource website (http://pga.gs.washington.edu/). Haplotype-tagging SNPs were identified using the programs for genomic applications (PGA) LDSelect Program (26) run for European Americans only (given the relative racial homogeneity of the LIBCSP population). This program has been shown to select a maximally informative set of common SNPs that distinguish 80% of common haplotypes and is based on the r2 linkage disequilibrium statistic (27). When multiple possible tag SNPs were identified for a bin, SNPs located in the exon, promoter and 3′ untranslated regions were given priority. Because of its low prevalence but functional importance, L162V was forced into the program. Based on this program, 14 PPARA SNPs were identified for genotyping (Table I).
Table I.
PPARA haplotype-tagging SNPs identified using LDSelect and the PGA European American population as the reference panel
| rs# | SNP location | Base pair change (major > minor) | Minor allele frequencya |
| rs4253730 | Intron 3 | A > G | 0.182 |
| rs4253760b | Intron 6 | T > G | 0.196 |
| rs4253705 | Intron 2 | T > C | 0.190 |
| rs135543 | Intron 2 | G > A | 0.283 |
| rs135542b | Intron 2 | A > G | 0.205 |
| rs4253649 | Not validated | C > G | 0.370 |
| rs4253758 | Intron 6 | T > C | 0.217 |
| rs4253699b | Intron 2 | T > C | 0.182 |
| rs4253655 | Intron 2 | G > A | 0.143 |
| rs4253681 | Intron 2 | T > C | 0.136 |
| rs4253755b | Intron 5 | G > A | 0.130 |
| rs4253706 | Intron 2 | G > A | 0.119 |
| rs4253623b | Intron 2 | A > G | 0.109 |
| rs1800206b | Exon 5 Leu > Val | C > G | 0.022 |
Sequence data on 23 European American Coriell samples are available on the University of Washington-Fred Hutchinson Center Research Center Variation Discovery Resource (PGA) website (http://pga.gs.washington.edu/).
SNPs included in analyses.
Genotyping.
Genotyping was conducted at Columbia University, New York, NY. All LIBCSP DNA samples are available on 96-well master plates. Approximately 10% of the samples on each plate were duplicates, and laboratory personnel were blinded to case–control and duplicate status. Genotyping was carried out using iPLEX technology (Sequenom, San Diego, CA) on a MassARRAY Compact Analyzer. This multiplex method uses the mass of the incorporated nucleotide for identification of genotype. For SNPs that could not be multiplexed (rs4253623 and rs4253699), Taqman (Applied Biosystems, Foster City, CA) assays were developed and were run on an ABI 7500 Real-Time PCR system.
Kappa statistics were estimated to determine concordance between blinded repeat samples on each plate, and only those SNPs with a minimum kappa statistic of 0.90 were included in analyses. Six of the 14 identified tag SNPs for PPARA met this criterion (Table II).
Table II.
Summary table of ORs for association between six PPARA polymorphisms and breast cancer risk by menopausal status in LIBCSP
| Genotype | Genotypea | MAFb | Cases, n (%)c | Controls, n (%)c | P for trendd | ORe | 95% CI |
| All women | |||||||
| rs135542 | AA | 0.224 | 602 (59.4) | 634 (59.3) | 0.90 | 1.00 | |
| AG | 370 (36.5) | 392 (36.7) | 1.01 | 0.84, 1.21 | |||
| GG | 41 (4.1) | 43 (4.0) | 1.01 | 0.65, 1.57 | |||
| AG + GG | 411 (40.6) | 435 (40.7) | 1.01 | 0.85, 1.21 | |||
| Total | 1013 | 1069 | |||||
| rs1800206 (L162V) | CC (L/L) | 0.054 | 927 (89.7) | 973 (89.7) | 0.72 | 1.00 | |
| CG (L/V) | 100 (9.7) | 109 (10.1) | 0.97 | 0.73, 1.30 | |||
| GG (V/V) | 7 (0.7) | 3 (0.3) | 2.44 | 0.63, 9.50 | |||
| L/V + V/V | 107 (10.4) | 112 (10.3) | 1.01 | 0.76, 1.34 | |||
| Total | 1034 | 1085 | |||||
| rs4253623 | AA | 0.123 | 811 (77.5) | 849 (77.0) | 0.77 | 1.00 | |
| AG | 218 (20.8) | 236 (21.4) | 0.97 | 0.79, 1.19 | |||
| GG | 17 (1.6) | 17 (1.5) | 0.98 | 0.49, 1.93 | |||
| AG + GG | 235 (22.5) | 253 (23.0) | 0.97 | 0.79, 1.19 | |||
| Total | 1046 | 1102 | |||||
| rs4253699 | TT | 0.220 | 624 (60.0) | 671 (61.5) | 0.52 | 1.00 | |
| CT | 358 (34.4) | 362 (33.2) | 1.06 | 0.88, 1.27 | |||
| CC | 58 (5.6) | 59 (5.4) | 1.07 | 0.73, 1.57 | |||
| CT + CC | 416 (40.0) | 421 (38.6) | 1.06 | 0.89, 1.27 | |||
| Total | 1048 | 1092 | |||||
| rs4253755 | GG | 0.120 | 803 (76.5) | 845 (77.4) | 0.64 | 1.00 | |
| AG | 231 (22.0) | 231 (21.2) | 1.05 | 0.85, 1.29 | |||
| AA | 16 (1.5) | 16 (1.5) | 1.05 | 0.52, 2.12 | |||
| AG + AA | 247 (23.5) | 247 (22.6) | 1.05 | 0.86, 1.29 | |||
| Total | 1050 | 1092 | |||||
| rs4253760 | TT | 0.186 | 675 (66.2) | 713 (67.2) | 0.41 | 1.00 | |
| GT | 293 (28.7) | 302 (28.5) | 1.02 | 0.84, 1.24 | |||
| GG | 52 (5.1) | 46 (4.3) | 1.25 | 0.83, 1.87 | |||
| GT + GG | 345 (33.8) | 348 (32.8) | 1.05 | 0.88, 1.26 | |||
| Total | 1020 | 1061 | |||||
| Postmenopausal | |||||||
| rs135542 | AA | 0.220 | 395 (59.0) | 397 (60.0) | 0.61 | 1.00 | |
| AG | 247 (36.9) | 239 (36.1) | 1.07 | 0.85, 1.35 | |||
| GG | 27 (4.0) | 26 (3.9) | 1.04 | 0.59, 1.81 | |||
| AG + GG | 274 (41.0) | 265 (40.0) | 1.07 | 0.86, 1.33 | |||
| Total | 669 | 662 | |||||
| rs1800206 (L162V) | CC (L/L) | 0.052 | 610 (89.3) | 609 (89.7) | 0.51 | 1.00 | |
| CG (L/V) | 68 (10.0) | 69 (10.2) | 1.01 | 0.71, 1.44 | |||
| GG (V/V) | 5 (0.7) | 1 (0.2) | 5.07 | 0.59, 43.71 | |||
| L/V + V/V | 73 (10.7) | 70 (10.3) | 1.06 | 0.75, 1.51 | |||
| Total | 673 | 679 | |||||
| rs4253623 | AA | 0.132 | 533 (77.4) | 522 (75.9) | 0.43 | 1.00 | |
| AG | 145 (21.0) | 150 (21.8) | 0.96 | 0.74, 1.24 | |||
| GG | 11 (1.6) | 16 (2.3) | 0.66 | 0.30, 1.45 | |||
| AG + GG | 156 (22.6) | 166 (24.1) | 0.93 | 0.72, 1.20 | |||
| Total | 689 | 688 | |||||
| rs4253699 | TT | 0.217 | 411 (59.8) | 425 (62.2) | 0.62 | 1.00 | |
| CT | 243 (35.4) | 220 (32.2) | 1.13 | 0.90, 1.42 | |||
| CC | 33 (4.8) | 38 (5.6) | 0.92 | 0.56, 1.49 | |||
| CT + CC | 276 (40.2) | 258 (37.8) | 1.10 | 0.88, 1.37 | |||
| Total | 687 | 683 | |||||
| rs4253755 | GG | 0.113 | 532 (76.3) | 535 (78.8) | 0.29 | 1.00 | |
| AG | 153 (22.0) | 135 (19.9) | 1.12 | 0.86, 1.46 | |||
| AA | 12 (1.7) | 9 (1.3) | 1.36 | 0.57, 3.27 | |||
| AG + AA | 165 (23.7) | 144 (21.2) | 1.14 | 0.88, 1.47 | |||
| Total | 697 | 679 | |||||
| rs4253760 | TT | 0.168 | 441 (65.0) | 454 (69.6) | 0.02 | 1.00 | |
| GT | 199 (29.4) | 177 (27.2) | 1.14 | 0.90, 1.46 | |||
| GG | 38 (5.6) | 21 (3.2) | 1.97 | 1.14, 3.43 | |||
| GT + GG | 237 (35.0) | 198 (30.4) | 1.23 | 0.98, 1.55 | |||
| Total | 678 | 652 |
The combined heterozygotes and rare homozygotes were modeled separately and compared with common homozygotes.
Minor allele frequency (MAF) calculated among controls.
May not add up to 100.0 due to rounding.
P-value for trend was calculated by coding each genotype as 0, 1 or 2 based on the number of risk alleles.
Adjusted for age, measured in 5-year intervals.
Statistical methods
Hardy–Weinberg equilibrium was tested among controls to ensure that assumptions of parametric statistical tests were met using a permuted version of the exact test in SAS/Genetics version 9.1 (Cary, NC) (28,29). Pairwise linkage disequilibrium (30,31) for the six assayed SNPs was determined using Haploview 4.0 (32). Unconditional logistic regressions including individual PPARA SNPs and all SNPs together were conducted using SAS. Main gene effects were modeled by using the full genotype model and by combining heterozygotes and rare homozygotes. All models were adjusted for age, the frequency-matched variable, and common homozygotes were the reference group for all analyses. Linear trend tests for allelic effects were also performed by coding each genotype as 0, 1 or 2 based on the number of risk alleles. Separate genetic models were also used to estimate effects among postmenopausal women only. We hypothesized a priori that the gene’s effect would be most pronounced in postmenopausal women because of the relationship between obesity and breast cancer incidence in these women (33,34).
In addition to conventional unconditional logistic regression modeling, hierarchical modeling using a semi-Bayesian approach was performed among all women and among postmenopausal women only. SAS IML commands developed by Witte et al. (35) were used to fit the multilevel models. Hierarchical models assumed that all SNPs were exchangeable. The first hierarchical model specified a τ2, or prior residual variance, of 0.169, whereas the second hierarchical model assumed a τ2 of 0.345. A residual variance equal to 0.345 specifies that the odds ratio (OR) will fall within a 10-fold range with 95% confidence, whereas a τ2 of 0.169 specifies a 5-fold range.
Confounders were chosen a priori using directed acyclic graphs. Race (White/non-White), family history of breast cancer (yes/no) and Jewish ethnicity (Jewish/non-Jewish) were examined as potential confounders based on the directed acyclic graphs. Covariates that resulted in a 10% or greater change in the beta coefficient of the genotype effect estimate were considered confounders. Using this criterion, no confounders were identified.
Product interaction terms were added to conventional logistic and Bayesian models for gene variants and aspirin use, defined as use at least once per week for 6 months or longer (among all women), and for BMI at reference and weight gain since age 50 (among postmenopausal women). Due to small sample sizes, heterozygotes and rare homozygotes were combined for all interaction models. Interactions were considered for each SNP separately with adjustment for all other SNPs and age. Weight gain models were also adjusted for BMI at age 50 to account for the potential influence of body mass on weight gain. Interaction contrast ratios and 95% confidence intervals (CIs) were calculated to assess departures from additive risks (36).
Haplotype reconstruction was performed using an expectation maximization algorithm (37). Expectation maximization haplotype inference uses an individual’s genotype data to impute the probability of having a certain haplotype pair (38). Haplotype-specific ORs and 95% CIs were estimated relative to all other haplotypes for all women and for postmenopausal women only. All haplotype analyses were conducted using unconditional logistic regression in SAS/Genetics.
Results
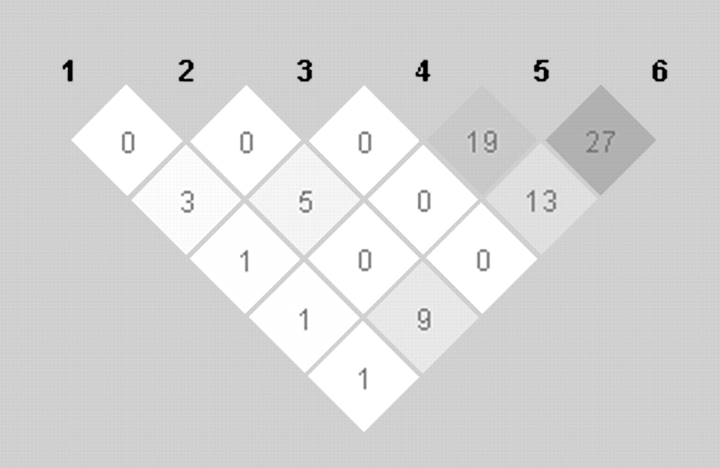
Six PPARA SNPs were included in analyses: rs135542, rs1800206, rs4253623, rs4253699, rs4253755 and rs4253760 (as ordered in Figure 1). All six SNPs were in Hardy–Weinberg equilibrium, and minor allele frequencies ranged from 5 to 22.4% (Table II). We found very low correlation between the PPARA polymorphisms (Figure 1), suggesting that the PGA population is an appropriate reference for tag SNP selection in the LIBCSP.
Fig. 1.
Linkage disequilibrium (r2) between six PPARA tag SNPs among LIBCSP controls.
As shown in Table II, PPARA polymorphism rs4253760 was associated with nearly a 100% relative increase in the risk of postmenopausal breast cancer (OR = 1.97 for rare versus common homozygotes; 95% CI: 1.14, 3.43) and showed evidence of linear trend (P = 0.02). Consistent findings were noted when all SNPs were assessed in one model and also for the hierarchical models (Table III).
Table III.
Odds Ratios (OR) and 95% Confidence Intervals (CI) for six PPARA polymorphisms and breast cancer among postmenopausal women in the LIBCSP for conventional and hierarchical models with varying priors
| SNP | Allelesa | Conventional b |
Conventional c |
Hierarchical d |
Hierarchical e |
||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | ||
| rs135542 | AG | 1.07 | 0.85, 1.35 | 1.05 | 0.82, 1.34 | 1.06 | 0.84, 1.34 | 1.06 | 0.83, 1.34 |
| GG | 1.04 | 0.59, 1.81 | 1.13 | 0.63, 2.02 | 1.13 | 0.69, 1.83 | 1.13 | 0.67, 1.91 | |
| rs1800206 | CG | 1.01 | 0.71, 1.44 | 0.93 | 0.60, 1.46 | 0.98 | 0.67, 1.43 | 0.96 | 0.64, 1.43 |
| GG | 5.07 | 0.59, 43.71 | 4.14 | 0.43, 39.79 | 1.29 | 0.58, 2.87 | 1.47 | 0.51, 4.27 | |
| rs4253623 | AG | 0.96 | 0.74, 1.24 | 1.15 | 0.86, 1.52 | 1.14 | 0.87, 1.49 | 1.14 | 0.86, 1.51 |
| GG | 0.66 | 0.30, 1.45 | 0.61 | 0.26, 1.43 | 0.83 | 0.46, 1.53 | 0.76 | 0.38, 1.52 | |
| rs4253699 | CT | 1.13 | 0.90, 1.42 | 0.97 | 0.73, 1.28 | 1.00 | 0.77, 1.29 | 0.99 | 0.76, 1.29 |
| CC | 0.92 | 0.56, 1.49 | 0.89 | 0.49, 1.62 | 1.02 | 0.64, 1.62 | 0.98 | 0.59, 1.64 | |
| rs4253755 | AG | 1.12 | 0.86, 1.46 | 1.06 | 0.74, 1.52 | 1.07 | 0.78, 1.47 | 1.06 | 0.76, 1.49 |
| AA | 1.36 | 0.57, 3.27 | 1.29 | 0.44, 3.76 | 1.20 | 0.62, 2.30 | 1.22 | 0.56, 2.69 | |
| rs4253760 | GT | 1.14 | 0.90, 1.46 | 1.17 | 0.85, 1.61 | 1.13 | 0.85, 1.49 | 1.15 | 0.85, 1.54 |
| GG | 1.97 | 1.14, 3.43 | 2.19 | 1.17, 4.13 | 1.71 | 1.05, 2.80 | 1.90 | 1.10, 3.28 | |
Minor alleles in bold.
Models included one SNP and age only.
All SNPs plus age in one model.
All SNPs exchangeable and prior τ2 = 0.169.
All SNPs exchangeable and prior τ2 = 0.345.
In general, hierarchical models produced more precise estimates compared with conventional analyses. This effect is most evident for the most unstable estimates, such as SNPs with a low prevalence in this population. For example, the OR for the association comparing rare to common homozygotes in rs1800206 was 4.14 (95% CI: 0.43, 39.79) among postmenopausal women, adjusting for all other PPARA SNPs (model labeled Conventionalb in Table III); using multilevel modeling, this OR decreased to 1.29 (95% CI: 0.58, 2.87; Hierarchicald).
Statistically significant departures from expectations for additive risks (α = 0.05) with aspirin use were estimated for rs135542 and rs4253699, but comparable departures from additivity were evident for all SNPs except rs1800206 (Table IV). Among postmenopausal women, there was no evidence of interaction by BMI at reference or by weight gain since age 50, both measured as three-level categorical variables, with no consistent pattern of elevated additive effects with increasing levels of obesity (results not shown).
Table IV.
ORs, interaction contrast ratios (ICRs) and 95% CI for six PPARA polymorphisms and breast cancer by aspirin use in all LIBCSP women under the dominant inheritance model
| Interactionab | ORc | 95% CI | ICR | 95% CI |
| rs135542 | ||||
| AA*(aspirin use)d | 0.92 | 0.69, 1.23 | ||
| (AG + GG)*(non-use) | 1.09 | 0.88, 1.36 | ||
| (AG + GG)*(aspirin use) | 0.56 | 0.39, 0.81 | −0.45 | −0.87, −0.04 |
| rs1800206 | ||||
| CC*(aspirin use) | 0.73 | 0.57, 0.92 | ||
| (CG + GG)*(non-use) | 1.05 | 0.71, 1.56 | ||
| (CG + GG)*(aspirin use) | 0.94 | 0.46, 1.89 | 0.16 | −0.59, 0.91 |
| rs4253623 | ||||
| AA*(aspirin use) | 0.64 | 0.49, 0.84 | ||
| (AG + GG)*(non-use) | 0.94 | 0.73, 1.22 | ||
| (AG + GG)*(aspirin use) | 1.07 | 0.71, 1.61 | 0.48 | −0.01, 0.98 |
| rs4263699 | ||||
| TT*(aspirin use) | 0.60 | 0.45, 0.80 | ||
| (CT + CC)*(non-use) | 0.85 | 0.66, 1.09 | ||
| (CT + CC)*(aspirin use) | 0.91 | 0.62, 1.32 | 0.46 | 0.08, 0.84 |
| rs4253755 | ||||
| GG*(aspirin use) | 0.65 | 0.50, 0.84 | ||
| (AG + AA)*(non-use) | 0.93 | 0.68, 1.27 | ||
| (AG + AA)*(aspirin use) | 1.08 | 0.67, 1.75 | 0.50 | −0.05, 1.05 |
| rs4253760 | ||||
| TT*(aspirin use) | 0.62 | 0.47, 0.82 | ||
| (GT + GG)*(non-use) | 0.92 | 0.70, 1.21 | ||
| (GT + GG)*(aspirin use) | 0.97 | 0.65, 1.44 | 0.42 | −0.01, 0.85 |
Interactions between aspirin use and each SNP were estimated using separate logistic regression models that also included main effect terms for the other PPARA SNPs. All ORs are estimated relative to a common referent group consisting of aspirin non-users with the referent genotype for each SNP. ICRs indicate departures from expectations for additive effects of aspirin use and each SNP on breast cancer risk.
Minor allele in bold.
Adjusted for age, measured in 5-year age intervals.
Aspirin use was defined as use at least once per week for 6 months or longer.
The haplotype reconstruction and analysis created 12 haplotypes with frequencies >1% from the six SNPs analyzed in all women and 14 haplotypes in the postmenopausal women. Haplotype distributions were similar between cases and controls among all women (data not shown) and among postmenopausal women only (Table V). Due to the low prevalence of selected haplotypes in this population, effect estimates were imprecise, particularly those for associations among postmenopausal women only. For example, although the OR for haplotype 10 among postmenopausal women (n = 30 cases and 11 controls) was elevated relative to all other haplotypes, the 5-fold width of the CI indicated substantial imprecision (OR = 5.02; 95% CI: 1.45, 17.39).
Table V.
ORs and frequencies by case–control status for 14 PPARA haplotypes relative to all other haplotypes among postmenopausal women in LIBCSP
| Haplotype numbera | rs135542 | rs1800206 | rs4253623 | rs4253699 | rs4253755 | rs4253760 | Control n (%) | Case n (%) | OR (95% CI)b |
| 1 | A | C | A | C | A | G | 55 (5.5) | 53 (5.3) | 1.01 (0.49, 2.09) |
| 2 | A | C | A | C | A | T | 18 (1.9) | 32 (3.2) | 2.03 (0.71, 5.82) |
| 3 | A | C | A | C | G | G | 18 (1.8) | 14 (1.4) | 0.97 (0.23, 4.18) |
| 4 | A | C | A | C | G | T | 55 (5.5) | 53 (5.2) | 0.89 (0.40, 1.99) |
| 5 | A | C | A | T | A | G | 25 (2.5) | 32 (3.2) | 1.64 (0.61, 4.46) |
| 6 | A | C | A | T | G | G | 19 (1.9) | 36 (3.6) | 2.83 (0.93, 8.65) |
| 7 | A | C | A | T | G | T | 414 (41.7) | 396 (39.2) | 0.92 (0.70, 1.20) |
| 8 | A | C | G | C | G | T | 13 (1.5) | 10 (1.0) | 0.77 (0.12, 4.91) |
| 9 | A | C | G | T | G | T | 104 (10.5) | 103 (10.2) | 0.97 (0.59, 1.59) |
| 10 | A | G | A | C | G | G | 11 (1.1) | 30 (3.0) | 5.02 (1.45, 17.39) |
| 11 | A | G | A | T | G | G | 13 (1.3) | 8 (0.8) | 0.47 (0.08, 2.83) |
| 12 | G | C | A | C | G | T | 15 (1.5) | 8 (0.8) | 0.42 (0.05, 3.55) |
| 13 | G | C | A | T | G | G | 11 (1.1) | 11 (1.1) | 1.07 (0.13, 8.67) |
| 14 | G | C | A | T | G | T | 173 (17.4) | 182 (18.0) | 1.19 (0.79, 1.80) |
Haplotypes with frequency ≥0.01, minor alleles in bold.
Adjusted for age, measured in 5-year age intervals.
Discussion
We found that the PPARA genetic polymorphism rs4253760 was associated with a 2-fold increase in the odds of postmenopausal breast cancer. This association persisted in the hierarchical models that were adjusted for the other five PPARA SNPs. This finding is consistent with our prior expectation of a more pronounced effect in postmenopausal women. rs4253760 is located in intron 6 and tags 10 SNPs (minor allele frequency > 10%) based on LDSelect and PGA data. This polymorphism may be correlated with the causal SNP in the PPARA gene, although it does not tag any non-synonymous coding polymorphisms and the haplotype analyses with the rare variant of rs4253760 are not supportive of a causal effect.
Our results also suggest that PPARA genetic polymorphisms may modify protective effects of aspirin use. In the LIBCSP population, aspirin use was associated with a 20% reduction in the odds of breast cancer (24) overall. In this study, inverse associations with non-steroidal anti-inflammatory drug use were evident only among women with referent genotypes of rs4253623, rs4263699, rs4253755 and rs4253760 or the homozygous variant genotype for rs135542.
Despite the additional computational complexity, hierarchical modeling offers two advantages over conventional logistic regression: (i) the shrinkage estimation method reduces type II error rate (39,40) and (ii) it reduces instability in the effect estimates due to multiple correlated exposures, such as multiple SNPs in the same model (40). For this study, two different τ2 values were considered: 0.169 and 0.345. The difference between the hierarchical and conventional models was most apparent for the fully specified conventional model, where each genotype was modeled simultaneously. Here, the τ2 0.169 models consistently produced estimates that were closer to the null and more precise than either the τ2 0.345 hierarchical models or the conventional logistical models.
It has become increasingly apparent that studies of a single polymorphism are not necessarily the best approach to identify deleterious variants (41). Haplotypes that use tag SNPs selected from bins comprehensively assess variation over the entire gene to identify cis–cis interactions, where tag SNPs are interacting on the same chromosome to increase disease risk. Our study implicates haplotype 10 in breast cancer incidence, although the low precision of the estimate makes interpretation difficult.
One limitation of these data is the low concordance of eight tag SNPs with their blinded repeats, which prevented us from including them in our analyses. Five of the requested SNPs were found in repeat regions of the PPARA gene and, therefore, would have been difficult to genotype successfully. The low concordances found for rs4253730, rs4253655 and rs135543 with their repeats are not easily explained and could be the result of genotyping error. Using 95% concordance rates rather than kappa statistics does not alter the number of SNPs eligible for inclusion. Two of the unexplained genotyping failures (rs135543 and rs4253655), however, have concordance rates of 92.0%.
Although these omissions reduced gene coverage, this study still provides more coverage than previous studies and analyses a well-characterized population. Only two studies have examined PPARA haplotypes to date, none of which examined cancer outcomes or included more than three SNPs (13,19). Flavell et al. (19) investigated three SNPs in connection to age of onset and progression of type II diabetes mellitus, whereas Doney et al. (13) explored the link between myocardial infarction risk among individuals with type II diabetes mellitus and two PPARA polymorphisms. Thus, even with reduced gene coverage, this study advances scientific knowledge of PPARA and its role in breast cancer incidence.
Reported weight at age 50 and other past exposures may be subjected to recall bias and non-differential misclassification since disease diagnosis has occurred before exposure ascertainment. This study examined body size as a potential modifier of the gene’s effect; therefore, for recall bias to be present, cases and controls would have to recall their weight differentially by PPARA genotype. Since women are unlikely to know their PPARA status, recall bias is unlikely to play a role in these analyses.
Although a benefit of our study is that it is population based, only 73% of cases and controls donated blood. However, the distributions of risk factors for breast cancer among all women in the study were comparable with those of women who donated blood (data not shown). Lastly, the ethnic distribution of our Long Island subjects differs from that of the American population as a whole, with 92.7% of our study population being Caucasian; thus, results from this study may not be readily applied to the USA population in general. Although the underlying prevalence of specific alleles and exposures may vary with ethnicity, it seems unlikely that the biological relations with breast cancer among participants in this study will differ from women in general. In fact, as expected, the minor allele frequencies differed only slightly between the PGA European American population and our Long Island women, with the greatest difference noted for rs4253755 (18.2 versus 22.0%, respectively). This difference is most likely due to PGA’s small sample (n = 23) for determining the allele frequencies in European Americans.
Lack of reproducibility among genetic studies has called into question the utility of association studies in genetic epidemiology (42). This is the first study to examine the association between PPARA polymorphisms and breast cancer development. While our use of semi-Bayesian techniques minimizes many issues inherent with small cell sizes, such as large but imprecise effects and low P-values, further replication is needed to confirm our findings.
In summary, although PPARA has been studied in cardiovascular disease, no studies have examined PPARA in connection to breast cancer. This study is the first to investigate its relationship to breast cancer risk and interaction by aspirin use and obesity in a large population-based sample. This study suggests that variants of PPARA modify the association between breast cancer and aspirin use. Research investigating PPARA’s possible involvement in the inflammatory pathway is needed. We found that among postmenopausal women carrying the homozygous alleles for rs4253760, the odds of breast cancer was nearly doubled. These findings warrant further investigation in an independent cohort.
Funding
National Cancer Institute and National Institute of Environmental Health Sciences (5T32CA72319, UO1CA/ES66572, P30ES009089 and P30ES10126); Breast Cancer Research Foundation.
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BMI
body mass index
- CI
confidence interval
- LIBCSP
Long Island Breast Cancer Study Project
- OR
odds ratio
- PGA
programs for genomic applications
- PPAR
peroxisome proliferator-activated receptor
- PPARA
peroxisome proliferator-activated receptor-alpha
- SNP
single-nucleotide polymorphism
References
- 1.Strachan T, et al. Human Molecular Genetics 3. London: Garland Science; 2004. [Google Scholar]
- 2.Kota BP, et al. An overview on biological mechanisms of PPARs. Pharmacol. Res. 2005;51:85–94. doi: 10.1016/j.phrs.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 3.van Raalte DH, et al. Peroxisome proliferator-activated receptor (PPAR)-alpha: a pharmacological target with a promising future. Pharm. Res. 2004;21:1531–1538. doi: 10.1023/b:pham.0000041444.06122.8d. [DOI] [PubMed] [Google Scholar]
- 4.Kersten S. Peroxisome proliferator activated receptors and obesity. Eur. J. Pharmacol. 2002;440:223–234. doi: 10.1016/s0014-2999(02)01431-0. [DOI] [PubMed] [Google Scholar]
- 5.Suchanek KM, et al. Peroxisome proliferator-activated receptor alpha in the human breast cancer cell lines MCF-7 and MDA-MB-231. Mol. Carcinog. 2002;34:165–171. doi: 10.1002/mc.10061. [DOI] [PubMed] [Google Scholar]
- 6.Evans RM, et al. PPARs and the complex journey to obesity. Nat. Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- 7.Gervois P, et al. A truncated human peroxisome proliferator-activated receptor alpha splice variant with dominant negative activity. Mol. Endocrinol. 1999;13:1535–1549. doi: 10.1210/mend.13.9.0341. [DOI] [PubMed] [Google Scholar]
- 8.Hanselman JC, et al. Expression of the mRNA encoding truncated PPAR alpha does not correlate with hepatic insensitivity to peroxisome proliferators. Mol. Cell. Biochem. 2001;217:91–97. doi: 10.1023/a:1007248007372. [DOI] [PubMed] [Google Scholar]
- 9.Flavell DM, et al. Variation in the PPARalpha gene is associated with altered function in vitro and plasma lipid concentrations in Type II diabetic subjects. Diabetologia. 2000;43:673–680. doi: 10.1007/s001250051357. [DOI] [PubMed] [Google Scholar]
- 10.Tai ES, et al. Association between the PPARA L162V polymorphism and plasma lipid levels: the Framingham Offspring Study. Arterioscler. Thromb. Vasc. Biol. 2002;22:805–810. doi: 10.1161/01.atv.0000012302.11991.42. [DOI] [PubMed] [Google Scholar]
- 11.Klos KL, et al. Consistent effects of genes involved in reverse cholesterol transport on plasma lipid and apolipoprotein levels in CARDIA participants. Arterioscler. Thromb. Vasc. Biol. 2006;26:1828–1836. doi: 10.1161/01.ATV.0000231523.19199.45. [DOI] [PubMed] [Google Scholar]
- 12.Chan E, et al. The V227A polymorphism at the PPARA locus is associated with serum lipid concentrations and modulates the association between dietary polyunsaturated fatty acid intake and serum high density lipoprotein concentrations in Chinese women. Atherosclerosis. 2006;187:309–315. doi: 10.1016/j.atherosclerosis.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Doney AS, et al. Association of common variation in the PPARA gene with incident myocardial infarction in individuals with type 2 diabetes: a Go-DARTS study. Nucl. Recept. 2005;3:4. doi: 10.1186/1478-1336-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai ES, et al. The L162V polymorphism at the peroxisome proliferator activated receptor alpha locus modulates the risk of cardiovascular events associated with insulin resistance and diabetes mellitus: the Veterans Affairs HDL Intervention Trial (VA-HIT) Atherosclerosis. 2006;187:153–160. doi: 10.1016/j.atherosclerosis.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 15.Balcerzyk A, et al. Synergistic effect between polymorphisms of PPARA and ABCA1 genes on the premature coronary artery disease. Acta Cardiol. 2007;62:233–238. doi: 10.2143/ac.62.3.2020810. [DOI] [PubMed] [Google Scholar]
- 16.Bosse Y, et al. The peroxisome proliferator-activated receptor alpha L162V mutation is associated with reduced adiposity. Obes. Res. 2003;11:809–816. doi: 10.1038/oby.2003.112. [DOI] [PubMed] [Google Scholar]
- 17.Evans D, et al. A polymorphism, L162V, in the peroxisome proliferator-activated receptor alpha (PPARalpha) gene is associated with lower body mass index in patients with non-insulin-dependent diabetes mellitus. J. Mol. Med. 2001;79:198–204. doi: 10.1007/s001090100189. [DOI] [PubMed] [Google Scholar]
- 18.Andrulionyte L, et al. Single nucleotide polymorphisms of the peroxisome proliferator-activated receptor-alpha gene (PPARA) influence the conversion from impaired glucose tolerance to type 2 diabetes: the STOP-NIDDM trial. Diabetes. 2007;56:1181–1186. doi: 10.2337/db06-1110. [DOI] [PubMed] [Google Scholar]
- 19.Flavell DM, et al. Peroxisome proliferator-activated receptor alpha gene variation influences age of onset and progression of type 2 diabetes. Diabetes. 2005;54:582–586. doi: 10.2337/diabetes.54.2.582. [DOI] [PubMed] [Google Scholar]
- 20.Bosse Y, et al. Combined effects of PPARgamma2 P12A and PPARalpha L162V polymorphisms on glucose and insulin homeostasis: the Quebec Family Study. J. Hum. Genet. 2003;48:614–621. doi: 10.1007/s10038-003-0087-2. [DOI] [PubMed] [Google Scholar]
- 21.Gammon MD, et al. The Long Island Breast Cancer Study Project: description of a multi-institutional collaboration to identify environmental risk factors for breast cancer. Breast Cancer Res. Treat. 2002;74:235–254. doi: 10.1023/a:1016387020854. [DOI] [PubMed] [Google Scholar]
- 22.Waksberg J. Sampling methods for random digit dialing. J. Am. Stat. Assoc. 1978;73:40–46. [Google Scholar]
- 23.Eng S, et al. Body size changes in relation to postmenopausal breast cancer among women on Long Island, New York. Am. J. Epidemiol. 2005;162:229–237. doi: 10.1093/aje/kwi195. [DOI] [PubMed] [Google Scholar]
- 24.Terry MB, et al. Association of frequency and duration of aspirin use and hormone receptor status with breast cancer risk. J. Am. Med. Assoc. 2004;291:2433–2440. doi: 10.1001/jama.291.20.2433. [DOI] [PubMed] [Google Scholar]
- 25.Gammon MD, et al. Environmental toxins and breast cancer on Long Island. I. Polycyclic aromatic hydrocarbon DNA adducts. Cancer Epidemiol. Biomarkers Prev. 2002;11:677–685. [PubMed] [Google Scholar]
- 26.Carlson CS, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson CS, et al. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 2003;33:518–521. doi: 10.1038/ng1128. [DOI] [PubMed] [Google Scholar]
- 28.Guo SW, et al. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 29.Schaid DJ, et al. Biased tests of association: comparisons of allele frequencies when departing from Hardy-Weinberg proportions. Am. J. Epidemiol. 1999;149:706–711. doi: 10.1093/oxfordjournals.aje.a009878. [DOI] [PubMed] [Google Scholar]
- 30.Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120:849–852. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weir B. Genetic Data Analysis II. Sunderland, MA: Sinauer associations Inc; 1996. [Google Scholar]
- 32.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Vainio H, et al. Sadie Jenkins Harmon Collection. Weight Control and Physical Activity. Lyon: IARC Press; 2002. [Google Scholar]
- 34.van den Brandt PA, et al. Pooled analysis of prospective cohort studies on height, weight, and breast cancer risk. Am. J. Epidemiol. 2000;152:514–527. doi: 10.1093/aje/152.6.514. [DOI] [PubMed] [Google Scholar]
- 35.Witte JS, et al. Hierarchical regression analysis applied to a study of multiple dietary exposures and breast cancer. Epidemiology. 1994;5:612–621. doi: 10.1097/00001648-199411000-00009. [DOI] [PubMed] [Google Scholar]
- 36.Hosmer DW, et al. Confidence interval estimation of interaction. Epidemiology. 1992;3:452–456. doi: 10.1097/00001648-199209000-00012. [DOI] [PubMed] [Google Scholar]
- 37.Excoffier L, et al. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 38.Zaykin DV, et al. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 39.Hung RJ, et al. Using hierarchical modeling in genetic association studies with multiple markers: application to a case-control study of bladder cancer. Cancer Epidemiol. Biomarkers Prev. 2004;13:1013–1021. [PubMed] [Google Scholar]
- 40.Witte JS. Genetic analysis with hierarchical models. Genet. Epidemiol. 1997;14:1137–1142. doi: 10.1002/(SICI)1098-2272(1997)14:6<1137::AID-GEPI96>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 41.Daly MJ, et al. High-resolution haplotype structure in the human genome. Nat. Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 42.Redden DT, et al. Nonreplication in genetic association studies of obesity and diabetes research. J. Nutr. 2003;133:3323–3326. doi: 10.1093/jn/133.11.3323. [DOI] [PubMed] [Google Scholar]