Abstract
DNA hypermethylation is a common epigenetic alteration in human prostate cancer and is considered to contribute to development of this disease. Accumulating data suggest that dietary factors may alter cancer risk by modifications of epigenetic processes in the cell. The present study was designed to investigate whether selenium (Se) would alter epigenetic events to regulate methylation-silenced genes in human prostate cancer cells. DNA methylation, histone modifications and gene expression were studied in LNCaP cells after selenite treatment using polymerase chain reaction, western blot analysis, chromatin immunoprecipitation assay and enzymatic activity assay. Our study shows that selenite treatment caused partial promoter DNA demethylation and reexpression of the π-class glutathione-S-transferase (GSTP1) in LNCaP cells in a dose- and time-dependent manner. Selenite treatment decreased messenger RNA levels of DNA methyltransferases (DNMTs) 1 and 3A and protein levels of DNMT1. Selenite also decreased histone deacetylase activity and increased levels of acetylated lysine 9 on histone H3 (H3-Lys 9), but decreased levels of methylated H3-Lys 9. Selenite treatment reduced levels of DNMT1 and methylated H3-Lys 9 associated with the GSTP1 promoter, but increased levels of acetylated H3-Lys 9 associated with this promoter. Additionally, selenite treatment decreased general DNA methylation and caused partial promoter demethylation and reexpression of the tumor suppressor adenomatous polyposis coli and cellular stress response 1, a gene involving tumor growth and metastasis. Our study demonstrates that Se can epigenetically modulate DNA and histones to activate methylation-silenced genes. These epigenetic modifications may contribute to cancer prevention by Se.
Introduction
Epigenetic alterations in multiple genes are believed to play a role in prostate carcinogenesis (1,2). Studies have demonstrated that hypermethylation of a number of genes involved in tumor suppression, hormonal response, cell cycle control, cell invasion and DNA damage repair is correlated with the pathologic grade and clinical stage of prostate cancer (2). The π-class glutathione-S-transferase (GSTP1) is one of the superfamily of glutathione-S-transferase genes that play an important role in protection against DNA damage from electrophilic compounds and oxidants. Studies have found that the GSTP1 promoter is frequently hypermethylated in prostate cancer, with frequencies ranging from 70 to 100% of prostate cancer and 50–70% of high-grade prostatic intraepithelial neoplasia (2–4), suggesting an association of silencing of this gene with both early and progression stages of prostate carcinogenesis. GSTP1 hypermethylation is also present in several prostate cancer cell lines, particularly the LNCaP cell line (3). The tumor supresssor adenomatous polyposis coli (APC) gene is also known to be frequently silenced in prostate cancer due to hypermethylation (5–7). Additionally, cellular stress response 1 (CSR1) encoding an antioxidant/tumor suppressor protein has been demonstrated to be hypermethylated in both human prostate cancer tissues and prostate cancer cell lines (8,9). These multiple silenced genes due to methylation may contribute to prostate cancer development and progression.
DNA hypermethylation is commonly associated with increased levels or altered function of DNA methyltransferases (DNMTs) (1,10). Additionally, histone modifications, particularly methylation and acetylation, may also be involved in transcriptional silencing of a number of genes in prostate cancer (2). Recent data suggest that some dietary supplements, including selenium (Se), may prevent cancer by modifications of epigenetic processes in the cell (11–13). Studies have shown that Se inhibited DNMT expression and activity (14,15). Thus, Se may regulate expression of some anticancer genes by epigenetic modifications of DNA and histones.
Se is essential for human health and a key trace element in maintenance of the activity of the antioxidant enzyme family of glutathione peroxidases against oxidative stress. The major dietary sources of Se for humans are from meats, fish, cereals, dairy products and plant foods (16). Among them, levels of Se are high in fish, moderate in meats and low in plant foods, except for Brazil nuts that contain the highest levels of Se. Chemical forms of Se include organic compounds such as selemethionine and selenocysteine and inorganic ones such as selenate and selenite. Selenomethionine is the dominant form of Se in foods, whereas inorganic Se compounds are the Se source for plants. Sodium selenate, sodium selenite, selenomethionine and selenized yeast are forms of Se in dietary supplements. Se deficiency has been linked to a variety of human disease, including cancer (17). Epidemiologic studies have found an inverse relationship between serum Se levels and cancer risk (18,19). Clinical studies demonstrated that Se supplementation by selenite or selenized yeast reduced certain types of cancer, particularly prostate cancer (20,21). Se and vitamin E are currently being evaluated for prostate cancer chemoprevention in the largest clinical trial, the Selenium and Vitamin E Cancer Prevention Trial, in the USA (22). Se also possesses toxicity when high levels are ingested. Therefore, Se can be beneficial or toxic to humans depending on the levels of intake. Both antioxidant activity and sublethal toxicity of Se have been proposed to be the underlying mechanisms of Se against cancer (23).
The aim of our study was to investigate whether sodium selenite would modify the expression of hypermethylation-silenced GSTP1, APC and CSR1 in human LNCaP prostate cancer cells and to analyze the underlying mechanisms. Our study demonstrates that non-toxic doses of selenite treatment inhibited the expression of DNMT and partially demethylated the promoters of these genes with resultant reexpression of their messenger RNAs (mRNAs). Selenite treatment also inhibited histone deacetylase (HDAC) activity and altered histone modifications. The results indicate that Se can modulate the expression of anticancer genes via epigenetic processes involving DNA methylation and histone modifications. The findings from our study suggest a novel epigenetic mechanism contributing to cancer chemoprevention by Se.
Materials and methods
Chemicals and antibodies
Sodium selenite, 5-aza-2′-deoxycytidine (5-Aza-dC), trichostatin A (TSA) and anti-β-actin antibody were purchased from Sigma Chemical Co. (St Louis, MO). Anti-DNMT1 antibody was purchased from Imgenex (San Diego, CA). Anti-acetyl-histone H3 lysine 9, anti-dimethyl-histone H3 lysine 9 and anti-histone H3 antibodies, CpGenome DNA Modification Kit and EZ ChIP Assay Kit were purchased from Millipore (Temecula, CA). Anti-GSTP1 antibody was purchased from Becton Dickinson and Company (Franklin Lakes, NJ). QIAquick PCR Purification Kit, DNeasy Tissue Kit and RNeasy Mini Kit were purchased from Qiagen (Valencia, CA). SuperScript II was purchased from Invitrogen (Carlsbad, CA).
Cell culture and treatment
Human prostate cancer cell line LNCaP was obtained from the American Type Culture Collection (Manassas, VA) and routinely maintained in 100 mm tissue culture dishes in RPMI 1640 supplemented with 5% heat-inactivated fetal bovine serum at 37°C in a humidified atmosphere of 95% air and 5% CO2. Cells were seeded at a density of 1 × 106 cells per dish and allowed to attach for 48 h before treatment with selenite, 5-Aza-dC for 7 or 14 days with media and treatment agents refreshed every 2 days. Cells were treated with only a single dose of TSA for 3 days.
Methylation-specific polymerase chain reaction
DNA was extracted from the cells using the DNeasy Tissue Kit and modified with sodium bisulfite using the CpGenome DNA Modification Kit according to the manufacturer’s instructions. Bisulfite-treated DNA was analyzed by methylation-specific polymerase chain reaction (MS-PCR) with primer pairs for the unmethylated or methylated promoters of GSTP1, APC and CSR1 as described previously (8,24). Polymerase chain reactions (PCRs) were run for 35 cycles with HotStar Taq polymerase Plus (Qiagen).
Reverse transcripton–polymerase chain reaction and real-time quantitative reverse transcripton–polymerase chain reaction
Total RNA was isolated using RNeasy Mini Kit. Reverse transcription reaction for first-strand complementary DNA synthesis was performed using SuperScript II. PCRs were run for 35 cycles using specific primers as described previously (24).
Real-time PCR was carried out in duplicate using iQ™ SYBR Green Supermix and iCycler iQ™ Real-time Detection System (Bio-Rad Laboratories, Hercules, CA). Primers of DNMT have been described previously (25). PCR was run for 40 cycles. β-Actin was used as an internal control for normalization of input RNA.
Western blot analysis
Cell pellets were lysed in 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol and 1.5 mM phenylmethylsulfonyl fluoride. HCl was added to a final concentration of 0.2 M. After incubation on ice for 30 min and centrifugation at 11 000g at 4°C for 10 min, the supernatants were used for western blot analysis. Cell lysates were electrophoresed in 16% sodium dodecyl sulfate–polyacrylamide gels and then transferred onto nitrocellulose membranes. After blocking in freshly prepared phosphate-buffered saline containing 3% non-fat dry milk at room temperature for 30 min, the membranes were incubated with antibodies against total histone H3 (1:2000), acetylated lysine 9 on histone H3 (H3-Lys 9) (1:5000) or dimethylated H3-Lys 9 (1:2000) at 4°C overnight followed by a goat anti-rabbit secondary antibody conjugated with horseradish peroxidase at 1:10 000 dilution in tris-tween buffered saline at room temperature for 1 h. GSTP1 and DNMT1 were analyzed as described previously (26). Anti-GSTP1 and DNMT1 antibodies were diluted at 1:2000 and 1:500, respectively. Protein bands were visualized on X-ray film using an enhanced chemiluminescence system (Pierce, Rockford, IL).
HDAC activity assay
The Fluor de Lys Fluorescent Assay System (Biomol, Plymouth Meeting, PA) was used to determine HDAC activity. Cell lysates (15 μg total protein) were incubated with the Fluor de Lys substrate in 96-well microplates at 37°C for 30 min. The Fluor de Lys developer was then added and incubated at room temperature for 10 min. Fluorescence was measured using a fluorescence microplate reader. The HDAC activity was presented as arbitrary fluorescence units.
Chromatin immunoprecipitation assay
LNCaP cells (1 × 106 cells) were treated with or without selenite for 7 days, and the chromatin immunoprecipitation (ChIP) assay was performed using the EZ ChIP Assay Kit. DNA cross-linking was performed by adding 1% formaldehyde into cell cultures at room temperature for 10 min, and glycine was then added (0.125 M final concentration) for 5 min to stop the cross-linking reaction. Cells were lysed with a lysis buffer with protease inhibitors and sonicated to shear genomic DNA to lengths between 300 and 800 bp. One-tenth of the cell lysate was used for input control and the rest was used for immunoprecipitation using antibodies against acetylated H3-Lys 9, dimethylated H3-Lys 9 or DNMT1. After collecting immunoprecipitates using protein G agarose columns, protein–DNA complexes were eluted and heated at 65°C to reverse the cross-linking. After digestion with proteinase K, DNA fragments were purified by spin columns and analyzed by PCR for 35 cycles in a sequence of 94°C for 30 s, 58°C for 30 s and 72°C for 1 min. Specific primer sets were designed to amplify a target sequence within the human GSTP1 promoter as described previously (24). PCR products were electrophoresed in a 1% agarose gel with ethidium bromide and visualized under ultraviolet light.
Dot blot analysis of DNA cytosine methylation
Genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen) according to the manufacturer’s instructions. Briefly, genomic DNA (1 μg) was denatured with 0.4 N NaOH at 100°C for 10 min, neutralized with ammonium acetate (pH 7.0) and blotted onto nitrocellulose membranes. Dot blots were incubated with an anti-5-methylcytosine antibody at 1:1000 dilution (Megabase Research, Lincoln, NE) followed by incubation with a horseradish peroxidase-conjugated secondary anti-rabbit-IgG antibody at 1:2000 dilution. The membranes were then treated with enhanced chemiluminescence detection reagents and exposed to Kodak autoradiograph films. DNA loading was determined by staining the membranes with 0.02% methylene blue.
Statistical analysis
Data are presented as means ± SDs with n = 3 or more and experiments were repeated at least three times. Student’s t-test was used to determine the statistical difference between data at the level of P < 0.05. For real-time PCR results, a 2-fold or more change in mRNA levels was considered to be significant.
Results
Demethylation and reexpression of GSTP1 by selenite treatment in LNCaP cells
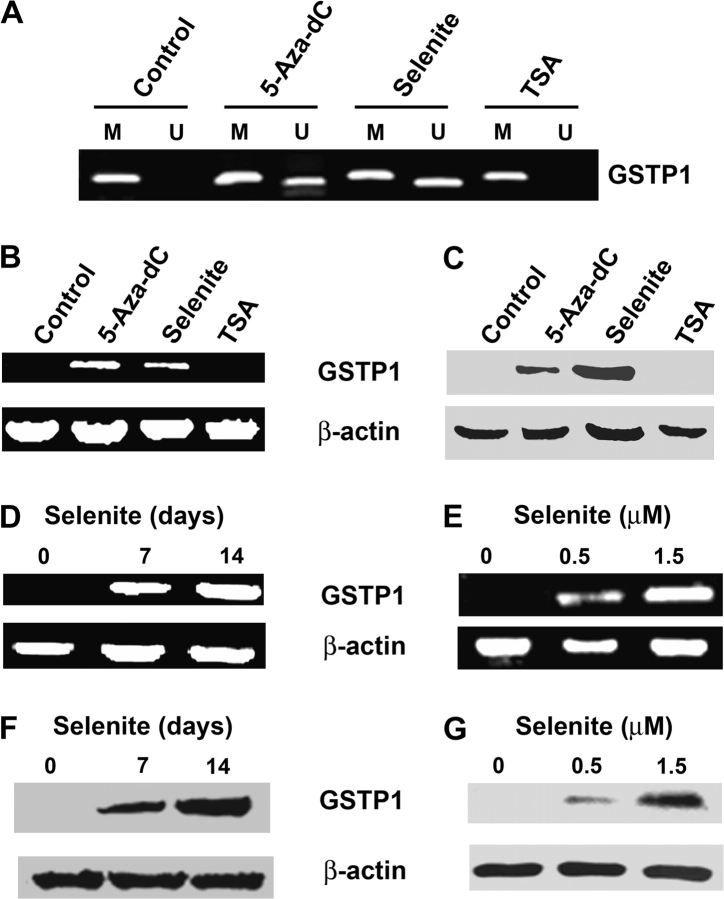
We first verified methylation and transcriptional silencing of GSTP1 in LNCaP cells using MS-PCR, reverse transcripton–polymerase chain reaction (RT–PCR) and western blot analyses. As shown in Figure 1, the GSTP1 promoter was completely methylated and the gene was silenced at both mRNA and protein levels. To determine that promoter hypermethylation is the underlying mechanism of GSTP1 silencing, LNCaP cells were treated with a DNMT inhibitor 5-Aza-dC. Cells were also assessed to determine whether treatment with sodium selenite would alter GSTP1 promoter methylation. HDAC inhibitor TSA was used as a negative control. LNCaP cells were treated with 1.5 μM selenite or 5 μM 5-Aza-dC for 7 days or 0.2 μM TSA for 3 days. The dose of selenite used in the study was a non-toxic dose, which caused slight cell growth inhibition with 14 and 16% decreases in cell number at day 7 and day 14, respectively (supplementary Figure 1 is available at Carcinogenesis Online). The doses of 5-Aza-dC and TSA used in the study had optimal effects on DNA methylation and histone acetylation, respectively, but caused significant cell killing (supplementary Figure 1 is available at Carcinogenesis Online). MS-PCR showed that treatment with 5-Aza-dC or selenite resulted in partial demethylation of the GSTP1 promoter, whereas TSA treatment did not change the methylation status of GSTP1 (Figure 1A). Consistent with the results of MS-PCR, reexpression of GSTP1 mRNA and protein occurred in cells treated with 5-Aza-dC or selenite, but not with TSA (Figure 1B and C). RT–PCR and western blot analyses demonstrated that regulation of GSTP1 expression by selenite was dose and time dependent after treatment with 0.5 and 1.5 μM selenite for 7 days or with 1.5 μM selenite for 7 or 14 days (Figure 1D–G). Selenomethionine treatment also resulted in promoter demethylation and mRNA and protein reexpression of GSTP1 (supplementary Figure 2 is available at Carcinogenesis Online).
Fig. 1.
Effects of selenite, 5-Aza-dC and TSA on promoter methylation and expression of GSTP1 in LNCaP cells. Cells were treated with 1.5 μM selenite or 5 μM 5-Aza-dC for 7 days or 0.2 μM TSA for 3 days. Genomic DNA, total RNA and total protein were isolated from cells and analyzed by MS-PCR, RT–PCR or western blot for detection of promoter methylation, mRNA and protein of GSTP1. (A) MS-PCR analysis of CpG island methylation in the GSTP1 promoter region. M, methylated; U, unmethylated. (B) RT–PCR analysis of mRNA levels of GSTP1. β-Actin was used as control for equal sample loading. (C) Western blot analysis of protein levels of GSTP1. (D and E) RT–PCR analysis of time- and dose-dependent effects of selenite on GSTP1 mRNA expression. (F and G) Western blot analysis of time- and dose-dependent effects of selenite on levels of GSTP1 protein. Cells were treated with 1.5 μM selenite for 7 or 14 days or with 0.5 or 1.5 μM selenite for 7 days. Data are representative of three independent experiments.
Inhibition of DNMT expression and HDAC activity by selenite in LNCaP cells
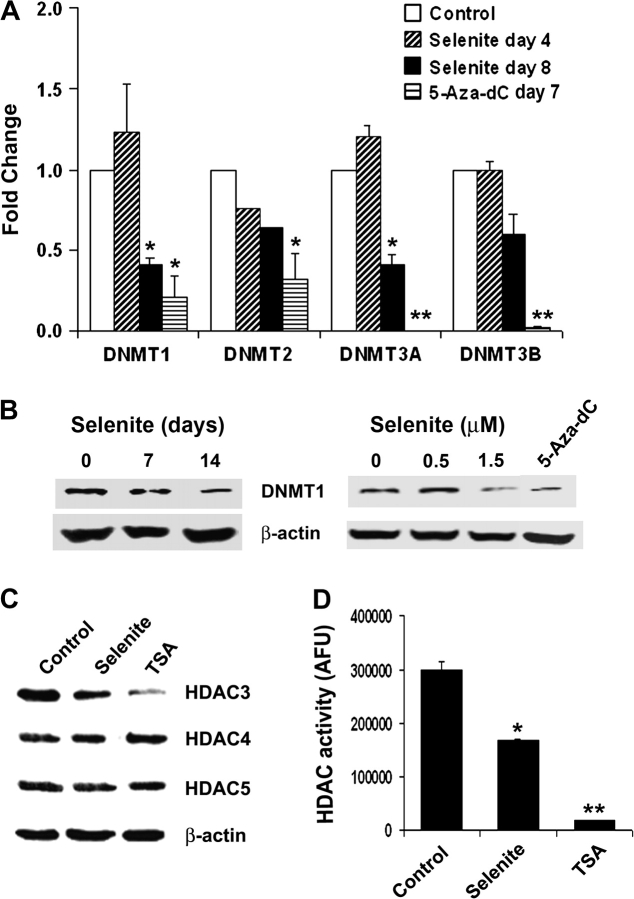
GSTP1 hypermethylation in prostate cancer is considered to be mediated by DNMT. To determine whether selenite treatment would alter expression of DNMTs, mRNA levels of DNMTs and protein levels of DNMT1 were analyzed by real-time RT–PCR and western blot analyses in LNCaP cells after treatment with selenite or 5-Aza-dC. mRNA levels of four major DNMT isoforms (DNMT1, 2, 3A and 3B) were measured by real-time RT–PCR. As shown in Figure 2A, mRNA levels of DNMT1 and 3A, but not DNMT2 and 3B, were significantly decreased in LNCaP cells treated with 1.5 μM selenite in a time-dependent manner, whereas cells treated with 5 μM 5-Aza-dC for 8 days showed a significant decrease in all four DNMTs. The inhibitory effect mediated by 5-Aza-dC was greater than selenite at the 1.5 μM dose tested. Western blot analysis showed that selenite treatment decreased protein levels of DNMT1 in a time- and dose-dependent manner (Figure 2B), concordant with changes in mRNA. Consistent with the result of real-time RT–PCR, 5-Aza-dC treatment resulted in a decrease in DNMT1 protein.
Fig. 2.
Effects of selenite, 5-Aza-dC and TSA on DNMTs or HDAC in LNCaP cells. (A) Real-time quantitative RT–PCR analysis of DNMT mRNA. Cells were treated with 1.5 μM selenite for 4 and 8 days or with 5 μM 5-Aza-dC for 7 days. *Indicates 2-fold or greater changes compared with the corresponding control. **Indicates 2-fold or greater changes compared with the corresponding control and selenite on day 8. (B) (left) Western blot analysis of time-dependent effect of selenite on DNMT1 protein levels in cells treated with 1.5 μM selenite for 7 days. (B) (right) Western blot analysis of dose-dependent effect of selenite on DNMT1 protein levels. Cells were treated with 0.5 or 1.5 μM selenite or 5 μM 5-Aza-dC as a positive control for 7 days. (C) Western blot analysis of protein levels of HDAC3, 4 and 5. Cells were treated with 1.5 μM selenite for 7 days or 0.2 μM TSA for 3 days. (D) Fluorometric analysis of HDAC activity. Cells were treated with 1.5 μM selenite for 7 days or 0.2 μM TSA for 3 days. *P < 0.05 compared with control. ** P < 0.05 compared with control or selenite. Data are mean ± SD (n = 3) or representative of three independent experiments.
Since histone acetylation, an important mechanism of regulation of gene expression, is in part regulated by HDAC, we also investigated whether selenite treatment altered HDAC expression and activity. As shown in Figure 2C, selenite treatment resulted in a slight decrease in HDAC3 protein after 7 day treatment at the 1.5 μM dose tested. TSA treatment resulted in a significant decrease in HDAC3 protein. The protein levels of HDAC4 and HDAC5 were not altered in cells treated with selenite or TSA. However, selenite treatment significantly inhibited HDAC activity (Figure 2D). Similar to selenite, TSA also inhibited HDAC activity and its inhibitory effect was greater than selenite at the 1.5 μM dose tested (Figure 2D).
Effects of selenite, 5-Aza-dC and TSA on histone methylation and acetylation and interactions of histone and DNMT1 with the GSTP1 promoter
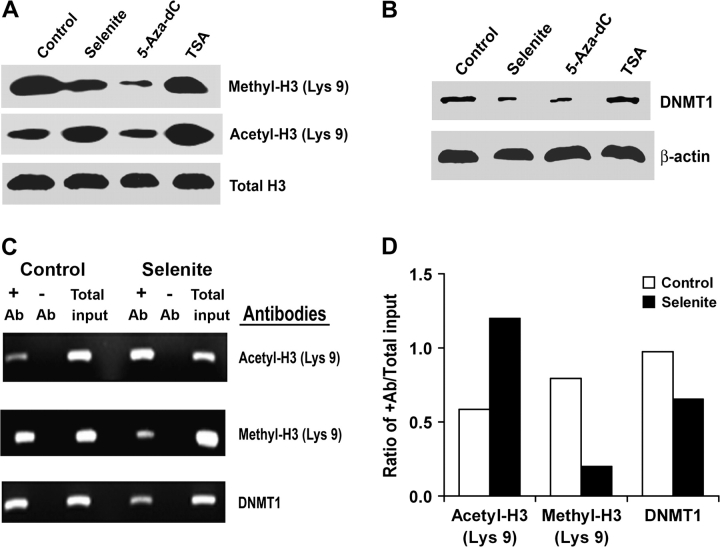
Methylation and/or deacetylation of histone proteins H3 and H4 are known to be involved in modification of chromatin structure and gene silencing. Deacetylation and methylation of H3-Lys 9 are associated with silencing of gene expression, whereas acetylation of H3-Lys 9 is involved in activation of gene expression (27). Therefore, we chose to analyze H3-Lys 9 to determine whether selenite would alter its methylation and acetylation states and to analyze its association with the GSTP1 promoter by western blot and the ChIP assay. The association of DNMT1 with the GSTP1 promoter was also analyzed. As shown in Figure 3A, selenite treatment decreased levels of methylated H3-Lys 9, but increased levels of acetylated H3-Lys 9. 5-Aza-dC treatment decreased the level of methylated H3-Lys 9, but did not change the level of acetylated H3-Lys 9. In contrast, TSA treatment increased the level of acetylated H3-Lys 9, but did not alter the level of methylated H3-Lys 9. Selenite, 5-Aza-dC or TSA treatment did not change protein levels of total histone 3. Selenite or 5-Aza-dC treatment also decreased levels of DNMT1 protein, but TSA had no effect on DNMT1 (Figure 3B). The ChIP assay showed that selenite treatment increased the level of acetylated H3-Lys 9 associated with the GSTP1 promoter, but decreased levels of methylated H3-Lys 9 and DNMT1 associated with the GSTP1 promoter (Figure 3C). Densitometry demonstrated that cells treated with selenite increased GSTP1 promoter-associated acetylated H3-Lys 9 by 2-fold and decreased GSTP1 promoter-associated methylated H3-Lys 9 by 3-fold, wheras GSTP1 promoter-associated DNMT1 was decreased by 30% compared with cells without selenite treatment (Figure 3D).
Fig. 3.
Effects of selenite, 5-Aza-dC and TSA on histone modifications, DNMT1 protein levels and interaction of histone or DNMT1 with the GSTP1 promoter in LNCaP cells. Cells were treated with 1.5 μM selenite and 5 μM 5-Aza-dC for 7 days or 0.2 μM TSA for 3 days. Total cellular protein or genomic DNA was isolated for western blot analysis or for ChIP assay. (A) Western blot analysis of protein levels of methylation and acetylation of Lys 9 on histone H3. (B) Western blot analysis protein levels of DNMT1. (C) ChIP assay of interaction of histone H3 or DNMT1 with the GSTP1 promoter. ChIP assay was performed with the anti-acetylated H3-Lys 9, anti-methylated H3-Lys 9 or anti-DNMT1 antibody. +Ab, with antibodies added; −Ab, no antibodies added; total input, total amount of sample used for ChIP assay. (D) Densitometric quantification of (C). Data are representative of three independent experiments.
Reduction of DNA methylation and reactivation of APC and CSR1 by selenite treatment in LNCaP cells
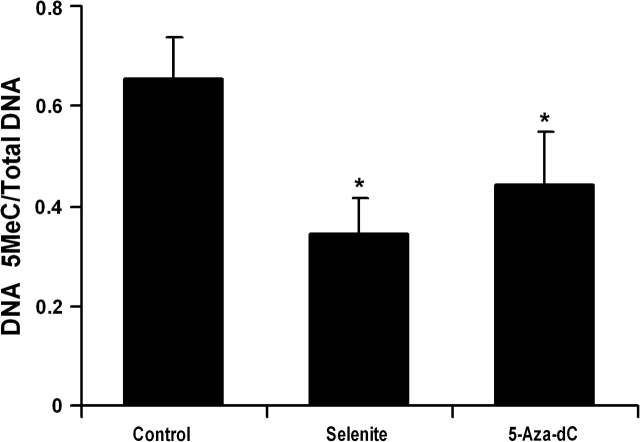
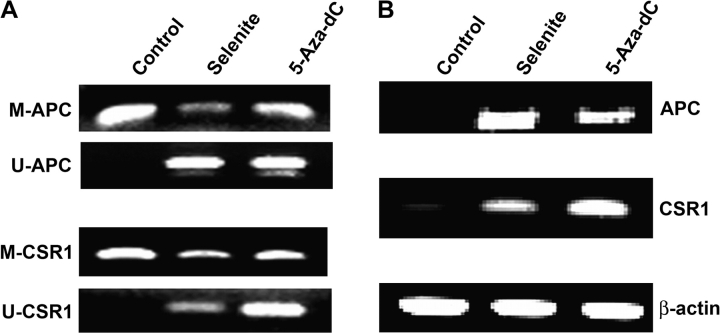
To further determine the effects of Se on general DNA methylation, total genomic DNA was isolated from LNCaP cells and analyzed by dot blot assay using an anti-5-methylated deoxycytosine antibody (supplementary Figure 3 is available at Carcinogenesis Online). As shown in Figure 4, treatment with 1.5 μM selenite or 5 μM 5-Aza-dC significantly decreased methylation of total DNA. Selenite showed a slightly greater effect than 5-Aza-dC at the dose tested. The results suggest that Se may have a broad effect on DNA methylation. To confirm this, we analyzed whether selenite treatment caused demethylation and reactivation of APC and CSR1, two genes also known to be silenced in LNCaP cells due to DNA hypermethylation (7,8). MS-PCR showed that treatment with selenite or 5-Aza-dC resulted in promoter demethylation of both APC and CSR1 (Figure 5A). Selenite had a slightly greater effect on APC than 5-Aza-dC, whereas 5-Aza-dC had a much greater effect on CSR1 than selenite. In agreement with MS-PCR results, RT–PCR showed that selenite induced a slightly higher level of APC mRNA expression, whereas 5-Az-dC induced a much greater expression of CSR1 mRNA (Figure 5B).
Fig. 4.
Effects of selenite and 5-Aza-dC on genomic DNA methylation in LNCaP cells. Cells were treated with 1.5 μM selenite or 5 μM 5-Aza-dC for 7 days and total genomic DNA was isolated and blotted onto a nitrocellulose membrane. DNA methylation at 5-cytosine was detected using an anti-5-methylcytosine (5MeC) antibody and sample loading was determined by staining total DNA with methylene blue. The intensity of individual dots was measured by densitometry and levels of 5-methylcytosine were normalized by total DNA. Data are mean ± SD (n = 3). *P < 0.05 compared with control.
Fig. 5.
Effects of selenite and 5-Aza-dC on promoter methylation and expression of APC and CSR1 in LNCaP cells. Cells were treated with 1.5 μM selenite or 5 μM 5-Aza-dC for 7 days. Genomic DNA and total RNA were isolated from cells and analyzed by MS-PCR or RT–PCR for detection of promoter methylation and mRNA of APC and CSR1. (A) MS-PCR analysis of CpG island methylation in the promoter region of APC and CSR1. M, methylated; U, unmethylated. (B) RT–PCR analysis of mRNA levels of APC and CSR1. β-Actin was used as control for equal sample loading. Data are representative of three independent experiments.
Discussion
Cancer is a disease associated with both genetic and epigenetic changes. Epigenetic gene regulation has been recognized to play a role in the etiology of cancer. DNA methylation and post-translational histone modifications are two important epigenetic events in regulation of gene expression and maintenance of cellular function. Changes in DNA methylation and/or histone modifications may contribute to cancer development. Abnormal DNA methylation is a hallmark of cancer and often mediates silencing of genes, particularly tumor suppressors, leading to cancer development and progression. A number of DNA methylation and histone-modifying agents are currently being evaluated in cancer chemoprevention and therapy (28). Recent data suggest that some dietary factors may be involved in epigenetic modifications to regulate cellular function and modify carcinogenesis. Se is an essential nutritional element and has been demonstrated to be a potential chemopreventive agent for prostate cancer. Two studies demonstrated that Se inhibited DNMT activity (14,15), suggesting that Se may have epigenetic effects on gene expression involved in prostate carcinogenesis.
In this study, we investigated epigenetic regulation of GSTP1, APC and CSR1 by selenite and elucidated the underlying mechanisms using the LNCaP prostate cancer cell line as a model system and sodium selenite as the form of Se. We used 5-Aza-dC, a well-characterized DNA methylation inhibitor, and TSA, a HDAC inhibitor, to compare with selenite. GSTP1 is a phase II detoxification enzyme that can protect the cell from damage by carcinogens including chemicals and oxidants. APC is a tumor suppressor well known for its role in colon carcinogenesis. CSR1 is a protein with antioxidant and tumor suppressor functions (8,9). These three genes are known to be silenced in prostate cancer due to abnormal promoter methylation and are thought to contribute to prostate development and progression (1,5,8). Our results demonstrate that 5-Aza-dC significantly decreased mRNA expression of DNMT1, 2, 3A and 3B, whereas selenite decreased only DNMT1 and 3A. 5-Aza-dC or selenite treatment decreased protein levels of DNMT1. The effect of selenite on DNMTs was dose and time dependent. These results indicate that selenite can downregulate expression of some DNMTs, results at least partially similar to those obtained with 5-Aza-dC treatment. The promoters of GSTP1, APC and CSR1 were demethylated and the genes were reexpressed at mRNA and/or protein levels in cells following treatment with selenite or 5-Aza-dC. Using GSTP1 as an example, we further demonstrate that the effects of selenite on GSTP1 were also dose and time dependent. In contrast, treatment with the HDAC inhibitor TSA changed neither methylation nor the expression of GSTP1. These results suggest that selenite may reactivate these genes, at least in part, by downregulation of DNMTs with resultant demethylation of their promoters, events similar to those observed with 5-Aza-dC. Additionally, Se has been demonstrated to inhibit DNMT enzymatic activity (14,15). Thus, inhibition of DNMT enzymatic activity is probably also to be involved in the effects of selenite on alterations of DNA methylation observed in our study. Our results suggest that DNMT1 is most likely the target of Se. It has been reported that Se upregulated the expression of GSTP1 mRNA in normal and hepatoma cells or GST activity in rat liver tissues, in which GSTP1 was unmethylated (29,30). We also tested the effect of selenite on ummethylated GSTP1 in PC3 prostate cancer cells and our results show that selenite upregulated GSTP1 mRNA levels in PC3 cells in a dose- and time-dependent manner (supplementary Figure 4 is available at Carcinogenesis Online). These results indicate that Se can regulate the expression of unmethylated GSTP1. Additionally, our study demonstrates that selenite decreased levels of 5-methylcytosine on total genomic DNA, suggesting that Se may have a broad effect on DNA methylation. This is supported by the results of demethylation of multiple genes in our study. Further studies are required to determine whether the epigenetic regulation of gene expression by Se is selective and different between normal and cancer cells.
A recent study by Ramachandran et al. (31) reported that Se did not alter methylation-mediated silencing of genes, including GSTP1, in LNCaP cells. Differences in time and dosage of Se treatment may account for different conclusions obtained in these two studies. In their study, LNCaP cells were treated with a single dose (10 μM) of selenite for 4 days only. Our study demonstrates that selenite treatment for 4 days did not cause significant changes in DNMT mRNA and reactivation of the silenced genes was observed on day 7, but not on day 4 (data not shown). Therefore, a 4 day treatment with Se is apparently not long enough to alter DNA methylation. Selenite is a highly toxic form of Se. We observed that treatment with 2 μM or higher doses of selenite resulted in significant cell death in LNCaP cells in our previous study (26). We also observed that cell killing by high toxic doses of selenite could mask the epigenetic effects of selenite (data not shown), suggesting that Se exerts its epigenetic effects at non-toxic levels. Thus, 10 μM selenite is apparently too toxic for analysis of epigenetic action of Se.
Chromatin structure is influenced by a variety of histone covalent modifications, including methylation and acetylation. These histone modifications occur at the N-terminal tails of the histone proteins and are epigenetic marks linked to transcriptional activation and repression of genes (32). Histone acetylation will result in an open chromatin structure associated with transcriptional activation, whereas histone deacetylation will lead to a closed chromatin structure with resultant transcriptional repression (32). Histone methylation can be associated with gene activation or repression, depending on methylation of specific lysine or arginine residues on H3 and H4 proteins (32). Post-translational modifications of histone methylation and acetylation may contribute to cancer development by modulation of the expression of tumor suppressor genes and oncogenes. Deacetylation and methylation of H3-Lys 9 are the most common histone covalent modifications involved in epigenetic repression of genes (27). HDACs play a key role in regulation of histone deacetylation. Our study demonstrates that selenite inhibited HDAC activity without significantly altering the levels of HDAC proteins. Additionally, selenite decreased levels of methylated H3-Lys 9, but increased levels of acetylated H3-Lys 9. Similar to selenite, TSA also inhibited HDAC activity and increased acetylated H3-Lys 9, but had no effect on histone methylation. These findings indicate that selenite inhibits HDAC activity to maintain H3-Lys 9 at a high level of acetylation and a low level of methylation, which favor transcriptional activation of genes. In contrast, 5-Aza-dC decreased methylation of H3-Lys 9, but had no effect on the acetylation of H3-Lys 9. These results suggest that selenite and 5-Aza-dC may inhibit methyltransferase to prevent histone methylation. Decreased levels of methylated H3-Lys 9 also favor activation of gene expression. The ChIP assay showed that selenite decreased the association of DNMT1 and methylated H3-Lys 9 with the GSTP1 promoter and increased the association of acetylated H3-Lys 9 with this promoter, indicating that selenite alters the functions of these proteins involved in epigenetic regulation in the cell. However, the decreased levels of DNMT1 associated with the GSTP1 promoter were much less than those of methylated H3-Lys 9, a 30% decrease for the former versus a 3-fold decrease for the latter. These results suggest that Se may have a greater effect on histone methylation than DNMT. Our study demonstrates that histone modifications by TSA alone were unable to reactivate methylation-silenced GSTP1, indicating that histone modifications alone are unable to reactivate a hypermethylated gene, but it can coordinate with DNA-demethylating agents to regulate gene expression and silencing (33). Unlike 5-Aza-dC and TSA, Se has dual actions involving both DNA methylation and modifications of histone methylation and acetylation. These combined effects of Se may have a synergistic effect on epigenetic events by Se itself or in combination with other methyltransferase or deacetylase inhibitors. Similar to our results, a recent study showed that a dietary chemopreventive agent isothiocyanate caused a similar dual action on DNA methylation and histone modifications in LNCaP cells (34). Another recent study demonstrated that treatment with genistein or lycopene at non-toxic concentrations partially demethylated the GSTP1 promoter and reactivated GSTP1 expression in human breast cancer cells (35). Both genistein and lycopene are known to have chemopreventive activity against prostate cancer (36). A study showed that GSTP1 silencing in prostate cancer might be associated with a loss of activating histone modifications (37). The results from this and our current studies suggest that Se may regulate the expression of unmethylated GSTP1 via histone modifications. These combined results implicate that some dietary chemopreventive agents, including Se, may prevent cancer by modifications of epigenetic processes in the cell.
In summary, our study is the first to demonstrate that Se can restore the expression of the hypermethylation-silenced genes GSTP1, APC and CSR1 in human prostate cancer cells by downregulation of DNMT and inhibition of HDAC activity. These genes are known to have anticancer activity by protection against oxidative damage, detoxification of carcinogenetic chemicals or tumor suppression. Our data suggest that epigenetic regulation of these anticancer genes by Se may play a role in prostate and other cancer chemoprevention. The findings of our study are of importance for understanding the anticancer mechanisms and clinical application of Se. Se is currently being evaluated as a prostate cancer preventive agent by the Selenium and Vitamin E Cancer Prevention Trial (22). It would be interesting to see whether Se supplements alter epigenetic events, including DNA methylation, post-translational histone modifications and expression of anticancer genes in human prostate tissues.
Supplementary material
Supplementary Figures 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (CA114281); Office of Research and Development, Biomedical Laboratory Research and Development Service, Department of Veterans Affairs.
Glossary
Abbreviations
- APC
adenomatous polyposis coli
- 5-Aza-dC
5-aza-2′-deoxycytidine
- CSR1
cellular stress response 1
- DNMT
DNA methyltransferase
- GSTP1
π-class glutathione-S-transferase
- HDAC
histone deacetylase
- H3-Lys 9
lysine 9 on histone H3
- mRNA
messenger RNA
- MS-PCR
methylation-specific polymerase chain reaction
- PCR
polymerase chain reaction
- RT–PCR
reverse transcripton–polymerase chain reaction
- Se
selenium
- TSA
trichostatin A
References
- 1.Nelson WG, et al. Abnormal DNA methylation, epigenetics, and prostate cancer. Front. Biosci. 2005;12:4254–4266. doi: 10.2741/2385. [DOI] [PubMed] [Google Scholar]
- 2.Li LC, et al. Epigenetic changes in prostate cancer: implication for diagnosis and treatment. J. Natl Cancer Inst. 2005;97:103–115. doi: 10.1093/jnci/dji010. [DOI] [PubMed] [Google Scholar]
- 3.Lee WH, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl Acad. Sci. USA. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou M, et al. Quantitative GSTP1 methylation levels correlate with Gleason grade and tumor volume in prostate needle biopsies. J. Urol. 2004;171:2195–2198. doi: 10.1097/01.ju.0000127728.71350.36. [DOI] [PubMed] [Google Scholar]
- 5.Ellinger J, et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008;71:161–167. doi: 10.1016/j.urology.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 6.Henrique R, et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin. Cancer Res. 2007;13:6122–6129. doi: 10.1158/1078-0432.CCR-07-1042. [DOI] [PubMed] [Google Scholar]
- 7.Yegnasubramanian S, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 8.Yu G, et al. CSR1 suppresses tumor growth and metastasis of prostate cancer. Am. J. Pathol. 2006;168:597–607. doi: 10.2353/ajpath.2006.050620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han HJ, et al. CSR, a scavenger receptor-like protein with a protective role against cellular damage caused by UV irradiation and oxidative stress. Hum. Mol. Genet. 1998;7:1039–1046. doi: 10.1093/hmg/7.6.1039. [DOI] [PubMed] [Google Scholar]
- 10.Patra SK, et al. DNA methyltransferase and demethylase in human prostate cancer. Mol. Carcinog. 2002;33:163–171. doi: 10.1002/mc.10033. [DOI] [PubMed] [Google Scholar]
- 11.Davis CD, et al. Frontiers in nutrigenomics, proteomics, metabolomics and cancer prevention. Mutat. Res. 2004;551:51–64. doi: 10.1016/j.mrfmmm.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Huang S. Histone methyltransferases, diet nutrients and tumor suppressors. Nat. Rev. Cancer. 2002;2:469–476. doi: 10.1038/nrc819. [DOI] [PubMed] [Google Scholar]
- 13.Davis CD, et al. DNA methylation, cancer susceptibility, and nutrient interactions. Exp. Biol. Med. 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- 14.Cox R, et al. A study of the mechanism of selenite-induced hypomethylated DNA and differentiation of Friend erythroleukemic cells. Carcinogenesis. 1986;7:2015–2018. doi: 10.1093/carcin/7.12.2015. [DOI] [PubMed] [Google Scholar]
- 15.Fiala ES, et al. Inhibition of DNA cytosine methyltransferase by chemopreventive selenium compounds, determined by an improved assay for DNA cytosine methyltransferase and DNA methylation. Carcinogenesis. 1998;19:597–604. doi: 10.1093/carcin/19.4.597. [DOI] [PubMed] [Google Scholar]
- 16.Combs GF., Jr Selenium in global food systems. Br. J. Nutr. 2001;85:517–547. doi: 10.1079/bjn2000280. [DOI] [PubMed] [Google Scholar]
- 17.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 18.Schrauzer GN, et al. Cancer mortality correlation studies. III. Statistical association with dietary selenium intakes. Bioinorg. Chem. 1977;7:35–56. doi: 10.1016/s0006-3061(00)80126-x. [DOI] [PubMed] [Google Scholar]
- 19.Li HJ, et al. A prospective study of plasma selenium levels and prostate cancer risk. J. Natl Cancer Inst. 2004;96:696–703. doi: 10.1093/jnci/djh125. [DOI] [PubMed] [Google Scholar]
- 20.Clark LC, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. J. Am. Med. Assoc. 1996;276:1957–1963. [PubMed] [Google Scholar]
- 21.Yu SY, et al. Protective role of selenium against hepatitis B virus and primary liver cancer in Qidong. Biol. Trace Elem. Res. 1997;56:117–124. doi: 10.1007/BF02778987. [DOI] [PubMed] [Google Scholar]
- 22.Klein EA, et al. SELECT: the next prostate cancer prevention trial. J. Urol. 2001;166:1311–1315. doi: 10.1016/s0022-5347(05)65759-x. [DOI] [PubMed] [Google Scholar]
- 23.Spallholz JE. On the nature of selenium toxicity and carcinostatic activity. Free Radic. Biol. Med. 1994;17:45–64. doi: 10.1016/0891-5849(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 24.Singal R, et al. Cytosine methylation represses glutathione S-transferase P1 (GSTP1) gene expression in human prostate cancer cells. Cancer Res. 2001;61:4820–4826. [PubMed] [Google Scholar]
- 25.Cui X, et al. Arsenic trioxide inhibits DNA methyltransferase and restores methylation-silenced genes in human liver cancer cells. Hum. Pathol. 2006;37:298–311. doi: 10.1016/j.humpath.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Zhao R, et al. Expression of p53 enhances selenite-induced superoxide radical production and apoptosis in human prostate cancer cells. Cancer Res. 2006;66:2296–2304. doi: 10.1158/0008-5472.CAN-05-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama J, et al. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatic assembly. Science. 2001;292:110–111. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 28.Piekarz RL, et al. Histone deacetylase inhibitors and demethylating agents: clinical development of histone deacetylase inhibitors for cancer therapy. Cancer J. 2007;13:30–39. doi: 10.1097/PPO.0b013e31803c73cc. [DOI] [PubMed] [Google Scholar]
- 29.‘tHoen PAC, et al. Induction of glutathione-S-transferase mRNA levels by chemopreventive selenocysteine Se-conjugates. Biochem. Pharmacol. 2002;63:1843–1849. doi: 10.1016/s0006-2952(02)00987-5. [DOI] [PubMed] [Google Scholar]
- 30.Liu JZ, et al. Dietary selenite modifies glutathione metabolism and 7,12-dimethylbenz(a)anthracene conjugation in rats. J. Nutr. 1994;124:172–180. doi: 10.1093/jn/124.2.172. [DOI] [PubMed] [Google Scholar]
- 31.Ramachandran K, et al. Methylation-mediated silencing of genes is not altered by selenium treatment of prostate cancer. Anticancer Res. 2007;27:921–926. [PubMed] [Google Scholar]
- 32.Rice JC, et al. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 33.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 2005;15:1–6. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Wang LG, et al. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol. Carcinog. 2007;46:24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- 35.King-Batoon A, et al. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ. Mol. Mutagen. 2008;49:36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 36.Klein EA. Chemoprevention of prostate cancer. Annu. Rev. Med. 2006;57:49–63. doi: 10.1146/annurev.med.57.121304.131435. [DOI] [PubMed] [Google Scholar]
- 37.Okino ST, et al. Chromatin changes on the GSTP1 promoter associated with its inactivation in prostate cancer. Mol. Carcinog. 2007;46:838–846. doi: 10.1002/mc.20313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.