Abstract
While numerous microRNAs (miRNAs) have been reported to alter their expression levels in human lung cancer tissues compared with normal tissues, the function of these miRNAs and their contribution to the long process of lung cancer development remains largely unknown. We applied a tobacco-specific carcinogen-induced cancer model to investigate the involvement of miRNAs in early lung cancer development, which could also provide information on potential, early biomarkers of lung cancers. Male F344 rats were first chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a carcinogen present in tobacco products, for up to 20 weeks. The expression profiles of miRNAs in rat lungs were then determined. As measured by miRNA microarrays and confirmed by Northern blot and real-time polymerase chain reaction analyses, NNK treatment reduced the expression of a number of miRNAs, such as miR-101, miR-126*, miR-199 and miR-34. Significantly, these miRNAs overlap with previously published reports on altered miRNA expression in human lung cancer samples. These miRNAs might, therefore, represent early-response miRNAs that signify the molecular changes associated with pulmonary tumorigenesis. Moreover, we identified cytochrome P450 (CYP) 2A3, a critical enzyme in rat lungs that activates NNK to render it carcinogenic, as a potential target of miR-126*. NNK treatment in rats repressed miR-126* but induced CYP2A3 expression, a mechanism that may potentiate the oncogenic effects of NNK.
Introduction
MicroRNAs (miRNAs) are a class of ∼22-nucleotide-long, non-coding RNAs widely expressed in eukaryotes and predominantly inhibit gene expression at the post-transcriptional level (1–4). miRNAs control a wide range of biological processes, including, for instance, metabolism, organogenesis, development, cell growth, cell death and cell fate determination (5,6). Furthermore, aberrant expression of miRNAs has been associated with human disease. For example, human miRNA expression signature can be used to distinguish between normal and cancerous lung tissues, with low let-7 expression and high miR-155 expression being correlated with poor prognosis for cancer patients (7–11). How miRNAs contribute to tumorigenesis, however, is largely unknown.
Lung cancer is the most common cause of cancer death in the world, with an overall 5 years survival rate of only 14% upon diagnosis (12,13). Approximately 90% of all lung cancers are directly attributable to smoking (14). In the USA, despite considerable efforts over the last 40 years to reduce the prevalence of tobacco use, >45 million (22%) adults are still smokers (15). Furthermore, former smokers are also at a higher risk of lung cancer than non-smokers. Development of alternate approaches to reduce lung cancer mortality, therefore, is a requisite public health issue, and our research explores two promising approaches, i.e. the identification of biomarkers for early detection and the development of effective chemopreventive agents.
Rodent models are invaluable tools for studying the initiation and progression of human disease, and we have been using these models to investigate the relationship between tobacco compounds and lung tumorigenesis (16). To mimic tobacco carcinogenesis, rodents are fed with carcinogens such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). NNK is a member of the nitrosamine family of compounds that, along with polycyclic aromatic hydrocarbons, are the most prevalent and potent carcinogens in tobacco products and smoke (17–19). Inside a cell, NNK is first metabolically activated by α-hydroxylation reactions to produce metabolites such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol, catalyzed by the cytochrome P450 (CYP) family of enzymes (20). In the liver, the major P450 enzymes for NNK include CYP1A2, CYP2A6 and CYP3A4 in humans, whereas in the lung, rat CYP2A3, mouse CYP2A5 or human CYP2A13 is the most efficient catalyst. NNK metabolites then form alkylating adducts with DNA, leading to genetic mutations. NNK induces tumors in multiple organs and is a systemic lung carcinogen in laboratory animals, and it is a group 1 human carcinogen classified by the International Agency for Research on Cancer (21–25).
Although dozens of miRNAs are differentially expressed in human lung cancer tissues compared with normal tissues (7–11), how and at what stages they contribute to cancer development remains unclear. Lung cancer takes decades to develop in smokers, and once diagnosed, patients have a very low 5 year survival rate (12,13). Thus, for prevention, diagnosis and treatment purposes, it is imperative to understand the early events accompanied with smoking, to which end animal models are essential. In this study, we examined alteration in miRNA expression at the early stages of lung cancer induction by obtaining miRNA expression profiles in the lungs of male F344 rats continuously fed with NNK for up to 20 weeks. Our goal was to generate a miRNA expression signature that could provide important, early diagnostic markers for lung cancers and to provide further insight into the etiology of NNK tumorigenesis.
Materials and methods
Animal studies
The study was approved by the University of Minnesota Institutional Animal Care and Use Committee. Seven-week-old male F344 rats were purchased from Charles River Laboratories (Kingston, NY). They were housed two animals per microisolator cage with corn cob bedding in the Research Animal Resources facility of the University of Minnesota under the following conditions: temperature 20–24°C and relative humidity 50 (10% and 12 h light–dark cycle). They were given NIH-07 diet (Harlan, Madison, WI) and tap water ad libitum. The animals were allowed 1 week for acclimatization to the facility and then randomly assigned into two groups for control or NNK treatment. Rats in the treatment group received 10 p.p.m. of NNK (synthesized as described in ref. 26) in the drinking water, and the control rats were given tap water. Aqueous NNK solution was prepared weekly and stored at 4°C, conditions under which NNK is known to be stable. Solutions were placed in the plastic water bottles of rat cages twice weekly, and water consumption was recorded. Mean body weights of rats were recorded at the beginning of the experiment and once a month thereafter. Rats were killed by CO2 overdose at 1, 5, 16 and 20 weeks, tissues were dissected and stored at −80°C. At these time points, no tumors were visible. For RNA isolation, frozen lung tissues were homogenized with a mortar and a pestle in liquid nitrogen. Total RNAs were then isolated using Trizol (Invitrogen, Carlsbad, CA). Quality and quantity of the RNAs were assessed by A260 and A280 nm reading.
Microarray experiments
Microarray technique was used to profile miRNA expression at a genome-wide scale. A miRNA probe set was purchased from Invitrogen, designed based on the Sanger miRBase Sequence Database, Release 9.0 (October 2006). The set contains ∼1140 oligonucleotides as probes, which are complementary to Caenorhabditis elegans, Drosophila, zebra fish, mouse, rat and human miRNAs, and also include a number of internal and negative control probes. The oligonucleotides were quadruply printed on Corning GAPSII-coated slides by the Microarray Facility at the University of Minnesota. For RNA labeling, 25 μg of total RNA was ligated to 0.5 μg of a synthetic linker, pCU-DY547 (Dharmacon, Lafayette, CO). To control the hybridization process, reference DNA oligonucleotides complementary to a subset of mammalian miRNAs were combined and labeled with a ULYSIS Alexa Fluor 647 Kit (Invitrogen). Labeled RNAs and DNAs were then mixed and hybridized to microarray slides (27). Afterward, slides were scanned with a ScanArray 5000 machine (Perkin Elmer, Waltham, MA), and BlueFuse (BlueGenome, Cambridge, UK) was used to quantify pixel intensities. Individual spots on the slides were further inspected to exclude abnormal spots from subsequent calculations.
Microarray data analyses
GeneSpring GX 7.3.1 (Agilent, Wilmington, DE) was used to normalize microarray data to allow for comparisons between different RNA samples hybridized to different slides. Signals at least 150% higher than background values were considered positive, and after background subtraction, normalized to the signal of a 28S ribosomal RNA probe printed on the same slide. Normalization to other control probes , such as U6 small nuclear RNA (snRNA) or several small nucleolar RNAs, yielded similar conclusions. A separate percentile normalization method (GeneSpring) was also employed. Gene expression changes were deemed valid if concurred by both gene-specific and percentile normalization methods. Excel was used to organize and present normalized data. P-values were calculated using two-tailed, heteroscedastic Student’s t-test from the normalized data.
Northern blotting
Approximately 30 μg of total RNAs was fractionated on a 7 M urea/10% polyacrylamide gel in 1× Tris-Borate-EDTA and transferred to a Hybond N+ membrane (GE Healthcare, Chalfont St. Giles, UK). The same membrane was probed, stripped and reprobed with 5′-end, 32P-labeled oligonucleotides complementary to target sequences (28). The oligonucleotide sequences are as follows: for miR-199a detection, 5′-GAACAGGTAGTCTGAACACTGGG-3′; miR-34b, 5′-GCAATCAGCTAATTACACTGCCTA-3′; miR-101, 5′-GCTTCAGTTATCACAGTACTGTA-3′ and U6 snRNA control, 5′-ACGAATTTGCGTGTCATCCTTGCG-3′. Results were analyzed using a Storm 840 PhosphorImager (GE Healthcare).
Real-time polymerase chain reaction
Reverse transcription and real-time polymerase chain reaction (PCR) to quantify miRNA and messenger RNA (mRNA) expression were performed using Qiagen's miScript system on an Applied Biosystems 7500 real-time PCR machine. Briefly, a poly(A) tail was first synthesized at the 3′ end of RNAs, which were subsequently reverse transcribed with a oligo(dT) linker. PCR was then performed with a universal linker primer and a miRNA-specific primer or with mRNA-specific primers. Primers for real-time PCR are as follows: for miR-199a detection, 5′-CCCAGTGTTCAGACTACCTGTTC-3′; miR-34b, 5′-TAGGCAGTGTAATTAGCTGATTG-3′; miR-101, 5′-AAGGTACAGTACTGTGATAACTGAA-3′; miR-126*, 5′-CATTATTACTTTTGGTACGCG-3′; U6, 5′-GGCAGCACATATACTAAAATTGGAA-3′; CYP2A3 mRNA, 5′-AGCATTGCGTGAGAGTAAAGGGA-3′ and 5′-CCCCTTCTCTGGCTCTACCTTTG-3′ and rat β-actin mRNA, 5′-TACTGCCCTGGCTCCTAGCA-3′ and 5′-GCCAGGATAGAGCCACCAATC-3′ (29). CT values were determined automatically. Relative expression levels were denoted by ΔCT, with the P-values calculated using two-tailed, heteroscedastic Student's t-test from the ΔCT values.
Reporter assays to validate CYP2A3 as a target of miR-126*
Approximately 210-nucleotide-long, 3′ untranslated region (UTR) of CYP2A3 mRNA (GenBank Accession Number J02852) was amplified from rat lung complementary DNA using primers 5′-GCGCTAGCTTTGCCACAATCCCACCAAAC-3′ and 5′-GCCTCGAGCCATAAATAATATCTACTTTTAT-3′, digested with NheI and XhoI and inserted downstream of a firefly luciferase gene to construct a reporter plasmid (30). Deletions in the miR-126*-binding site were made by the Quikchange method (Stratagene, La Jolla, CA) and verified by sequencing. We used a short hairpin RNA to produce miR-126* in transfected cells. To generate the short hairpin RNA, two primers, 5′-TGTAATACGACTCACTATAGCGTACCAAAGCTAATAATGTGTGAAGC-3′ and 5′-ACGCGTACCAAAAGTAATAATGTCATCTGTGGCTTCACACATTATT-3′, were annealed and extended in a PCR reaction. After gel purification, the PCR product was in vitro transcribed by T7 RNA polymerase (Promega, Madison, WI). The resulting RNA was then dephosphorylated by Calf Intestinal Phosphatase (New England BioLabs, Ipswich, MA) and purified using Trizol again. The precursor of human miR-30a (31) was used as a negative control and was identically prepared. 293T cells, maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum under 5% CO2 at 37oC, were typically transfected with 1–2 ng of the CYP2A3 reporter plasmid, 1 ng of pRL-CMV (Promega, as internal control) and 50–100 ng of RNA using Lipofectamine 2000 reagent (Invitrogen). Two days after transfection, luciferase activities were measured by the Dual-Luciferase Assay System (Promega).
Detection of CYP2A3 by western blotting
Approximately 100 mg of frozen rat lung tissue was pulverized in a mortar containing liquid nitrogen with a pestle. Pulverized tissue was then transferred to a 7 ml Kontes glass homogenizer and dounced in 1 ml lysis buffer containing 50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 150 mM NaCl, 15 mM MgCl2, 0.1% TritonX-100 and 8 M urea. Homogenate was clarified by centrifugation. Approximately 60 μg of total protein was fractionated on a 4–12% Bis–Tris mini gel (Invitrogen), transferred to a membrane and probed with a CYP2A6 antibody that also recognizes other CYP2A family members (BD Biosciences, San Jose, CA), stripped and then probed with an actin antibody (Cell Signaling, Danvers, MA) for loading controls. Western blot signals were quantified using ImageJ (National Institutes of Health).
Results
Short-term, chronic treatment of rats with NNK
The goal of this study was to identify miRNA expression changes associated with the early-stage neoplastic transformation in the lungs. We used a well-studied rodent cancer model to mimic lung cancer induction in humans. Male F344 rats were systemically and chronically fed with a low concentration of NNK, a known carcinogen present in tobacco products and smoke, in the drinking water for up to 20 weeks. There is no significant difference in water consumption or body weights between the control and NNK groups during these 20 weeks (32). Under these conditions, DNA adducts form and accumulate in cells (32), but it would take up to 2 years for the rats to fully develop tumors in multiple organs, including the lungs (16). Rats were killed at week 1, 5, 16 and 20 after the initiation of NNK treatment. We then isolated total RNA from the lungs. We were able to extract good-quality RNAs from a total of 13 control animals: four 1-week rats, two 5-week rats, three 16-week rats and four 20-week rats, as well as from 16 NNK-treated animals: four 1-week rats, five 5-week rats, two 16-week rats and five 20-week rats.
Microarray analyses of miRNA expression
We employed custom microarray technique to profile global miRNA expression in the lungs. For the above total 29 RNA samples, we performed four sets of RNA labeling and microarray hybridization experiments. To minimize bias, due to our conducting the experiments on different days, with different reagents, and on different print batches of microarray slides, we always processed at least one group of control samples and at least one group of NNK-treated samples from different time points simultaneously, to generate a data set. Because of the relatively small number of animals used in this study, time-dependent changes in miRNA expression could not be confidently resolved. As a result, we focused on the comparisons between control and NNK treatments; a change in miRNA expression was defined as being consistently observed in the majority of the time points, e.g. at least three, usually four, of the total four time points, with data of good quality given more weight. From each individual data set, we acquired a list of miRNAs that might increase or decrease their expression in response to NNK. We then identified a common list of miRNAs that consistently changed their expression levels in NNK-treated rats, based on the total four data sets of biological replicates. An alternative approach would be to combine all four data sets to compare miRNA expression between control and NNK-treated samples. For this study of a small size of samples, the latter approach was disproportionately affected by the occasionally abnormal data points, so its results are not presented here.
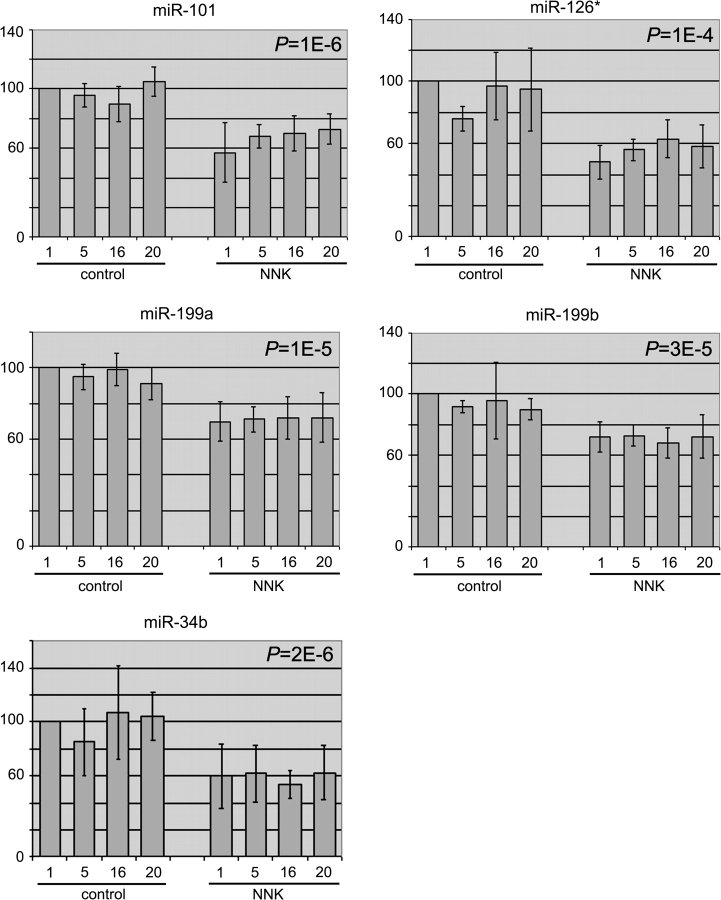
At the conclusion of our microarray experiments, we found that most miRNAs largely maintained their normal expression levels during NNK treatment up to 20 weeks. Nonetheless, a small number of miRNAs, i.e. miR-101, miR-126*, miR-199a, miR-199b and miR-34b, was downregulated in NNK-treated rats when compared with controls. Figure 1 summarizes the expression of these miRNAs, as normalized by the percentile method. Normalization to 28S ribosomal RNA or U6 snRNA reached similar conclusions (supplementary Figures 1–6 are available at Carcinogenesis Online). Reduction in miRNA expression was already apparent in rats treated with NNK for only 1 week, and the expression levels were mostly constant through 20 weeks of treatment, with the extent of reduction being 30–50% of control levels. Changes in the expression of miR-199* and miR-126, which are processed from the same precursor RNAs as miR-199 and miR-126*, respectively (1), were not sufficiently consistent (data not shown), so they were not examined further for this study.
Fig. 1.
Relative miRNA expression determined by microarray studies. Expression levels of the indicated miRNAs in the lungs of 1-, 5-, 16- and 20-week, control and NNK-treated rats were compared with those in the control, 1-week rats, which were arbitrarily set at 100 (y-axis). Shown are the averages, standard deviations (error bars) and P-values.
Confirmation of microarray results by Northern blot and real-time PCR analyses
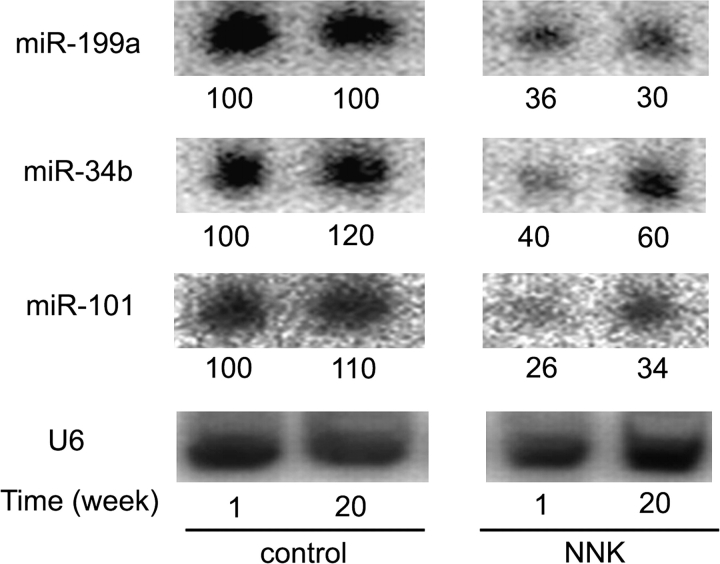
We first used Northern blotting to determine the relative expression levels of individual miRNAs. Figure 2 shows that this method confirmed the reduced expression of miR-199a (as a representative of both miR-199a and miR-199b), miR-34b and miR-101 in the lungs of NNK-treated rats. miR-34b expression varied in NNK-treated rats, but the differences were minor and probably due to uncertainty in quantifying weak Northern blot signals or variations of individual animals. Signals for miR-126* were too weak to quantify accurately (data not shown).
Fig. 2.
Northern blot validation of microarray results. Total RNAs pooled from two rats at 1 or 20 weeks of the same treatments were separated on a denaturing gel and transferred to a membrane. The membrane was then probed for miRNAs or the control U6 snRNA. Results were analyzed using a PhosphorImager. Levels of the indicated miRNAs, after normalized to the U6 signals and compared with those of the control, 1-week RNA arbitrarily set at 100, were shown below the phosphorimages.
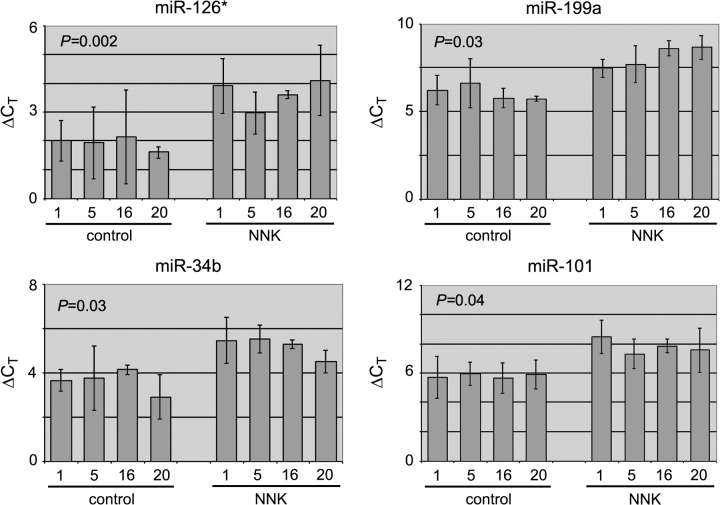
We also preformed reverse transcription and real-time PCR to measure miRNA expression in the lungs. Real-time PCR yielded values for cycle threshold, or CT, an inverse indicator of RNA abundance. As indicated by the elevated ΔCT values in NNK-treated samples compared with control samples, NNK reduced the expression of miR-101, miR-126*, miR-199a and miR-34b (Figure 3). We also examined the expression of two other miR-34 family members, miR-34a and miR-34c, by real-time PCR. We found that their expression was also reduced, although with only marginal, statistical significance (supplementary Figure 7 is available at Carcinogenesis Online). Future studies with more animals will establish whether all miR-34 family members are reduced by NNK treatment. Overall, the degree of NNK-induced reduction in miRNA expression as determined by real-time PCR appeared to be larger than that by microarray or Northern blot analysis, assuming that the PCR amplification rates were ideally two per cycle. This phenomenon is well documented and due, at least in part, to the fact that real-time PCR method is more sensitive and has a much wider dynamic range. Based on our microarray, Northern blot and real-time PCR studies, we conclude that miR-34b, miR-101, miR-126* and miR-199 have lowered expression levels very early in the lungs of male F344 rats chronically treated with the tobacco-specific carcinogen NNK.
Fig. 3.
Real-time PCR analyses of miRNA expression. RNA samples were reverse transcribed and then amplified using miRNA-specific primers or a primer for the normalization control, U6 snRNA. ΔCT values (y-axis) for the miRNAs were calculated as CT-miRNA−CT-U6. Averages, standard deviations (error bars) and P-values from two to three biological replicates are shown.
Identification of CYP2A3 as a potential target of miR-126*
miRNAs function primarily by binding to the complementary sequences in the 3′ UTRs of their target mRNAs to inhibit translation in animal cells (1–4). As an effort to understand how changes in miRNA expression contributed to NNK-induced tumorigenesis, we searched miRBase (33) for computer program-predicted targets of the aforementioned miRNAs. We noted that the human and frog CYP2A13 genes are prominently predicted to be targeted by miR-126*. CYP2A13, a member of the CYP family of enzymes, is a major P450 enzyme in the respiratory system that catalyzes the α-hydroxylation reactions of NNK and its metabolites, which activate these chemicals and render them mutagenic (20). Therefore, CYP2A13 is critically involved in eliciting the biological effects of NNK. The orthologues of CYP2A13 in rodents are CYP2A3 in rats and CYP2A4 and CYP2A5 in mice. Although miRBase does not predict CYP2A3, CYP2A4 and CYP2A5 as targets of miR-126*, the 3′ UTRs of their mRNAs nevertheless contain conserved sequences that are partially complementary to miR-126* (Figure 4A). In a reporter assay performed on transiently transfected 293T cells, miR-126* repressed the expression of a firefly luciferase gene containing the 3′ UTR of CYP2A3 mRNA (Figure 4B, left columns). Deleting much of the predicted miR-126*-binding site in the 3′ UTR abrogated the inhibitory effect of miR-126* on reporter expression (Figure 4B, right columns), demonstrating the specificity of the interaction between CYP2A3 mRNA and miR-126*.
Fig. 4.
CYP2A3 as a potential target of miR-126*. (A) Sequence alignment between miR-126* and its partially complementary 3′ UTRs of CYP2A family members from several species. Underlined nucleotides were deleted in the mutant CYP2A3 reporter construct; a relatively large deletion was necessary to disrupt the interaction between miR-126* and the 3′ UTR. (B) Inhibition of reporter expression by miR-126*. 293T cells were cotransfected with a reporter construct expressing the firefly luciferase linked to the wild-type or mutant CYP2A3 3′UTR, pRL-CMV and short hairpin RNA that expressed a control miRNA or miR-126*. Relative reporter expression is represented by firefly luciferase activities divided by the Renilla luciferase activities expressed from pRL-CMV, set at 100% for the control RNA transfections. Shown are the averages and standard deviations (error bars) of three to four experiments. The P-value was calculated using two-tailed, paired Student’s t-test. (C) CYP2A3 mRNA expression in the lungs of 1-, 5-, 16- and 20-week, control and NNK-treated rats, as measured by real-time PCR. The β-actin mRNA was the normalization control, and ΔCT values (y-axis) were calculated as CT-CYP2A3−CT-actin. Averages and standard deviations (error bars) are shown. To assess the differences in CYP2A3 mRNA expression based on the ΔCT values in control and NNK-treated, 16- and 20-week rats, two-tailed, heteroscedastic Student’s t-test was performed to yield a P-value of 0.003. (D) Protein expression in the lungs of individual, control and NNK-treated rats, as detected by western blotting. The ratios between CYP2A3 and actin band intensities are listed on top. Shown is a representative of three experiments that gave similar results.
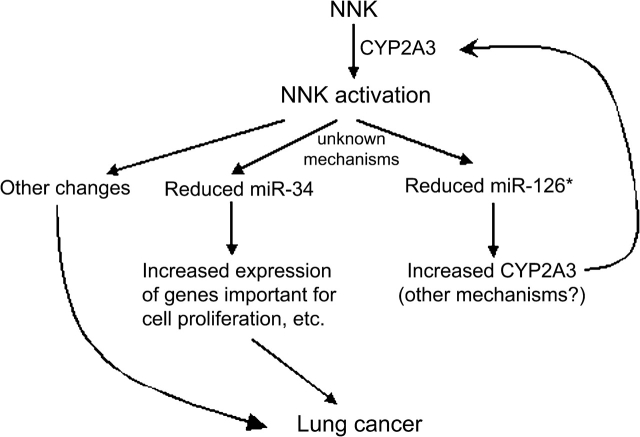
If miR-126* represses CYP2A3 expression, and NNK reduces miR-126* levels, then CYP2A3 expression should increase in NNK-treated rats. Because miRNA-mediated inhibition of mRNA translation sometimes leads to the subsequent degradation of target mRNAs (34,35), we first measured CYP2A3 mRNA levels in the lungs of control and NNK-treated rats by real-time PCR. As shown by the lower ΔCT values in Figure 4C, CYP2A3 mRNA was elevated in 16- and 20-week, NNK-treated rats. We then analyzed CYP2A3 protein expression in the same tissue samples by western blotting. Because a specific antibody for CYP2A3 is not available, we used a commercial CYP2A6 antibody that cross-reacts with other members of the CYP2A family, with the understanding that CYP2A3 is the principal P450 2A enzyme in rat lungs (20). Figure 4D shows that CYP2A3 protein levels were higher in NNK-treated rats than that in control rats. These results are consistent with the putative role of miR-126* in repressing the expression of CYP2A3. A model is presented in Figure 5 to rationalize our findings in this study and is explained below.
Fig. 5.
A model to explain some of the consequences of NNK exposure. When administered to rats, NNK is activated by CYP2A3 in the lungs, which then reduces the expression of certain miRNAs such as miR-126* and miR-34. CYP2A3 is itself a target of miR-126*, so lowered miR-126* contributes to the increased production of CYP2A3, further activating NNK in a positive feedback loop. As miR-34 represses the expression of many genes involved in cell cycle control, apoptosis, etc., loss of miR-34 deregulates those important genes. These proposed molecular changes, together with others, eventually lead to the development of lung cancer.
Discussion
Several studies have compared miRNA profiles in well-developed human lung cancer and normal lung tissues and reported differential expression of numerous miRNAs in cancerous tissues (7–11). Here, we examined miRNA expression changes associated with NNK treatment that mimicked the early stages of lung cancer development, using an animal model in which male F344 rats were chronically administered with NNK, a potent, tobacco-specific carcinogen, for up to 20 weeks. miRNAs identified in this study, i.e. miR-34b, miR-101, miR-126* and miR-199 (Figures 1–3), were likewise found to be reduced in human cancers (10,11). As the length of time required for pulmonary adenocarcinoma development in our model is >100 weeks (16), the aforementioned miRNAs might be the early-response genes in lung cancer induction and could indicate early biomarkers for lung cancer. Alteration in the expression of miR-199*, miR-126 and the let-7 family of miRNAs, while identified in published reports (7–10), was not consistently observed in this study. On the other hand, we failed to detect any clear changes in the expression of other prominent miRNAs such as miR-155. There could be several reasons for the omissions. First, our ability to identify all the significantly changed miRNAs might be limited by the relatively small number of animals we examined, which is far lower than the number of human tissues tested in previous studies (9,10). Many miRNAs might not change their expression levels until later stages in pulmonary tumorigenesis. miRNA expression changes could arise from gene amplification or deletion, which might not have occurred in the rats by the end of our study. Lastly, there are dozens of carcinogens besides NNK in tobacco products that presumably also effect miRNA expression.
How miR-34b, miR-101, miR-126* and miR-199 contribute to lung cancer development is a question that necessitates extensive, future studies. The finding that miR-126* may regulate the expression of CYP2A3 (Figure 4) suggests that the early changes in miRNA expression indeed have important biological consequences. Rat CYP2A3 is believed to be the principal catalyst in the lungs of NNK α-hydroxylation, the primary bioactivation pathway for NNK (20). In concordance with reduced miR-126* expression, enhanced CYP2A3 expression in rats following NNK treatment, as demonstrated here, could theoretically reinforce NNK genotoxicity (Figure 5). Of course, we cannot exclude the possibility that the induction of CYP2A3 is controlled by multiple mechanisms. Another miRNA with clearly interesting implications is miR-34. A transcription target of the tumor suppressor p53, miR-34 inhibits the expression of a large number of genes involved in DNA damage response, cell cycle progression and apoptosis (11,36–40). p53 mRNA expression did not change in our NNK-treated rats (data not shown), but p53 function is known to be regulated at multiple levels. Regardless of the mechanisms leading to the reduction in miR-34 expression, loss of miR-34 could upregulate many genes that control important processes such as cell proliferation and cell death, which probably, over the long term, contribute to lung cancer formation (Figure 5).
In summary, our findings present several exciting avenues for further research. miRNA expression analysis of NNK-treated rats resulted in the identification of a number of miRNAs whose expression changes are consistent with those associated with human lung cancer, providing important etiological clues into the temporal sequence of miRNA expression related to NNK tumorigenesis and on the stages at which the functions of these miRNAs might begin to interface with lung tumorigenesis. The identification of CYP2A3 as a potential target of miR-126* suggests a mechanism by which NNK exerts and augments its genotoxic potentials in the lung. Our results further indicate that our animal system is well suited for modeling the molecular changes associated with human lung cancer progression. Future research will follow miRNA expression till the formation of lung cancer in rodents and study the functions of miRNAs in lung cancer development, as well as investigate the possibility that some of these miRNAs and the target genes or pathways regulated by the miRNAs might constitute early diagnostic markers for human lung cancers.
Supplementary material
Supplementary Figures 1–7 can be found at http://carcin.oxfordjournals.org/
Funding
National Institute on Drug Abuse (DA 011806); United States Department of Defense (W81XWH-07-1-0183) to Y.Z.
Acknowledgments
We thank Dr Stephen Hecht for advice, Dr Sabita Roy for the use of her real-time PCR machine, Dr Ann Fallon for allowing us to borrow her luminometer and Dr Fekadu Kassie for assistance with the animal study. We would also like to thank the laboratory of Dr Sharon Murphy and Jeannette Zinggeler Berg for the generous gift of CYP2A6 antibody and assistance with western analysis.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CYP
cytochrome P450
- miRNA
microRNA
- mRNA
messenger RNA
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- PCR
polymerase chain reaction
- snRNA
small nuclear RNA
- UTR
untranslated region
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Engels BM, et al. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 3.Du T, et al. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 4.Valencia-Sanchez MA, et al. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Carthew RW. Gene regulation by microRNAs. Curr. Opin. Genet. Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 8.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 11.Bommer GT, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 12.Spira A, et al. Extensive-stage small-cell lung cancer. Semin. Surg. Oncol. 2003;21:164–175. doi: 10.1002/ssu.10034. [DOI] [PubMed] [Google Scholar]
- 13.Parkin DM, et al. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 14.International Agency for Research on Cancer. Tobacco Smoke and Involuntary Smoking. IARC Monographs on the Evaluation of Carcinogenic Risk to Humans. Vol. 83. Lyon: IARC Press; 2004. pp. 53–119. [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control. Cigarette smoking among adults–United States, 2006. MMWR Morb. Mortal. Wkly Rep. 2007;56:1157–1161. [PubMed] [Google Scholar]
- 16.Rivenson A, et al. Induction of lung and exocrine pancreas tumors in F344 rats by tobacco-specific and Areca-derived N-nitrosamines. Cancer Res. 1988;48:6912–6917. [PubMed] [Google Scholar]
- 17.Spiegelhalder B, et al. Tobacco-specific nitrosamines. Eur. J. Cancer Prev. 1996;5:33–38. [PubMed] [Google Scholar]
- 18.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific nitrosamines. Chem. Res. Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 19.Harris JE. Smoke yields of tobacco-specific nitrosamines in relation to FTC tar level and cigarette manufacturer: analysis of the Massachusetts Benchmark Study. Public Health Rep. 2001;116:336–343. doi: 10.1093/phr/116.4.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jalas JR, et al. Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem. Res. Toxicol. 2005;18:95–110. doi: 10.1021/tx049847p. [DOI] [PubMed] [Google Scholar]
- 21.Hecht SS, et al. Tobacco-specific nitrosamines, an important group of carcinogens in tobacco and tobacco smoke. Carcinogenesis. 1988;9:875–884. doi: 10.1093/carcin/9.6.875. [DOI] [PubMed] [Google Scholar]
- 22.Preston-Martin S, et al. Epidemiological evidence for the role of nirtroso compounds in human cancer. Cancer Surv. 1989;8:459–473. [PubMed] [Google Scholar]
- 23.Hoffmann D, et al. Advances in tobacco carcinogenesis. In: Cooper CS, Grover PI, editors. Handbook of Experimental Pharmacology. Heidelberg, Germany: Springer-Verlag; 1990. pp. 63–102. [Google Scholar]
- 24.Bartsch H, et al. Environmental exposure to N-nitroso compounds (NNOC) and precursors: an overview. Eur. J. Cancer Prev. 1996;5:11–18. [PubMed] [Google Scholar]
- 25.Magee PN. Nitrosamines and human cancer: introduction and overview. Eur. J. Cancer Prev. 1996;5:7–10. doi: 10.1097/00008469-199609001-00002. [DOI] [PubMed] [Google Scholar]
- 26.Hecht SS, et al. Effects of alpha-deuterium substitution on the mutagenecity of 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) Carcinogenesis. 1983;4:305–310. doi: 10.1093/carcin/4.3.305. [DOI] [PubMed] [Google Scholar]
- 27.Thomson JM, et al. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y, et al. Both natural and designed microRNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 29.Kasahara T, et al. Increased expression of cyclin-dependent kinase-interacting protein p21 during tamoxifen-induced hepatocarcinogenesis in female rats. J. Health Sci. 2005;51:185–190. [Google Scholar]
- 30.Zeng Y, et al. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng Y, et al. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lao Y, et al. Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 2007;20:235–245. doi: 10.1021/tx060207r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 35.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 36.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corney DC, et al. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 38.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.