Abstract
Therapeutic strategies based on modulation of microRNA (miRNA) activity hold great promise due to the ability of these small RNAs to potently influence cellular behavior. In this study, we investigated the efficacy of a miRNA-based therapy for liver cancer. We demonstrate that hepatocellular carcinoma (HCC) cells exhibit reduced expression of miR-26a, a miRNA that is normally expressed at high levels in diverse tissues. Expression of this miRNA in liver cancer cells in vitro induces cell-cycle arrest associated with direct targeting of cyclins D2 and E2. Systemic administration of this miRNA in a mouse model of HCC using adeno-associated virus (AAV) results in inhibition of cancer cell proliferation, induction of tumor-specific apoptosis, and dramatic protection from disease progression without toxicity. These findings suggest that delivery of miRNAs that are highly expressed and therefore tolerated in normal tissues but lost in disease cells may provide a general strategy for highly-specific miRNA-based therapies.
Introduction
MicroRNAs (miRNAs) are a diverse class of highly conserved small RNA molecules that function as critical regulators of gene expression in multicellular eukaryotes and some unicellular eukaryotes (Kato and Slack, 2008). miRNAs are initially transcribed as long primary transcripts (pri-miRNAs) that undergo sequential processing by the RNase III endonucleases Drosha and Dicer to yield the mature ~20–23 nucleotide species (Kim, 2005). Mature miRNAs associate with the RNA-induced silencing complex (RISC) and interact with sites of imperfect complementarity in 3' untranslated regions (UTRs) of target mRNAs. Targeted transcripts subsequently undergo accelerated turnover and translational repression (Valencia-Sanchez et al., 2006). Importantly, the ability of individual miRNAs to regulate hundreds of transcripts allows these RNAs to coordinate complex programs of gene expression and thereby induce global changes in cellular physiology. Indeed, a growing body of evidence has documented that miRNAs provide functions essential for normal development and cellular homeostasis and accordingly, dysfunction of these molecules has been linked to several human diseases (Kloosterman and Plasterk, 2006).
Over the last five years, a particularly important role for miRNAs in cancer pathogenesis has emerged. Virtually all examined tumor types are characterized by globally abnormal miRNA expression patterns (Calin and Croce, 2006). Profiles of miRNA expression are highly informative for tumor classification, prognosis, and response to therapy. Moreover, recent results have documented a functional contribution of specific miRNAs to cellular transformation and tumorigenesis. For example, miRNAs are known targets of genomic lesions that frequently activate oncogenes and inactivate tumor suppressors in cancer cells such as amplification, deletion, and epigenetic silencing (Calin et al., 2004; Lujambio and Esteller, 2007; Zhang et al., 2006). Additionally, miRNAs provide critical functions downstream of classic oncogenic and tumor suppressor signaling pathways such as those controlled by Myc and p53 (Chang et al., 2008; He et al., 2007b; O'Donnell et al., 2005). Finally, functional studies have directly documented the potent pro- and anti-tumorigenic activity of specific miRNAs both in vitro and in vivo (Ventura and Jacks, 2009).
As a consequence of the important functions provided by miRNAs in cancer cells, potential therapeutic approaches that target this pathway have recently attracted attention. Although significant focus in this area has been directed towards antisense-mediated inhibition of oncogenic miRNAs (Love et al., 2008; Stenvang et al., 2008), several lines of evidence suggest that miRNA replacement represents an equally viable if not more efficacious strategy. Although specific miRNAs are often overexpressed in cancer cells, most miRNAs are downregulated in tumors (Gaur et al., 2007; Lu et al., 2005). Global miRNA repression enhances cellular transformation and tumorigenesis in both in vitro and in vivo models (Kumar et al., 2007), underscoring the pro-tumorigenic effects of miRNA loss-of-function. Moreover, we have previously demonstrated that hyperactivity of Myc, a common occurrence in diverse tumor types, leads to widespread miRNA repression (Chang et al., 2008). Enforced expression of individual miRNAs in lymphoma cells transformed by Myc or other oncogenes dramatically suppresses tumorigenesis. These observations suggest that re-expression of even a single miRNA in tumor cells could provide significant therapeutic benefit. Supporting this notion, two recent reports demonstrated that viral delivery of let-7 miRNAs suppressed tumor growth in a mouse model of lung adenocarcinoma (Esquela-Kerscher et al., 2008; Kumar et al., 2008). Importantly, let-7 directly targets KRAS, the initiating oncogene in this tumor model (Johnson et al., 2005). Thus, the utility of miRNAs as more general anti-cancer therapeutics in situations where they do not target the initiating oncogene remains to be studied.
Hepatocellular carcinoma (HCC) is the third-leading cause of death from cancer and the fifth most common malignancy worldwide (Roberts, 2008; Thorgeirsson and Grisham, 2002). HCC is often diagnosed at an advanced stage when it is no longer amenable to curative therapies. Highly active drug-metabolizing pathways and multi-drug resistance transporter proteins in tumor cells further diminish the efficacy of current therapeutic regimens for this cancer type. Alternative approaches are therefore needed to overcome these barriers to successful therapy. Fortunately, the liver is well-suited for such alternative strategies since it is easily targeted by both viral and non-viral gene and small-molecule delivery systems (Alexander et al., 2008; Pathak et al., 2008). In this regard, gene therapy vectors based on adeno-associated virus (AAV) are particularly promising. Recent advances in AAV vector technology include a self-complementary genome which enhances therapeutic gene expression and non-human primate AAV serotypes which facilitate efficient transduction following vascular delivery. Significantly, these improvements allow 90–100% transduction of hepatocytes and long-term gene expression without toxicity following a single systemic administration of recombinant virus (McCarty et al., 2003; Nakai et al., 2005; Rodino-Klapac et al., 2007; Wang et al., 2003). Due to their small size, regulatory RNAs are especially amenable to AAV-mediated delivery.
In this study, we tested the hypothesis that miRNAs can be used as general anti-cancer therapeutics through their effects on tumor cell proliferation and death even when they do not target the initiating oncogene. We reasoned that the most therapeutically useful miRNAs would be expressed at low levels in tumors but would be highly expressed, and therefore tolerated, in normal tissues. miR-26a fulfills these criteria, exhibiting high expression in normal adult liver but low expression in both human and murine liver tumors. miR-26a directly downregulates cyclins D2 and E2 and induces a G1 arrest of human liver cancer cells in vitro. AAV-mediated miR-26a delivery potently suppresses cancer cell proliferation and activates tumor-specific apoptosis in vivo, resulting in dramatic suppression of tumor progression without toxicity. These findings provide proof-of-concept support for systemic delivery of tumor-suppressing miRNAs as a powerful and highly specific anti-cancer therapeutic modality.
Results
Downregulation of putative anti-tumorigenic miRNAs in Myc-induced liver tumors
We previously demonstrated that Myc activation in B cell lymphoma models results in downregulation of a large cohort of miRNAs including miR-15a/16, miR-26a, miR-34a, miR-150, and miR-195 (Chang et al., 2008). Enforced expression of these specific miRNAs dramatically inhibits B cell lymphomagenesis. In order to extend these findings to a solid tumor model and to investigate the potential use of these miRNAs as anti-cancer therapeutics, we studied their expression in a previously described model of liver cancer in which mice harboring a tetracycline (tet)-repressible MYC transgene (tet-o-MYC) are crossed with mice expressing the tet-transactivator protein (tTA) driven by the liver activator promoter (LAP) (Beer et al., 2004; Felsher and Bishop, 1999; Shachaf et al., 2004). Upon removal of doxycycline (dox), bi-transgenic animals express MYC specifically in the liver and subsequently develop liver tumors resembling HCC with complete penetrance.
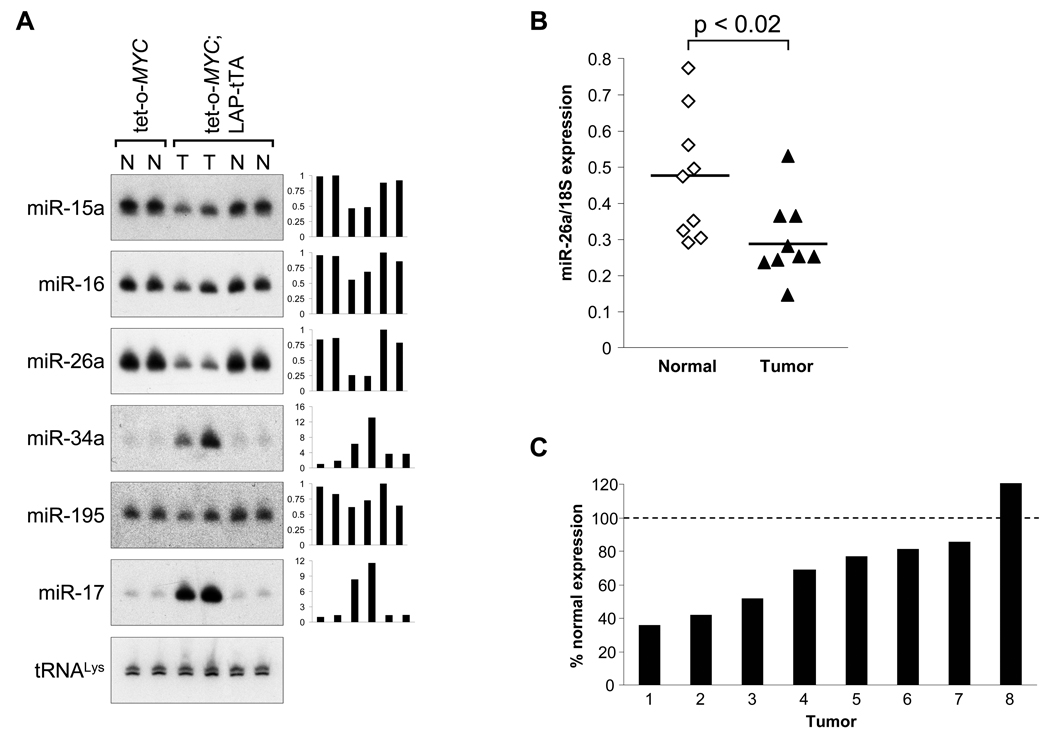
We sought miRNAs that are highly expressed and therefore tolerated in normal tissues but are expressed at reduced levels in tumors. Such miRNAs might exhibit anti-proliferative and pro-apoptotic effects that are restricted to cancer cells. Northern blotting revealed that miR-26a, and to a lesser extent miR-15a and miR-16, fulfilled these criteria, exhibiting high expression in normal liver from adult mice lacking the LAP-tTA transgene (harboring an inactive tet-o-MYC transgene) but low expression in liver tumors from tet-o-MYC; LAP-tTA bi-transgenic animals (Figure 1A). Notably, these miRNAs did not exhibit reduced expression in normal-appearing liver from bi-transgenic animals, consistent with the previous demonstration that Myc levels are only minimally increased in non-tumor tissue in these mice (Shachaf et al., 2004). Additional miRNAs with anti-tumorigenic activity in B lymphoma cells were expressed at approximately equivalent levels in normal liver and tumors (miR-195, Figure 1A) or were not detectable in these tissues (miR-150, data not shown). miR-34a was strongly upregulated in liver tumors (Figure 1A), perhaps reflecting its regulation by p53 which is retained and active in some tumors in these animals (Beer et al., 2004; He et al., 2007b). As expected, miR-17 was expressed at high levels in liver tumors, consistent with our previous demonstration that the miR-17-92 cluster is directly transactivated by Myc (O'Donnell et al., 2005).
Figure 1. Dysregulated expression of miRNAs in mouse and human liver tumors.
(A) Northern blot analysis of miRNA expression in normal liver (N) or tumor tissue (T) from mice of the indicated genotypes. Graphs depict relative quantification of miRNA levels normalized to tRNALys abundance.
(B) qPCR analysis of miR-26a expression in human HCC and normal liver biopsies. miRNA abundance was normalized to 18S rRNA expression. p value calculated by two-tailed t test.
(C) miR-26a expression in individual HCC tumors relative to expression in paired normal liver samples.
Because miR-26a exhibited the most dramatic downregulation in liver tumors in bi-transgenic animals, we further examined its expression in a panel of human HCC samples. Consistent with the mouse tumor data, miR-26a was expressed at statistically-significantly lower levels in human tumors compared to normal human liver tissue (Figure 1B). Paired normal liver biopsy material was available for a subset of the HCC samples. 7/8 of these samples showed reduced miR-26a expression relative to the associated normal liver control (Figure 1C). Based on these results from human and mouse liver tumors, we selected miR-26a for functional studies to evaluate its anti-tumorigenic properties and potential therapeutic utility for liver cancer in vitro and in vivo.
miR-26a expression induces a G1 arrest in human liver cancer cells
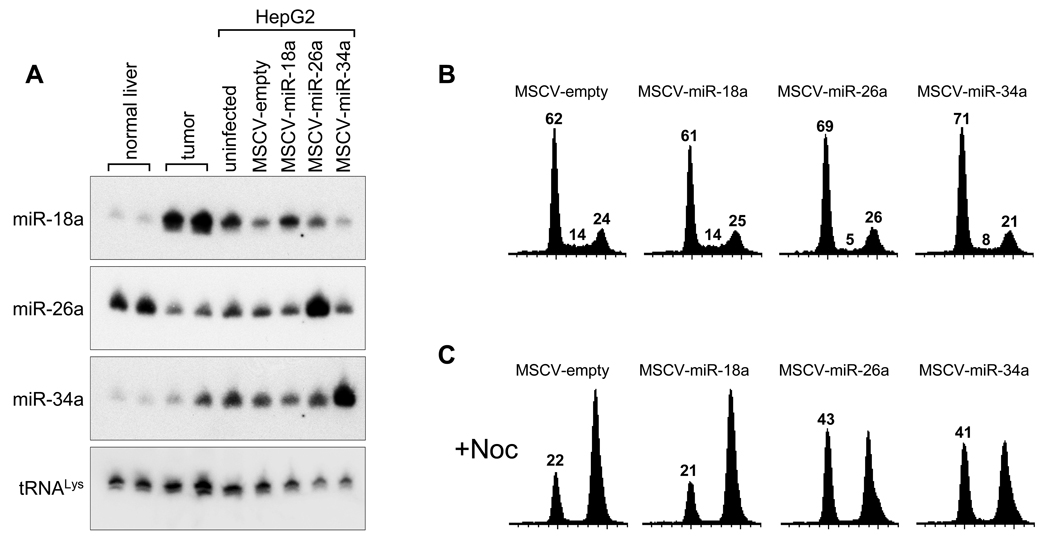
As an initial test of the anti-proliferative properties of miR-26a in liver cancer cells, we utilized a murine stem cell virus (MSCV)-derived retroviral construct to enforce expression of this miRNA in HepG2 cells. As controls for this experiment, we used viruses that express miR-18a, a component of the pro-tumorigenic miR-17-92 cluster, and miR-34a, which is known to have potent anti-proliferative and pro-apoptotic activity in other cell lines (Bommer et al., 2007; Chang et al., 2007; He et al., 2007a; Raver-Shapira et al., 2007; Tarasov et al., 2007). Northern blotting demonstrated that miR-26a expression levels in HepG2 cells closely mirror expression levels in tumors from tet-o-MYC; LAP-tTA animals (Figure 2A). Infection with MSCV-miR-26a results in enforced expression of this miRNA at a level comparable to that observed in normal liver tissue. Flow cytometric analysis of retrovirally-infected cell populations revealed fewer cells in S phase and increased numbers of cells in G1 following infection with MSCV-miR-26a or MSCV-miR-34a compared to cells infected with MSCV-empty or MSCV-miR-18a, suggesting that miR-26a and miR-34a induce a G1 arrest (Figure 2B). To more accurately quantify the numbers of cells arrested in G1, we treated cells with the microtubule-destabilizing agent nocodazole which traps cycling cells in M phase. Cell populations with enforced miR-26a or miR-34a expression were characterized by significantly increased numbers of cells remaining in G1 (Figure 2C), confirming that these miRNAs arrest the cell-cycle at this stage.
Figure 2. miR-26a induces a G1 arrest in human hepatocellular carcinoma cells.
(A) Northern blots documenting miRNA expression levels in normal liver and tumors from tet-o-MYC; LAP-tTA mice and in uninfected or retrovirally-infected HepG2 cells. tRNALys served as a loading control.
(B) Cell-cycle profiles of retrovirally-infected HepG2 cells as determined by propidium-iodide (PI) staining and flow cytometry. Numbers over each histogram indicate the percentage of cells in G1, S, and G2 cell-cycle phases.
(C) Cell-cycle profiles of retrovirally-infected HepG2 cells following treatment with nocodazole (Noc). Numbers over each histogram indicate the percentage of cells remaining in G1.
miR-26a directly represses expression of cyclin D2 and cyclin E2
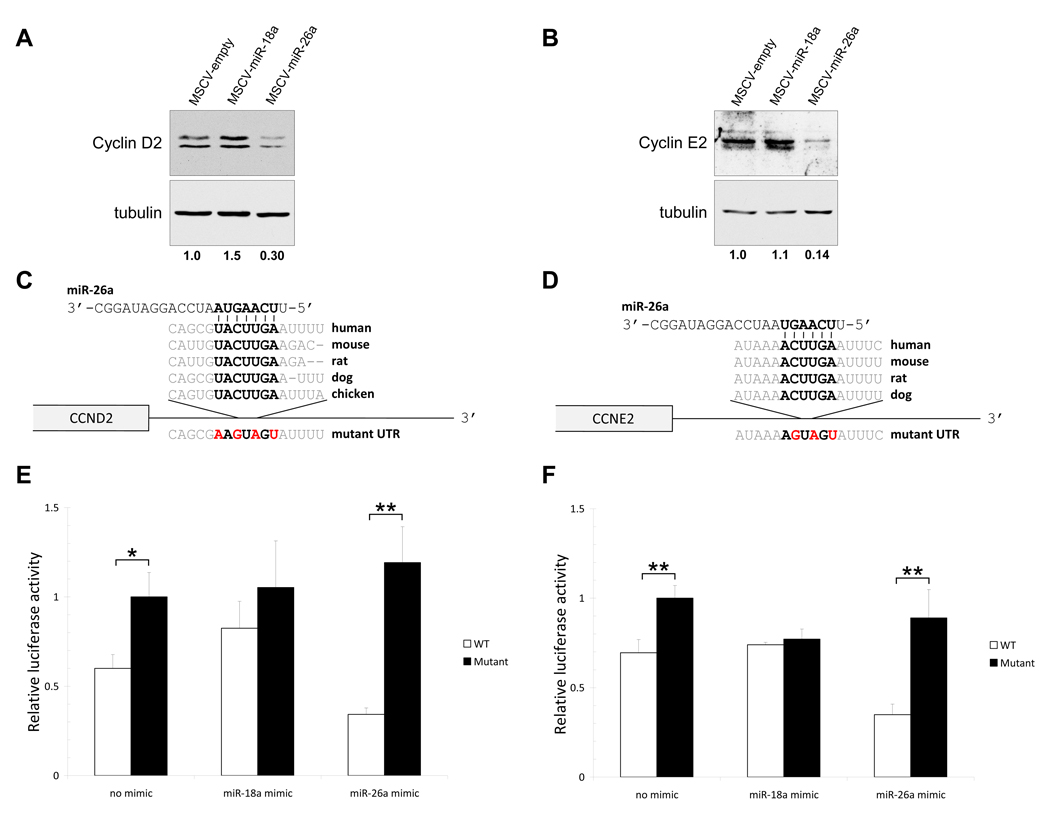
To investigate the mechanisms through which miR-26a induces a G1 cell-cycle arrest, we examined predicted targets of this miRNA using the Targetscan algorithm (Grimson et al., 2007). This analysis predicts that miR-26a regulates cyclin E1 (CCNE1), cyclin E2 (CCNE2), cyclin D2 (CCND2), and cyclin-dependent kinase 6 (CDK6), all of which play a critical role in transition through the G1-S checkpoint (Vermeulen et al., 2003). Western blotting was used to determine if miR-26a represses any of these putative targets in retrovirally-infected cell populations. Although infection with MSCV-miR-26a did not affect the abundance of cyclin E1 and CDK6 (data not shown), significantly reduced levels of cyclin D2 and cyclin E2 were observed in cells with enforced miR-26a expression (Figure 3A,B).
Figure 3. miR-26a negatively regulates cyclins D2 and E2.
(A–B) Western blots documenting abundance of cyclins D2 and E2 in retrovirally-infected HepG2 cells. Relative quantification of band intensities, normalized to tubulin levels, are shown below blots.
(C–D) Sequence and evolutionary conservation of the miR-26a binding sites in the 3' UTRs of transcripts encoding cyclin D2 (CCND2) and cyclin E2 (CCNE2). Mutations introduced into luciferase reporter constructs are shown in red.
(E–F) Relative firefly luciferase activity derived from CCND2 (E) and CCNE2 (F) 3' UTR reporter constructs following transfection into HepG2 cells alone or in combination with miR-18a or miR-26a synthetic miRNA mimics. All values were normalized to renilla luciferase activity produced from a co-transfected control plasmid. Error bars represent standard deviations from 3 independent transfections. *, p<0.05; **, p<0.01 (two-tailed t test).
Both CCND2 and CCNE2 have a single predicted miR-26a binding site in their 3' UTRs which are highly conserved in mammals and, in the case of CCND2, present in chicken (Figure 3C,D). To verify that these transcripts are directly regulated by miR-26a, reporter plasmids were constructed in which portions of the 3' UTRs encompassing the predicted binding sites, with or without mutations that would disrupt miRNA interaction, were cloned downstream of a luciferase open reading frame. When introduced into HepG2 cells, constructs with intact miR-26a binding sites were expressed at significantly lower levels than the mutant constructs, consistent with direct functional interaction of endogenously-expressed miR-26a with these sites (Figure 3E,F). Co-transfection of reporter plasmids with a synthetic miR-26a mimic further repressed luciferase activity produced from wild-type, but not mutant, reporter constructs. Demonstrating the specificity of these effects, co-transfection with a miR-18a mimic actually diminished downregulation of the wild-type reporters, possibly by competing for limiting miRNA pathway components and thereby relieving the transcript from repression. These data document that miR-26a directly represses expression of cyclin D2 and cyclin E2, providing one mechanism through which this miRNA arrests cell-cycle progression.
MYC is not a target of miR-26a
Having demonstrated that miR-26a potently arrests proliferation of hepatocellular carcinoma cells in vitro, we next initiated a series of experiments to assess whether systemic delivery of this miRNA could be used as a therapeutic strategy for this tumor type in vivo. The tet-o-MYC; LAP-tTA liver cancer model represents an ideal setting for these studies. Because these mice were constructed with a human MYC transgene that includes its 3' UTR (Felsher and Bishop, 1999), we first confirmed that miR-26a does not regulate MYC itself. MYC is not a predicted target of miR-26a according to several commonly used algorithms including Targetscan, miRanda, and PicTar (Betel et al., 2008; Grimson et al., 2007; Krek et al., 2005). Manual inspection of the human MYC 3' UTR further documents the absence of even a minimal hexamer complementary to the miR-26a seed sequence. Western blotting confirms that retroviral expression of miR-26a in HepG2 cells does not affect Myc protein abundance (Figure S1). Thus, miR-26a does not target the initiating oncogene in tet-o-MYC; LAP-tTA mice, supporting the use of this model for assessing the general tumor suppressing properties of miR-26a in vivo.
Development of an AAV vector system to simultaneously express a miRNA and eGFP
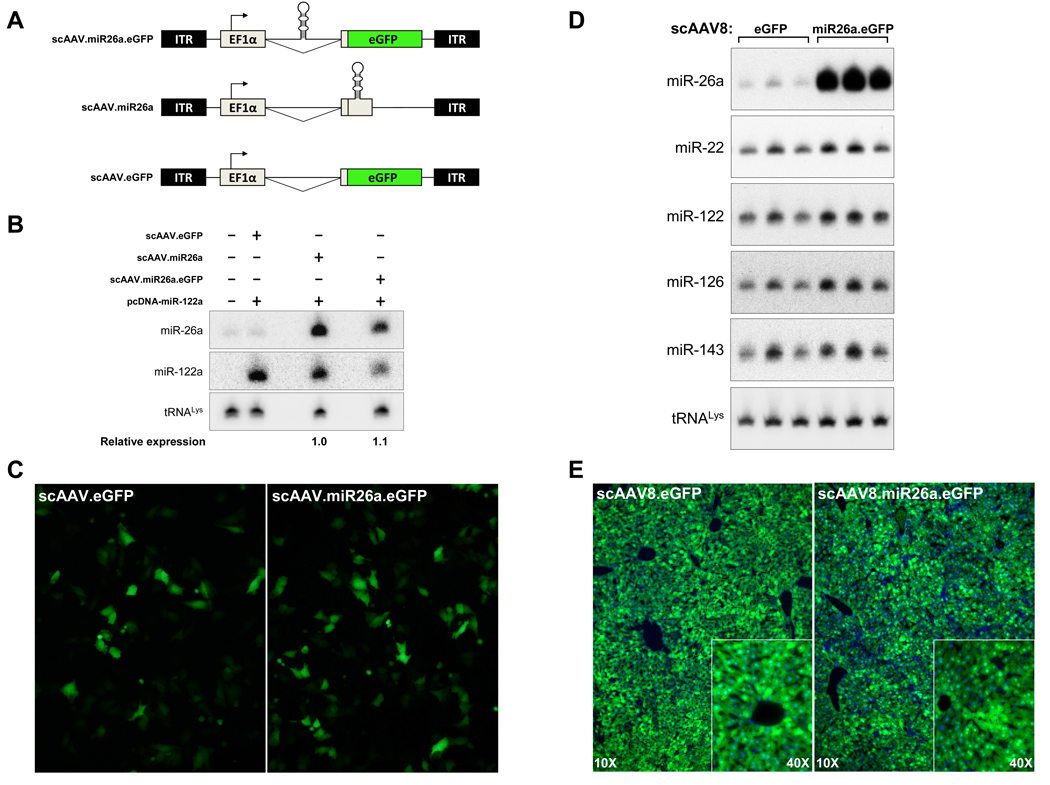
The development of self-complementary AAV (scAAV) vectors and the availability of AAV serotypes for improved transduction of specific target tissues has expanded the usefulness of this virus for therapeutic gene delivery (Gao et al., 2002; McCarty, 2008; McCarty et al., 2003; Nakai et al., 2005; Wang et al., 2003). In particular, these advances allow highly efficient transduction of hepatocytes following systemic administration of scAAV8 vectors. We therefore constructed a scAAV vector system to evaluate the therapeutic potential of miR-26a in tet-o-MYC; LAP-tTA mice. To allow facile assessment of target tissue transduction, the vector included enhanced green fluorescent protein (eGFP) driven by the ubiquitously expressed elongation factor 1 alpha (EF1α) promoter. Moreover, since miRNAs are frequently embedded within introns of both protein-coding and noncoding primary transcripts, we cloned miR-26a into the short intron which is part of the EF1α promoter unit (Wakabayashi-Ito and Nagata, 1994), thus allowing simultaneous production of eGFP and miR-26a from a single transcript (scAAV.miR26a.eGFP; Figure 4A). We confirmed that this vector efficiently expresses both miR-26a and eGFP by transient transfection of HeLa cells. Northern blotting demonstrated that the scAAV.miR26a.eGFP vector produced an equivalent amount of mature miRNA as a control vector in which the miRNA was in an exonic context (scAAV.miR26a; Figure 4A,B). Fluorescent microscopy of transfected cells similarly documented equivalent eGFP expression from scAAV.miR26a.eGFP and a control vector lacking intronic miR-26a sequences (scAAV.eGFP; Figure 4A,C).
Figure 4. Development of an AAV vector system to simultaneously express a miRNA and eGFP.
(A) Schematic representation of scAAV vectors used in this study depicting locations of inverted terminal repeats (ITRs), elongation factor 1 α promoter (EF1α), miRNA (shown in hairpin form), and enhanced green fluorescent protein (eGFP) open reading frame.
(B) Northern blot of transiently-transfected HeLa cells demonstrating equivalent levels of miR-26a when expressed from an intronic (scAAV.miR26a.eGFP) or exonic (scAAV.miR26a) context. Co-transfection with a miR-122a expression plasmid (pcDNA-miR-122a) provided a control for transfection efficiency while tRNALys levels documented equal loading.
(C) Fluorescent microscopy showing eGFP expression in HeLa cells transiently-transfected with the indicated AAV vectors.
(D) Northern blots showing expression of miRNAs in livers 21 days following administration of the indicated AAV vectors.
(E) Fluorescent microscopy showing efficient transduction of hepatocytes, as indicated by eGFP expression, 21 days following AAV administration.
scAAV.eGFP and scAAV.miR26a.eGFP were then packaged with the AAV8 serotype for in vivo delivery. 1×1012 vector genomes (vg) per animal were administered with a single tail-vein injection and liver tissue was harvested three weeks later for analysis of miRNA and eGFP expression. As expected, mice transduced with scAAV8.miR26a.eGFP exhibited high-level expression of miR-26a in the liver (Figure 4D). Fluorescent microscopy documented over 90% transduction of hepatocytes with both vectors (Figure 4E). Importantly, it has previously been demonstrated that AAV8-mediated delivery of some short-hairpin RNA (shRNA) constructs induces acute liver toxicity due to competitive inhibition of the miRNA pathway (Grimm et al., 2006). scAAV8.miR26a.eGFP administration does not cause these effects, as demonstrated by normal levels of endogenously-expressed miRNAs in transduced livers (Figure 4D), an absence of any acute inflammation, fibrosis, or overt histologic evidence of toxicity (Figure S2), and maintenance of normal levels of serum markers of liver function (ALT, AST, alkaline phosphatase, total and direct bilirubin) (Table S1). These data demonstrate that scAAV8 provides an effective, non-toxic means to deliver miRNAs to the liver.
Therapeutic delivery of miR-26a suppresses tumorigenesis in tet-o-MYC; LAP-tTA mice
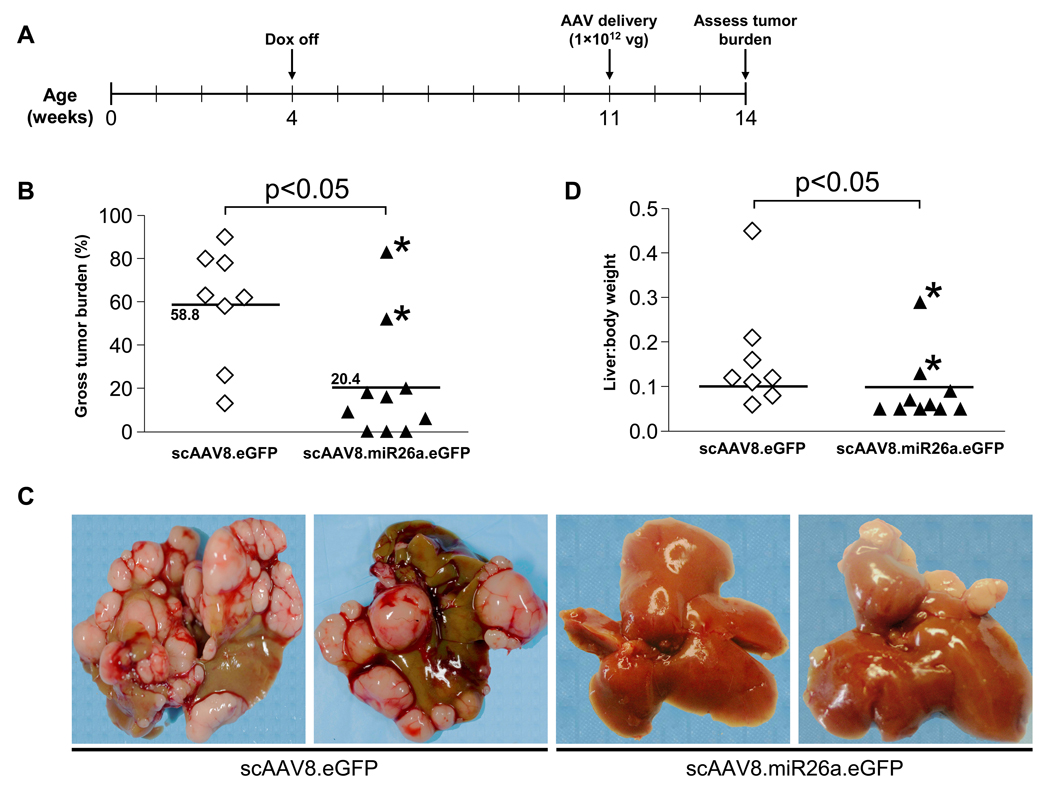
To assess the therapeutic efficacy of miR-26a delivery for liver cancer, we administered scAAV8.miR26a.eGFP, or scAAV8.eGFP as a negative control, to tumor-bearing tet-o-MYC; LAP-tTA mice. Animals were taken off dox at 4 weeks of age and virus was administered at 11 weeks of age, a time-point at which animals typically have multiple small to medium size tumors (Figure 5A, Figure S3). Three weeks later, animals were sacrificed and tumor burden was assessed. 6 out of 8 mice treated with control virus developed fulminant disease in which the majority of liver tissue was replaced with tumor tissue (Figure 5B,C, Figure S4). In contrast, 8 of 10 scAAV8.miR26a.eGFP-treated animals were dramatically protected, exhibiting only small tumors or a complete absence of tumors upon gross inspection (Figure 5B, p < 0.05). Liver:body weight ratios were significantly lower as well in scAAV8.miR26a.eGFP-treated versus scAAV8.eGFP-treated animals (p < 0.05), further documenting tumor suppression (Figure 5D).
Figure 5. AAV-mediated miR-26a delivery suppresses tumorigenesis in tet-o-MYC; LAP-tTA mice.
(A) Time-line of miR-26a therapeutic delivery experiment.
(B) Gross tumor burden of livers from miR-26a-treated and control animals, as determined by quantification of tumor area using the ImageJ software package. The mean tumor burden in each treatment group is indicated by horizontal lines. Data points highlighted by asterisks represent animals that exhibited low AAV transduction efficiency (see Fig. S5). p value calculated by two-tailed t-test.
(C) Representative images of livers from miR-26a-treated and control animals.
(D) Liver:body weight ratios of miR-26a-treated and control animals. A chi-square statistic was used to compare the fraction of animals in each treatment group with a liver:body weight ratio above 0.1 (indicated by horizontal line). Data points highlighted by asterisks represent animals that exhibited low AAV transduction efficiency (see Fig. S5).
Aggressive liver tumors arose in 2 of 10 scAAV8.miR26a.eGFP-treated mice. To investigate the cause of these treatment failures, we assessed AAV transduction efficiency by scoring GFP positive hepatocytes and by quantifying transduced vector genomes by qPCR. Interestingly, these mice exhibited significantly lower transduction efficiency than the successfully treated animals (Figure S5). This strongly suggests that the development of disease in these animals was a result of technical failure rather than biologic resistance to miR-26a–mediated tumor suppression. We conclude that efficient transduction of hepatocytes with scAAV8.miR26a.eGFP uniformly diminished disease progression in this model.
Delivery of miR-26a reduces cancer cell proliferation and induces tumor-specific apoptosis
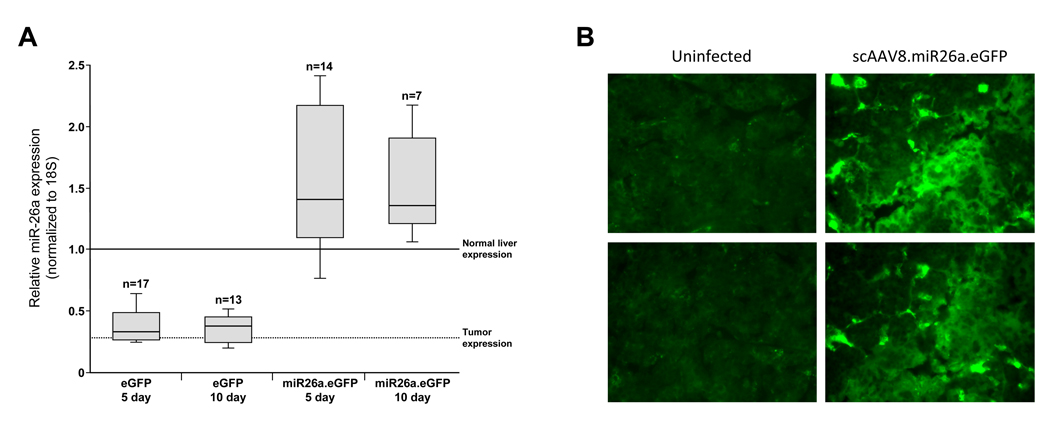
To ascertain the cellular mechanisms underlying miR-26a–mediated tumor suppression, we first confirmed that scAAV8.miR26a.eGFP effectively delivered the miRNA to tumor cells. Quantitative real-time PCR (qPCR) was used to measure miR-26a expression in tumors 5 or 10 days after vector administration (dox off at 4 weeks, virus administered at 11 weeks; 21–30 tumors analyzed per treatment condition). These studies revealed restoration of physiologic miR-26a expression in scAAV8.miR26a.eGFP-treated tumors whereas miRNA levels were unchanged following administration of scAAV8.eGFP (Figure 6A). Fluorescence microscopy also documented GFP expression in tumor cells, further demonstrating AAV transduction (Figure 6B).
Figure 6. AAV-mediated transduction of tumor cells in tet-o-MYC; LAP-tTA mice.
(A) Expression of miR-26a in tumors in AAV-transduced animals. qPCR was used to measure miRNA abundance in tumors 5 or 10 days following administration of scAAV8.eGFP (eGFP) or scAAV8.miR26a.eGFP (miR26a.eGFP). All values were normalized to 18S rRNA expression. Normal liver expression and tumor expression lines were derived from qPCR analysis of miR-26a levels in samples shown in Figure 1A. Each box represents the range of expression observed. The ends of the boxes represent the 25th and 75th percentiles, the bars indicate the 10th and 90th percentiles, and the median is depicted by a horizontal line within the boxes.
(B) Fluorescence microscopy of tumor sections demonstrating GFP expression in tumor cells in AAV-transduced animals.
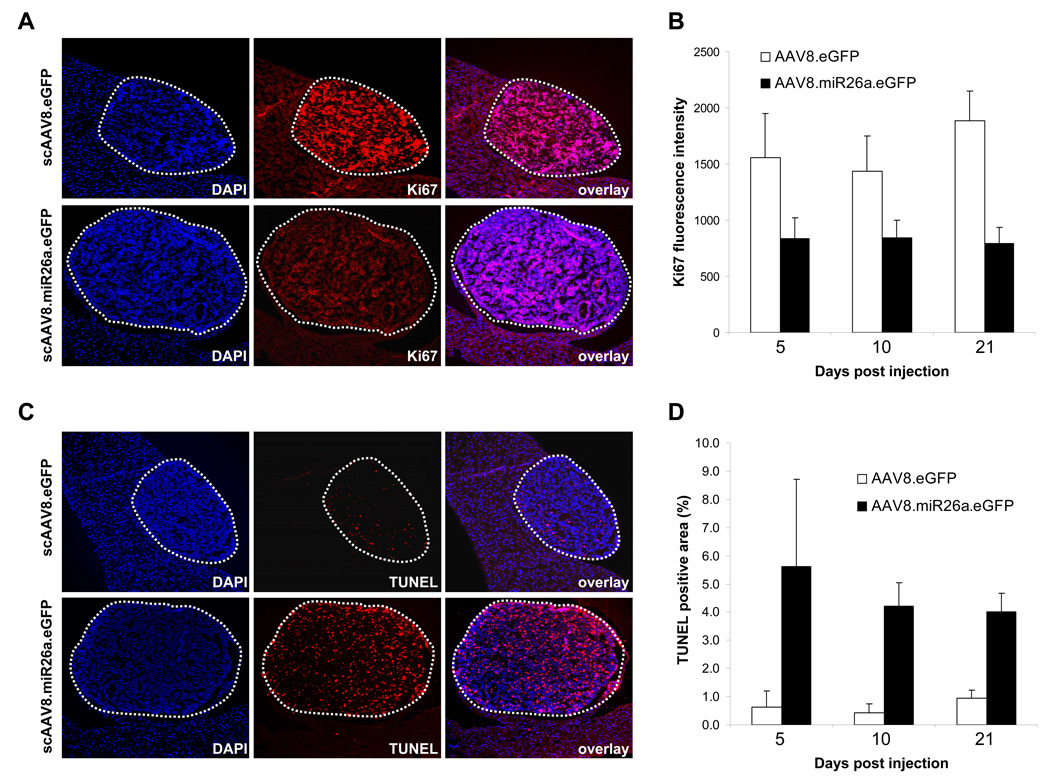
We next examined cellular proliferation in liver tissue 5, 10, and 21 days following vector administration. Ki67 staining revealed rapid proliferation of tumor cells at all time-points in scAAV8.eGFP-treated animals (Figure 7A,B, Figures S6–S7). Consistent with our earlier demonstration that expression of miR-26a arrests the cell-cycle in HepG2 cells, we observed markedly reduced Ki67 staining in tumors following administration of scAAV8.miR26a.eGFP. No difference in Ki67 staining of non-tumor tissue was apparent between the treatment groups, although baseline Ki67 signal was low in normal liver reflecting the slow proliferative rate of adult hepatocytes.
Figure 7. miR-26a delivery induces tumor-specific cell-cycle arrest and apoptosis.
(A) Representative DAPI and Ki67-stained sections from miR-26a-treated and control tet-o-MYC; LAP-tTA animals 5 days after AAV administration showing tumors (outlined with dotted line) and adjacent normal-appearing liver.
(B) Quantification of Ki67 staining in tumors from miR-26a-treated and control animals. Olympus Slidebook 4.2 was used to quantify the Ki67 fluorescence intensity in tumors in 3–5 randomly chosen fields per animal (n=2–4 animals per treatment per timepoint). The mean Ki67 fluorescence intensity per condition is plotted with error bars representing standard deviations.
(C) Representative DAPI and TUNEL-stained sections from miR-26a-treated and control animals 5 days after AAV administration. Tumors are outlined with dotted lines.
(D) Quantification of TUNEL staining in tumors from miR-26a-treated and control animals. ImageJ was used to quantify the TUNEL positive area in tumors in 6 randomly chosen fields per animal (n=2–4 animals per treatment per time-point). The mean TUNEL positive area per condition is plotted with error bars representing standard deviations.
Treatment-induced apoptosis was also assessed by TUNEL staining. Apoptotic cells were rare in normal liver tissue and present at low levels in tumors from scAAV8.eGFP-treated animals at all time points (Figure 7C,D, Figures S6–S7). Remarkably, AAV-mediated delivery of miR-26a potently induced apoptosis specifically in tumor cells without measurably causing death of normal hepatocytes. These data are consistent with our earlier findings that scAAV8.miR26a.eGFP administration caused no measurable liver toxicity in normal animals (Figure S2 and Table S1). To further investigate the specificity of miR-26a-induced apoptosis, we performed TUNEL staining on a broader panel of tissues including those with high proliferative indices (testis, spleen) and low proliferative indices (heart, lung, kidney, pancreas) (Figure S8). scAAV8.miR26a.eGFP administration did not cause a measurable increase in the frequency of apoptotic cells in any of these tissues. Together with the Ki67 data, these results document that miR-26a-mediated tumor suppression is associated with rapid and sustained inhibition of cancer cell proliferation and highly-specific induction of tumor cell death.
Discussion
Since the initial discovery of a functional RNA interference (RNAi) system in mammals, significant effort has been devoted to the development of therapeutics that utilize this pathway (de Fougerolles et al., 2007). While progress has been made towards the design and delivery of short interfering (si) and short hairpin (sh) RNAs for therapeutic gene silencing, accumulating evidence indicates that modulation of miRNA activity also represents an attractive strategy. miRNAs potently influence cellular behavior through the regulation of extensive gene expression networks (Baek et al., 2008; Selbach et al., 2008). Therapeutic modulation of a single miRNA may therefore affect many pathways simultaneously to achieve clinical benefit. Thus far, most translational in vivo studies targeting miRNAs have aimed to inhibit miRNA function through the use of antisense reagents such as antagomirs, locked nucleic acid (LNA) oligomers, and other modified oligonucleotides (Elmen et al., 2008; Esau et al., 2006; Krutzfeldt et al., 2005). While the in vivo use of synthetic oligonucleotide inhibitors is promising and will no doubt remain a fruitful area of investigation, the therapeutic delivery of miRNAs has certain advantages, especially in cancer. It has been demonstrated that most tumors are characterized by globally diminished miRNA expression (Gaur et al., 2007; Lu et al., 2005) and that experimental impairment of miRNA processing enhances cellular transformation and tumorigenesis (Kumar et al., 2007). Additionally, common oncogenic lesions can result in widespread miRNA repression (Chang et al., 2008). Thus, miRNA delivery might allow the therapeutic restitution of physiological programs of regulation lost in cancer and other disease states.
Therapeutic miRNA delivery may have unique technical advantages as well. First, the risk of off-target gene silencing is likely to be lower than that associated with artificial RNAi triggers since physiologic gene expression networks have evolved to accommodate the regulatory effects of endogenous miRNAs. Second, as compared to siRNAs or shRNAs that target a single transcript, the regulation of hundreds of targets in multiple pathways by miRNAs may reduce the emergence of resistant clones in diseases such as cancer since many simultaneous mutations would be required to subvert the effects of miRNA expression. At the same time, however, miRNA-based therapies will require thorough pre-clinical validation as these broad effects may in some cases have toxic consequences. Finally, it has been previously shown that the miRNA biogenesis pathway can be competitively inhibited by the expression of certain shRNAs, resulting in toxic effects following delivery of these transcripts (Grimm et al., 2006). This may be due to inefficient processing and/or nuclear-cytoplasmic transport of shRNA sequences which are not evolutionarily adapted for precise handling by this pathway. Supporting this notion, shRNA-associated toxicity can be mitigated by placing a shRNA into a miRNA-like context (McBride et al., 2008). In this study we demonstrated high expression of an exogenously supplied natural miRNA without toxic effects on endogenous miRNA biogenesis.
The results described herein demonstrate for the first time that therapeutic delivery of a miRNA can result in tumor suppression even in a setting where the initiating oncogene is not targeted. This establishes the principle that miRNAs may be useful as anti-cancer agents through their ability to broadly regulate cancer cell proliferation and survival. Furthermore, this study design involved the treatment of existing tumors with a miRNA, a paradigm closely related to the clinical scenarios in which such therapies would be employed. Finally, we demonstrate highly specific effects of miRNA delivery on tumor cells without affecting surrounding normal tissue. Although the molecular basis of this specificity requires further investigation, it is likely that the high physiologic expression of miR-26a in normal hepatocytes confers tolerance to exogenous administration of this miRNA. In contrast, the specific reduction of miR-26a in neoplastic cells and their sensitivity to its restored expression underscores the contribution of loss-of-function of this miRNA to tumorigenesis in this setting. It is noteworthy that large scale cloning efforts have documented expression of miR-26a in most mouse and human tissues (Landgraf et al., 2007), while in situ hybridization data from zebrafish has documented ubiquitous expression with especially high levels in the head, spinal cord, and gut (Wienholds et al., 2005). The widespread expression of this miRNA is consistent with our observation that systemic AAV-mediated delivery of miR-26a is well-tolerated by many tissues. Overall, our demonstration that miR-26a delivery potently suppresses even a severe, multifocal model of carcinogenesis in the absence of measurable toxicity provides proof-of-principle that the systemic administration of miRNAs may be a clinically viable anti-cancer therapeutic strategy.
In this study, we elected to use an AAV-based vector system, an especially attractive platform for regulatory RNA delivery (Giering et al., 2008; Grimm and Kay, 2007; McCarty, 2008). When delivered in viral vectors, miRNAs are continually transcribed, allowing sustained high level expression in target tissues. The use of tissue-specific promoters could restrict this expression to particular cell types of interest. Compared to retroviral delivery systems, DNA viruses such as AAV carry substantially diminished risk of insertional mutagenesis since viral genomes persist primarily as episomes (Schnepp et al., 2003). Further, the availability of multiple AAV serotypes allows efficient targeting of many tissues of interest (Gao et al., 2002; McCarty, 2008). Finally, the general safety of AAV has been well documented, with clinical trials using this platform already underway (Carter, 2005; Maguire et al., 2008; Park et al., 2008). Despite these advantages, prior studies have achieved mixed results when attempting to use AAV vectors to transduce HCC cells in vivo (Peng et al., 2000; Shen et al., 2008). Our successful use of AAV to treat an animal model of HCC may indicate that tumor cells are highly sensitive to restored expression of miRNAs, resulting in strong tumor suppression even with a relatively low number of vector genomes introduced into each cell. The use of self-complementary vectors may have further enhanced tumor cell transduction and therapeutic miRNA expression in the present study. Finally, while our findings are most consistent with miR-26a influencing cancer cell proliferation and survival through a cell-autonomous mechanism, transduction of tumor-associated stromal cells may have also contributed to the observed therapeutic effects through a yet unknown mechanism.
We have previously documented that miR-26 family members suppress tumorigenesis in c-Myc-driven B lymphoma cells (Chang et al., 2008) and now extend these findings to the tet-o-MYC; LAP-tTA HCC model. Our demonstration that a genetically complex human liver cancer cell line is also susceptible to miR-26a-mediated anti-proliferative effects suggests that the tumor suppressive activities of this miRNA are not limited to Myc-initiated malignancies. Several additional lines of evidence further support this notion. Work from our group and others has revealed a role for miR-26a in the p53 tumor suppressor network as this miRNA is upregulated in a p53-dependent manner following DNA damage (Chang et al., 2007; Xi et al., 2006). Additionally, a profiling study of human anaplastic thyroid cancers (ATC) identified miR-26a as consistently downregulated and demonstrated that transient transfection of this miRNA significantly impairs proliferation of ATC cells in vitro (Visone et al., 2007). Moreover, the miR-26a-1-encoding locus at 3p21.3 is contained within a sub-megabase interval that is frequently deleted in small cell lung carcinomas, renal cell carcinomas, and breast carcinomas (Kashuba et al., 2004). These observations suggest broad anti-tumorigenic properties of miR-26 family members in diverse settings. Nevertheless, if future work reveals that the effectiveness of miR-26 delivery is restricted to settings of Myc dysregulation, therapeutic delivery of this miRNA may still be beneficial for a large number of cancer subtypes since hyperactivity of Myc is one of the most common attributes of human cancer cells.
Although miR-26a delivery confers dramatic tumor protection, it is likely that many equally or more effective miRNAs with therapeutic potential remain to be functionally characterized. The approach employed in this study provides an experimental framework to identify additional favorable candidates. We suggest that the most promising miRNAs will, like miR-26a, be highly expressed in a wide variety of normal tissues, be underexpressed in the disease state being studied, and, when evaluated using in vitro or in vivo models, demonstrate specific phenotypic effects in disease cells while sparing normal cells. While there clearly remains significant work to be done both in identifying such miRNAs and optimizing their controlled delivery, our findings highlight the therapeutic promise of this approach.
Methods
Cell culture
HEK293, HEK293T, and HeLa cells were cultured in high glucose (4.5g/L) DMEM supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. HepG2 cells were cultured in EMEM supplemented with 10% FBS, penicillin, and streptomycin.
RNA isolation, Northern blotting, and qPCR
Total RNA was isolated from cultured cells or tissue using Trizol (Invitrogen) according to the manufacturer’s protocol. Northern blotting was performed as previously described (Hwang et al., 2007) using Ultrahyb-oligo buffer (Ambion) and oligonucleotide probes perfectly complementary to the mature miRNA sequences. All membranes were stripped and re-probed for tRNALys as a loading control. qPCR for miR-26a and 18S rRNA was performed using pre-designed Taqman primers and probes (ABI) according to manufacturer’s instructions.
Cell-cycle profiling
5×105 retrovirally-infected HepG2 cells were plated after selection. 24 hours later, cells were harvested for analysis by propidium iodide (PI) staining and flow cytometry as described previously (Hwang et al., 2007). For M-phase trapping experiments, 100 ng/mL nocodazole was added for 24 hours prior to harvesting floating and adherent cells for PI staining.
Western blotting
Antibodies for immunoblotting were as follows: anti-c-Myc mouse monoclonal (clone 9E10; Zymed), anti-cyclin E1 rabbit polyclonal (Abcam), anti-cyclin E2 rabbit polyclonal (Cell Signaling), anti-cyclin D2 rabbit polyclonal (Cell Signaling), anti-CDK6 mouse monoclonal (clone DCS83; Cell Signaling), and anti-a-tubulin mouse monoclonal (clone DM1A; Calbiochem).
Luciferase reporter assays
2.5×105 HepG2 cells were plated in triplicate wells of a 24-well plate and transfected 16 hours later with 100 ng of the indicated pGL3 3' UTR reporter construct and 5 ng of phRL-SV40 (Promega) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Where indicated, miR-18a or miR-26a mimics (Dharmacon) were co-transfected at 25 nM final concentration. 24 hours after transfection, cells were lysed and assayed for firefly and renilla luciferase activity using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to renilla luciferase activity for each transfected well. Data depicted is representative of three independent experiments performed on different days.
Vector delivery
Tet-o-MYC; LAP-tTA mice were maintained on dox-containing food (100 mg/kg) until 4 weeks of age. At 11 weeks of age, AAV was administered at a dose of 1012 vg per animal by tail vein injection (200 uL total volume) using a 30 gauge ultra-fine insulin syringe. The Research Institute at Nationwide Children's Hospital Animal Care and Use Committee approved all housing and surgical procedures.
Analysis of Liver Function Tests
Four month old C57/BL10 mice (n=5) were dosed with 1012 vg of scAAV8.miR26a.eGFP by tail vein injection. Blood was collected prior to vector administration and at 3 weeks post injection from the submandibular vein. Serum analysis was performed by Ani Lytics (Gaithersburg, MD).
Immunohistochemistry
Tissues were collected in 10% buffered formalin, embedded in paraffin, and stained in hematoxylin and eosin following standard procedures. For Ki67 detection, slides were microwaved for 15 min in 10 mM sodium citrate (pH 6.0), cooled for 20 min at 25°C, and washed three times (5 minutes each) with PBST (PBS, 0.1%Tween 20). The tissue was permeabilized by incubating the slides in 1% Triton X-100 in PBS at 4° C for 30 min and then washed again three times in PBST. After blocking for 1 hour at 25° C in blocking buffer (PBS containing 10% goat serum and 0.2% Triton X-100), slides were incubated overnight in a humidity chamber with a mouse anti-human Ki67 monoclonal antibody (BD Biosciences) diluted 1:100 in blocking buffer. Following another three PBST washes, slides were incubated with Alexa 594-conjugated goat anti-mouse secondary antibody at a 1:200 dilution (Molecular Probes). Slides were washed and nuclei counter-stained with DAPI. The intensity of the Ki67 signal was quantified using the Olympus Slidebook 4.2 software. TUNEL staining was performed with the TMR Red In situ cell death detection kit (Roche) according to manufacturer’s instructions. Quantification of TUNEL positive area was performed using ImageJ software. For GFP visualization, tissues were fixed in 4% paraformaldehyde followed by an overnight incubation in 30% sucrose incubation and then embedded in OCT compound.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Bishop for tet-o-MYC mice, Terri Shaffer and Lucia Rosas for help with virus administration, and the staff at Nationwide Children’s Hospital Viral Vector Core facility. This work was supported by US National Institutes of Health grant R01CA120185 (to J.T.M.), the Sol Goldman Center for Pancreatic Cancer Research (J.T.M.), and the Research Institute at Nationwide Children’s Hospital (J.R.M.). J.T.M. is a Rita Allen Foundation Scholar and a Leukemia and Lymphoma Society Scholar. K.A.O. is a fellow of the Damon Runyon Cancer Research Foundation. R.R.C. is supported by the Medical Scientist Training Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional methods are provided in Supplemental Online Materials
References
- Alexander IE, Cunningham SC, Logan GJ, Christodoulou J. Potential of AAV vectors in the treatment of metabolic disease. Gene Ther. 2008;15:831–839. doi: 10.1038/gt.2008.64. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, Arvanitis C, Attardi LD, Feng S, Ruebner B, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2:e332. doi: 10.1371/journal.pbio.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BJ. Adeno-associated virus vectors in clinical trials. Hum Gene Ther. 2005;16:541–550. doi: 10.1089/hum.2005.16.541. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- Grimm D, Kay MA. RNAi and Gene Therapy: A Mutual Attraction. Hematology Am Soc Hematol Educ Program. 2007;2007:473–481. doi: 10.1182/asheducation-2007.1.473. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007a;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007b;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kashuba VI, Li J, Wang F, Senchenko VN, Protopopov A, Malyukova A, Kutsenko AS, Kadyrova E, Zabarovska VI, Muravenko OV, et al. RBSP3 (HYA22) is a tumor suppressor gene implicated in major epithelial malignancies. Proc Natl Acad Sci U S A. 2004;101:4906–4911. doi: 10.1073/pnas.0401238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Slack FJ. microRNAs: small molecules with big roles - C. elegans to human cancer. Biol Cell. 2008;100:71–81. doi: 10.1042/BC20070078. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love TM, Moffett HF, Novina CD. Not miR-ly small RNAs: big potential for microRNAs in therapy. J Allergy Clin Immunol. 2008;121:309–319. doi: 10.1016/j.jaci.2007.12.1167. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Esteller M. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle. 2007;6:1455–1459. [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh EN, Jr, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Park K, Kim WJ, Cho YH, Lee YI, Lee H, Jeong S, Cho ES, Chang SI, Moon SK, Kang BS, et al. Cancer gene therapy using adeno-associated virus vectors. Front Biosci. 2008;13:2653–2659. doi: 10.2741/2872. [DOI] [PubMed] [Google Scholar]
- Pathak A, Vyas SP, Gupta KC. Nano-vectors for efficient liver specific gene transfer. Int J Nanomedicine. 2008;3:31–49. [PMC free article] [PubMed] [Google Scholar]
- Peng D, Qian C, Sun Y, Barajas MA, Prieto J. Transduction of hepatocellular carcinoma (HCC) using recombinant adeno-associated virus (rAAV): in vitro and in vivo effects of genotoxic agents. J Hepatol. 2000;32:975–985. doi: 10.1016/s0168-8278(00)80102-6. [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Roberts LR. Sorafenib in liver cancer--just the beginning. N Engl J Med. 2008;359:420–422. doi: 10.1056/NEJMe0802241. [DOI] [PubMed] [Google Scholar]
- Rodino-Klapac LR, Janssen PM, Montgomery CL, Coley BD, Chicoine LG, Clark KR, Mendell JR. A translational approach for limb vascular delivery of the micro-dystrophin gene without high volume or high pressure for treatment of Duchenne muscular dystrophy. J Transl Med. 2007;5:45. doi: 10.1186/1479-5876-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepp BC, Clark KR, Klemanski DL, Pacak CA, Johnson PR. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J Virol. 2003;77:3495–3504. doi: 10.1128/JVI.77.6.3495-3504.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Shen Z, Wong OG, Yao RY, Liang J, Kung HF, Lin MC. A novel and effective hepatocyte growth factor kringle 1 domain and p53 cocktail viral gene therapy for the treatment of hepatocellular carcinoma. Cancer Lett. 2008;272:268–276. doi: 10.1016/j.canlet.2008.03.064. [DOI] [PubMed] [Google Scholar]
- Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18:89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- Wakabayashi-Ito N, Nagata S. Characterization of the regulatory elements in the promoter of the human elongation factor-1 alpha gene. J Biol Chem. 1994;269:29831–29837. [PubMed] [Google Scholar]
- Wang Z, Ma HI, Li J, Sun L, Zhang J, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12:2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.