Abstract
Expression of brain-derived neurotrophic factor (BDNF) is developmentally regulated in prefrontal cortex (PFC). The underlying molecular mechanisms, however, remain unclear. Here, we explore the role of microRNAs (miRNAs) as post-transcriptional inhibitors of BDNF. A sequential approach involving in silico, miRNA microarray, in situ hybridization and qRT–PCR studies identified a group of 10 candidate miRNAs, segregating into five miRNA families (miR-30a-5p/b/c/d, miR-103/107, miR-191, miR-16/195, miR-495), which exhibited distinct developmental and lamina-specific expression in human PFC. Luciferase assays confirmed that at least two of these miRNAs, miR-30a-5p and miR-195, target specific sequences surrounding the proximal polyadenylation site within BDNF 3′-untranslated region. Furthermore, neuronal overexpression of miR-30a-5p, a miRNA enriched in layer III pyramidal neurons, resulted in down-regulation of BDNF protein. Notably, a subset of seven miRNAs, including miR-30a-5p, exhibited an inverse correlation with BDNF protein levels in PFC of subjects age 15–84 years. In contrast, the role of transcriptional mechanisms was more apparent during the transition from fetal to childhood and/or young adult stages, when BDNF mRNA up-regulation was accompanied by similar changes in (open chromatin-associated) histone H3-lysine 4 methylation at BDNF gene promoters I and IV. Collectively, our data highlight the multiple layers of regulation governing the developmental expression of BDNF in human PFC and suggest that miRNAs are involved in the fine-tuning of this neurotrophin particularly in adulthood.
INTRODUCTION
MicroRNAs (miRNAs) are evolutionary conserved small non-coding RNAs that are known to post-transcriptionally inhibit protein coding genes, by affecting their translation and/or mRNA stability (1). They are derived from longer precursor molecules, are incorporated to the RNA-induced silencing complex (RISC) and interact with complementary regions mainly within the 3′ untranslated region (3′-UTR) of their target mRNAs (2). Evidence from the early days of miRNA research, and up to the present day, has suggested that the expression of some miRNAs is highly regulated in a temporal- and region-specific manner and that they participate in divergent biological processes (3).
In the vertebrate nervous system, miRNAs have been shown to play an important role during development (4) and in regulation of synaptic plasticity (5). A subset of miRNAs are abundantly expressed in the human brain (6,7) and have been implicated in numerous brain diseases (8–13). However, very little is known about their expression and function in the human prefrontal cortex (PFC), a brain area responsible for high order cognitive functions, which displays delayed maturation and is disrupted in patients with psychiatric disease (14,15).
Brain-derived neurotrophic factor (BDNF) plays a prominent role during cortical development and maturation (16), and alterations in BDNF expression have been reported in a plethora of neuropsychiatric diseases (17). Interestingly, pyramidal neurons—the primary source of BDNF in cerebral cortex—express high levels of DICER, an RNAse III endoribonuclease and key molecule for miRNA biogenesis, as well as components of RISC, such as eIF2c (18). Furthermore, the 3′-UTR of human BDNF is predicted according to computational analysis to include numerous miRNA target sites that show a high degree of conservation between different mammalian species (19). These findings, taken together, point to a potential role for miRNAs in the control of cortical BDNF expression. However, to date, this hypothesis has not yet been tested.
Here, we present evidence that multiple miRNAs, including a subset of the miR-30 family, are involved in fine-tuning of BDNF expression specifically during late maturation and aging of human PFC. Our findings suggest that BDNF expression in human cerebral cortex is regulated by a complex system of small RNAs, which in turn display lamina-specific enrichment and are differentially regulated during development. These findings provide the first evidence for the miRNA pathway acting as a key regulator of BDNF expression during maturation and aging of human PFC.
RESULTS
In silico analysis of putative miRNA target sites within BDNF 3′-UTR
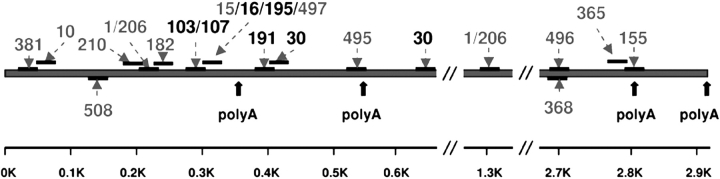
Potential miRNA target sites within the 3 kb of BDNF 3′-UTR were identified by combining three in silico tools: TargetScan 3.1 (19), Pictar (20,21) and RNAhybrid (22). Altogether, 17 distinct target sites—all of which appear to be highly conserved in various mammalian species (human, chimp, dog, mouse, rat)—were found, which potentially could interact with 26 different miRNAs (Fig. 1 and Supplementary Material, Table S3).
Figure 1.
BDNF 3′-UTR contains numerous predicted target sites for miRNAs expressed in human cerebral cortex. Map of BDNF 3′-UTR (human) showing potential miRNA target sites conserved across mammalian species (see also Supplementary Material, Table S3); target sites for miRNAs expressed in moderate to high levels in human cerebral cortex (Supplementary Material, Fig. S1) are shown in black whereas the remaining predicted sites are shown in gray. Notice that miRNAs expressed in moderate to high levels are located in the vicinity of the two proximal poly-A sites in the BDNF 3′-UTR.
Next, we used an array-based approach to measure the expression of these 26 candidate miRNAs and identified 10 miRNA species that were present at moderate or high levels in the adult human parietal cortex; for the remaining miRNAs, levels were very low or indistinguishable from background (Supplementary Material, Fig. S1A). We then compared our array results with published microarray data on adult human PFC (11). Remarkably, all but one (miR-495) showed a similar order of expression in prefrontal and parietal cortex (Supplementary Material, Fig. S1B).
Notably, the target site(s) for each of the 10 expressed miRNAs—which segregate into five different miRNA families (miR-103/107, miR-191, miR-16/195, miR-30a-5p/b/c/d, miR-495)—were in close vicinity to the two proximal (out of four total) BDNF 3′-UTR polyadenylation sites (Fig. 1); in adult cerebral cortex, the bulk of BDNF transcript extends beyond these two proximal polyA sites (23,24). This would suggest that a large fraction of BDNF transcript could be targeted by the miRNAs listed above.
Laminar and cellular specificity of miRNAs expressed in PFC
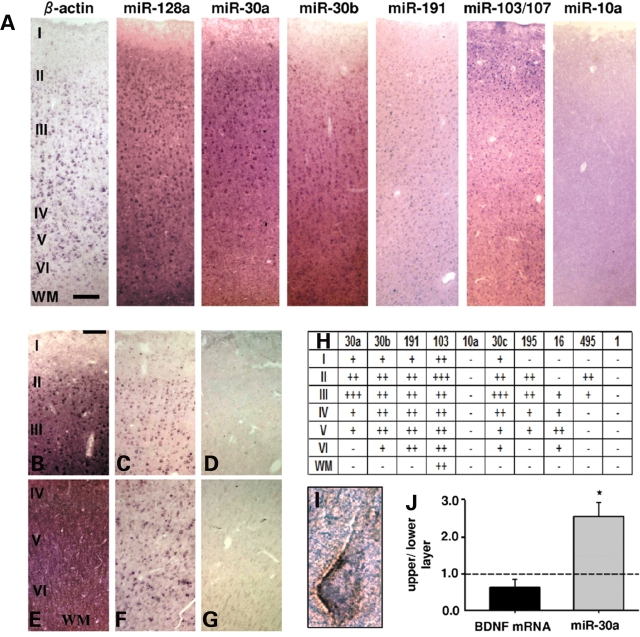
We wanted to examine the laminar and cellular expression pattern of the predicted miRNAs in adult human PFC; BDNF transcript is found in putative pyramidal neurons positioned within layers II–VI (25,26). Cellular labeling was weak or not discernible from background in sections processed by in situ hybridization with locked nucleic acid (LNA-ISH) for miR-1 and miR-10a (Fig. 2A and H and Supplementary Material, Fig. S2), two miRNAs expressed at very low or non-detectable levels, respectively, according to our microarray data (Supplementary Material, Fig. S1A). In contrast, miR-128a, a reportedly pan-neuronal miRNA marker (27), also detected at high levels in our microarray analysis (data not shown), was robustly expressed throughout the full thickness of PFC (Fig. 2A). Therefore, we conclude that LNA-ISH is applicable to human postmortem brain tissue, which is in accordance to previous reports (28). Next, we studied prefrontal expression patterns of the 10 predicted miRNAs with 8 probes (due to one-base-differences, a single probe was applied for miR-103/107 and for miR-30a/d; see also Supplementary Material, Table S2). Each of the 8 probes revealed a distinct laminar expression (Fig. 2A and H and Supplementary Material, Fig. S2). For example, miR-30a showed robust labeling in the upper cortical layers, including a subset of large, putative pyramidal neurons primarily residing in layer III (Fig. 2A and B). In contrast, labeling in PFC layers V and VI was very weak or non-detectable (Fig. 2A and E). To further confirm these lamina-specific differences, we assayed miR-30a levels by qRT–PCR separately for the upper (including layers II and III) and lower (mainly V and VI) layers of the cortex. Indeed, levels of miR-30a were ∼2.5-fold higher in the upper when compared with the deeper layers (Fig. 2J). In contrast, neither BDNF mRNA (Fig. 2J) nor protein levels (data not shown) showed significant differences between the upper and lower layers; the latter finding may not be too surprising, however, given that BDNF protein produced in pyramidal neuron somata could potentially be distributed via their processes to other cortical layers and neuronal populations further removed from the site of synthesis (17).
Figure 2.
Lamina and cellular expression pattern of selected miRNAs in human PFC. (A) Representative images (from 2–6 replicates) of six-layered PFC (adult) sections processed by LNA-ISH with miRNA-specific probes, and β-actin as control. Notice lamina-specific expression patterns, including enrichment of miR-30a in layers II and III. (B–G) Additional images of upper (B–D) and deeper (E–G) cortical layers for miR-30a LNA-ISH (B and E) together with β-actin (C and F) and no probe negative control (D and G). (H) Table summarizing relative expression of 10 miRNAs across PFC layers I–VI, and underlying white matter (WM) (+++high, ++moderate, +weak, −indistinguishable from background). Note that miR-103/107 probe detects both miRNAs due to a single nucleotide difference at their 3′ end. (I) Double labeled layer III pyramidal neuron from section processed for Neurofilament-H immunohistochemistry (brown) after miR-30a LNA in situ (purple). (J) Bar graphs show qRT–PCR data for miR-30a and BDNF mRNA from dissected tissue corresponding to upper (layer II and III) and lower (layer V and VI) cortical layers from five adult samples (see Materials and Methods) shown as relative ratios (upper to lower). Notice the significant approximately 2.5-fold enrichment of miR-30a in upper layers. Bar in (A) = 200 µm, in (B–G) = 100 µm. Image (I) taken at 63 × 10 magnification.
In addition, expression in pyramidal neurons was further confirmed in sections processed by LNA-ISH followed by immunolabeling for a neurofilament epitope selectively expressed by a subset of pyramidal neurons (30) (Fig. 2I) and by RT–PCR from laser-capture dissected pyramidal neurons (Supplementary Material, Fig. S2). Expression of miR-495 was highly restricted and limited to a subpopulation of cells positioned in and around layer II (Fig. 2H and Supplementary Material, Fig. S2). Cellular labeling for miR-16 in tissue sections was weak and mostly confined to deeper portions of cortical gray matter (Supplementary Material, Fig. S2). It is possible that the high levels of miR-16 in mature erythrocytes (31) could have contributed to the comparatively high but variable levels of expression in whole tissue homogenates assayed by array (Supplementary Material, Fig. S1).
Among all miRNAs tested, only miR-103/107 was enriched in upper cortical layers and was also expressed in white matter. This particular finding is in good agreement with a recent report showing the same laminar enrichment in superior and middle temporal gyrus (13). Intriguingly, however, none of the miRNAs included in our study showed discernable layer specificity in mouse cerebral cortex (Supplemental Fig. 2 and data not shown), suggesting that miRNA expression in cerebral cortex shows important differences between human and rodent. In conclusion, miR-30a and other members of the miR-30 family, and several additional miRNAs predicted to interact with the 3′-UTR of BDNF are abundantly expressed in adult human PFC, with distinct laminar specificity.
Validation of human BDNF 3′-UTR miRNA target sites
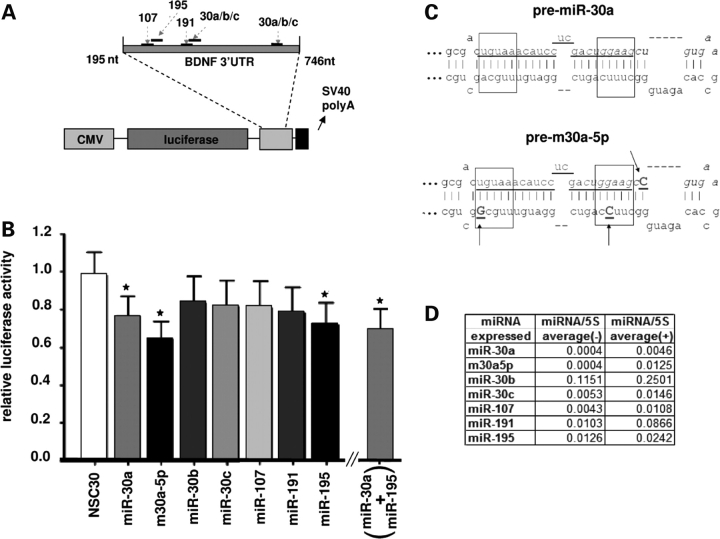
To determine whether the candidate miRNAs described above target the 3′-UTR of human BDNF mRNA, we constructed a luciferase reporter plasmid with a 551 bp fragment of human BDNF 3′-UTR containing all the highly predicted miRNA target sites fused to the 3′ end of the luciferase gene (Fig. 3A). We utilized CMV-driven vectors that encode for each of the following miRNA precursors—miR-30a, miR-30b, miR-30c, miR-107, miR-191 and miR-195 (Supplementary Material, Fig. S3). Since the endogenous levels of these miRNAs varied, transfection of these plasmids in HEK293 cells resulted in various degrees of overexpression of the mature miRNAs (Fig. 3D). It should be noted that, in these assays, miR-16 and miR-495 were not included because of uncertain (miR-16), or highly restricted (miR-495) expression in the cortex. Furthermore, because of high sequence similarity (one single nucleotide difference), miR-30d and miR-103 are predicted to have very similar effects on BDNF 3′-UTR with miR-30a and miR-107 respectively; hence these miRNAs were not tested separately.
Figure 3.
A region of human BDNF 3′-UTR is targeted by a subset of miRNAs. (A) Map of the luciferase reporter vector containing the 551 bp portion of human BDNF 3′-UTR (nt 195 to 746 from beginning of 3′-UTR, nucleotides 1500–2051, Genbank ID NM_170735), which includes target sites for the miRNAs assayed in (B). Bar graph in (B) shows results from reporter assay, expressed as luciferase units normalized to β-galactosidase units and relative to a reference control (EGFP expressing vector not containing any miRNA sequence). Bars represent the calculated means by ANOVA REML model plus standard error. Asterisks depict statistically significant differences (after post-hoc Tukey correction) compared with the (miR-30a based) non-silencing seed sequence mutant (NSC30); P = 0.010 (miR-30a); 0.0005 (m30a-5p); 0.0136 (miR-195); 0.0207 (miR-30a+miR-195). n = 2–6 independent replicates. Notice the significant reduction in reporter activity after transfection with miR-30a, m30a5p and miR-195 expressing vectors. (C) Predicted secondary structure of the (top) wild-type pre-miRNA 30a and (bottom) modified precursor designed to preferentially express miR-30a-5p (m30a-5p in (B)). Sequence underlined represents mature miR-30a-5p miRNA, which remains unaltered in the modified precursor. Arrows demarcate nucleotides modified from wild-type. See text for further details. (D) Table summarizing expression changes of mature miRNAs after transfection of HEK293 cells with appropriate vectors (averaged from two independent experiments); (+) labels transfected cells, (−) non-transfected controls.
Due to the fact that miR-30a was enriched in layer III pyramidal neurons (Fig. 2), which are a critical component of prefrontal cortical neuronal networks (29), and one of the major sites of BDNF synthesis (25), we anticipated that this miRNA might be particularly important for the post-transcriptional inhibition of BDNF. Additionally, in silico analysis of the interaction between miR-30 miRNAs and BDNF 3′-UTR indicated a more favorable thermodynamic interaction of miR-30a compared with miR-30b and miR-30c, resulting from a higher 3′ end complementarity of miR-30a with the 3′-UTR (Supplementary Material, Fig. S3). Indeed, results from the luciferase assay showed that overexpression of miR-30a, but not miR-30b and miR-30c, lead to a significant decrease in activity of the reporter (Fig. 3B). Among the remaining miRNAs tested, only miR-195 induced a significant reduction in luciferase activity (Fig. 3B). Importantly, no changes in reporter activity were observed after transfection of a miR-30a-based non-silencing control (‘NSC30’, Fig. 3B), in which the seed sequence (nt 2–7, see Fig. 4D for details) was mutated in order to disrupt complementarity with BDNF 3′-UTR target sites. Furthermore, the NSC30 mature sequence was not predicted to target the 551 bp of BDNF 3′-UTR, according to RNA hybrid software (data not shown).
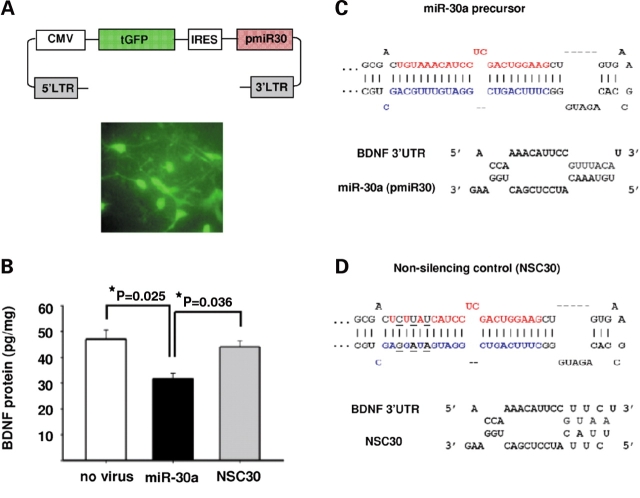
Figure 4.
MiR-30a mediates translational inhibition of BDNF in neurons. Neuronal cultures from rat forebrain were infected with lentiviruses that contained constructs co-expressing GFP and precursor miRNAs. (A) Top: Map of the lentiviral vector used; Bottom: Representative image of neuronal culture infected with the GFP-expressing lentiviral vector shown above. (B) Bar graphs showing BDNF protein levels (mean ± SEM) for cultures infected with miR-30a (n = 3), or miR-NSC30 (n = 3) (see text for details), and non-infected (‘no virus’, n = 2) cultures. Notice the significant decrease in BDNF protein in miR-30a overexpressing cultures. P-values after post-hoc Tukey/ANOVA. (C and D) Top: Secondary structures of (C) miR-30a precursor and (D) miR-NSC30 which contains a 3 base substitution in the seed sequence of miR-30a-5p. Mature miR-30a-5p is depicted in red, miR-30a-3p in blue and the nucleotide changes in the non-silencing control, NSC30, are shown in black and underlined. Bottom: Predicted interactions between the first target site in BDNF 3′-UTR (see text for details) and either (C) wild-type miR-30a-5p or (D) non-silencing precursor (‘NSC30’).
Notably, processing of the miR-30a precursor can lead to two miRNAs, miR-30a-5p and miR-30a-3p (32,33). Interestingly, both our microarray (Supplementary Material, Fig. S1A) and ISH experiments (Fig. 2A and data not shown) indicate that expression of miR-30a-5p is much higher than miR-30a-3p in the human cerebral cortex. Therefore, we modified the miR-30a precursor sequence by replacing one or two Watson–Crick base pairings with G-U wobbles at the 5′ end and vice versa for the 3′ end (marked by arrows in Fig. 3C). The rationale was to destabilize the 5′ end relative to the 3′ end of the precursor, which is expected to shift relative levels of mature miRNA towards the 5′ end product (34). Indeed, transfection with this modified miR-30a precursor (m30a-5p, see Fig. 3B–D) resulted in a 3-fold increase in levels of mature miR-30a-5p, compared with transfection with the wild-type form of the precursor (Fig. 3D). There was a ∼36% reduction in reporter activity in cells transfected with m30a-5p, compared with ∼24% in cells transfected with wild-type miR-30a precursor (Fig. 3B). Finally, it is worth mentioning that there are two potential miR-30 interaction sites within the BDNF 3′-UTR, although the second site (Figs 1 and 3A) is likely to be less functional due to the presence of a G-U wobble in the seed sequence (first site, bp 397–402; second site, bp 680-685 of BDNF 3′-UTR, see Supplementary Material, Table S3) (35). Therefore, we wanted to demonstrate that the first canonical site is sufficient to mediate the inhibitory effect of miR-30a. To this end, we transfected HeLa cells with a luciferase vector containing a 70 bp BDNF 3′-UTR sequence, which included the first target site (nucleotides 1653–1722, Genbank ID NM_170735). There was a consistent, ∼35% decrease in reporter activity in cells transfected with miR-30a precursor relative to a scrambled precursor control (n = 3 independent experiments, data not shown).
Furthermore, we co-transfected both miR-30a and miR-195, which independently reduced luciferase activity, with half the amount needed to exert the observed significant inhibitory effect (375 ng of each per well), a concentration that for miR-30a was unable to induce significant repression (data not shown). The combination of these two miRNAs even in such lower concentrations resulted in a significant (∼31%) reduction in luciferase activity (Fig. 3B). This level of inhibition was not, however, significantly higher than the independent inhibitory effects of miR-30a (∼24%) and miR-195 (∼28%), so that the possibility of a synergistic effect remains to be clarified. These results indicate that a subset of miRNAs expressed in PFC, including miR-30a, can exert an inhibitory interaction with BDNF 3′-UTR sequences.
miR-30a negatively regulates BDNF protein in neurons
The validation of miR-30a:BDNF 3′-UTR interaction with luciferase reporter assays in two different cell lines, and its higher relative expression in neurons that are able to synthesize BDNF compared with some of the other candidate miRNAs, including miR-195, suggest that miR-30a is more likely to be a potential regulator of neuronal BDNF expression. To pursue this further, we used a lentivirus-based system to overexpress miR-30a precursor in cultured neurons (derived from E14.5 rat forebrain progenitor cells, see Materials and Methods) (Fig. 4A) and then assayed BDNF protein by ELISA. A lentivirus expressing miR-NSC30 (Fig. 4D) was also included in these experiments. Transduction efficiencies were verified with GFP expression and included a majority of cells (on average, 60%) (Fig. 4A). Cultures transduced with miR-30a showed a 2–4-fold increase in mature miR-30a levels, as measured by qRT–PCR (Supplementary Material, Fig. S4). There was a significant, ∼30% decrease in BDNF protein levels in neuronal cultures overexpressing miR-30a (Fig. 4B). In contrast, the control—which was identical to miR-30a except for three mutations in the seed sequence (Fig. 4C and D)—did not alter neuronal BDNF levels. Notably, miRNAs exhibiting partial complementarity to their mRNA target (such as miR-30a:BDNF 3′-UTR, Fig. 4C), predominantly block translation, but additional mechanisms involving mRNA decay have also been reported (1). Therefore, we assayed BDNF mRNA in our cultures, and no changes were observed (Supplementary Material, Fig. S4). We conclude that miR-30a exerts an inhibitory effect on BDNF translation in neurons.
Expression of selected miRNAs in PFC shows inverse correlation with BDNF protein during late adolescence and adulthood
Our experiments ex vivo described above strongly suggest that miR-30a-5p regulates BDNF protein levels, via interaction with a conserved sequence located in the proximal portion of BDNF 3′-UTR. Based on this observation, it could be possible that there is an inverse relationship between BDNF and miR-30a levels in (human) PFC tissue. Furthermore, it has been shown that when multiple miRNAs target a specific transcript, synergistic effects could lead to more robust target regulation when compared with each miRNA separately (36). Given this potential cooperativity in miRNA targeting and the limitations of our luciferase assay to address this issue, we wanted to investigate if their combined effects on BDNF levels during various stages of PFC development could be physiologically relevant.
Towards this end, we first used qRT–PCR to assay expression levels of the following miRNAs using seven sets of primers (with confirmed sequence specificity, see Materials and Methods) for qRT–PCR (miR-30a,b,c,d, miR-103/107, miR-191 and miR-195) in 37 PFC specimens (BA 10) across a wide age range, from the second trimester of pregnancy to 84 years. Tissue levels for these miRNAs were variable across the lifespan, although 5/7 miRNAs were defined by a significant increase in specimens from individuals older than 41 years, in comparison to either specimens younger than 15 years (miR-30c, miR-30d, miR-191, miR-195) (Fig. 5A and Supplementary Material, Fig. S5), or—in the case of miR-30a—in late adolescent to young adult specimens (ages 15-41) (Fig. 5A and B). Importantly, PMI, brain pH, gender and other postmortem confounds (See also Materials and Methods and Supplementary Material, Table S1) had no significant effect on miRNA expression (data not shown). Furthermore, miR-128a, a pan-neuronal miRNA not predicted to interact with BDNF, showed a progressive decrease during the course of PFC development (Fig. 5A and Supplementary Material, Fig. S5), which contrasts the observed increases in BDNF-related miRNAs (Fig. 5A and B and Supplementary Material, Fig. S5). Given these highly dynamic differences in miRNA levels during the course of PFC development, and especially in the mature PFC, we asked whether these changes relate to BDNF protein content.
Figure 5.
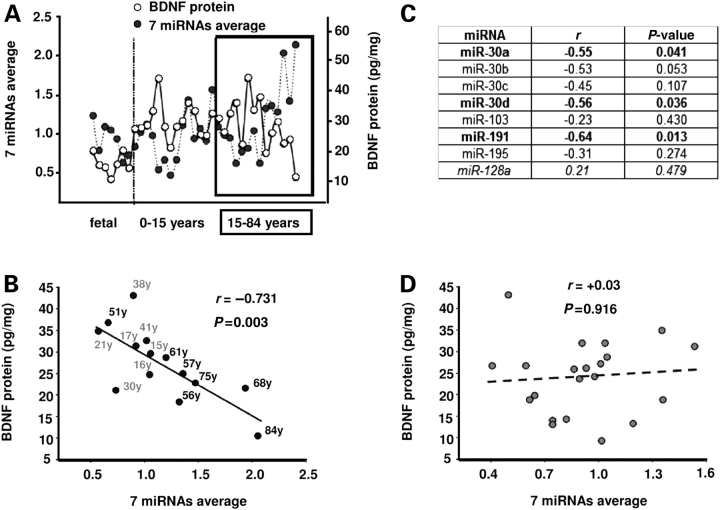
Expression patterns of selected miRNAs and BDNF in PFC across the lifespan. (A) Heatmap showing relative expression levels presented as quartiles (from higher to lower: red, yellow, green, blue) of (i) control miR-128a not predicted to target BDNF, (ii) 7 microRNAs each predicted to target BDNF and expressed in human PFC (data from qRT–PCR and normalized to 5S rRNA), (iii) average of weighted means of all 7 miRNAs shown in (ii), (iv) BDNF protein as measured by ELISA and (v) BDNF mRNA as measured by qRT–PCR and normalized to 18S rRNA. All samples from human PFC (gray matter, BA10) ranging in age from 21 estimated weeks of gestation (ewg) to 84 years (y). Brain Hemisphere (H), gender (G) and tissue pH (pH) are also shown for each sample. Notice the age-related expression changes of the BDNF-related miRNAs, including the decline in miR-30a levels in late adolescent—young adult group (15–41 years of age), which are distinct from the control miR-128a. (B–D) Bar graph summarizing the developmental expression data shown in (A) for miR-30a (B), BDNF protein (C), and BDNF mRNA levels (D). Graphs represent weighted means ± SEM. (n = 7–15 per age group) normalized to 5S rRNA for miRNAs and to 18S rRNA for BDNF mRNA, for (x-axis) four different age groups ranging from fetal to adult ages (n = 7–15/age group). Notice again the robust decline in miR-30a levels in the group of samples 15–41 years old and the significant increases in BDNF mRNA after birth (0–15 year) and in BDNF protein both after birth and between 15 and 41 years of age (B). *P < 0.05, (ANOVA post hoc Tukey).
To address this question, we measured BDNF protein by ELISA in the same postmortem specimens. Our data showed that both BDNF protein and mRNA are up-regulated during the early stages of postnatal PFC development, yet appear to be discordant during late adolescence and adulthood (Fig. 5A, C and D). In addition, there was, as expected, a positive correlation between BDNF mRNA and protein, in the entire developmental cohort (r = 0.379, P = 0.025) and, independently, in samples <15 years of age (r = 0.525, 0.021). However, there was no significant correlation in samples more than 15 years of age (data not shown).
Strikingly, in mature (15–84 years old) PFC a highly robust, inverse correlation between the expression of the 7 miRNAs as a group and BDNF protein levels (Fig. 6A and B) was observed. Furthermore, there was an inverse correlation between PFC BDNF levels for the same age group (15–84 years old), and for three miRNAs independently (miR-30a, miR-30d and miR-191) (Fig. 6C). These statistical associations were highly specific, because the (neuronal enriched) miR-128a—which is not predicted to target BDNF—had no correlation to BDNF protein levels in any age group (Fig. 6C and data not shown). In addition, no correlation was detected between BDNF mRNA and miRNA expression levels (data not shown). Taken together, these findings suggest that the orchestrated developmental expression of a group of miRNAs including miR-30a might exert an inhibitory effect on BDNF translation especially in the mature PFC.
Figure 6.
Inverse correlation between BDNF protein and BDNF-related miRNAs in late adolescent and adult PFC. (A) Plot showing case-by-case the relative changes in BDNF protein (white circles) when compared with weighted average of all seven miRNA probes (black) across the life span (n = 35). Notice the consistent inverse relationship between BDNF and miRNA average from late adolescence until old age (15–84 years, box in graph). (B) Inverse correlation between BDNF protein and average from weighted means of the seven miRNAs for 15–84-year-old samples (y, year, 15–41 shown in gray, rest shown in black); r, Pearson coefficient. Notice, also, declining BDNF, and increased miRNA levels in six out of the seven oldest samples. (C) Table showing correlations between miRNAs and BDNF protein in late adolescent and adult (15–84 years old) PFC. Notice independent significant inverse correlations between miR-30a, miR-30d and miR-191 with BDNF protein (all shown in bold), and lack of inverse relationship between BDNF and a control miRNA (miR-128a, shown in italics) not predicted to target BDNF. (D) No correlation between BDNF protein and average from weighted means of the seven miRNA probes for fetal to 15-year-old samples.
Transcriptional mechanisms regulating BDNF expression in immature PFC
In contrast to the significant findings in adults, there were no significant correlations between BDNF protein and the 7 miRNAs as a group, or individually, in subjects less than 15 years old (Fig. 6D and data not shown). However, we noticed that levels of BDNF mRNA were increased up to 3-fold after birth, consistent with an earlier report (26), but did not change significantly thereafter (Fig. 5D). These findings raise the possibility that BDNF levels at these earlier stages of PFC development are less dependent on miRNA-mediated post-transcriptional regulation and instead are regulated on the level of gene expression.
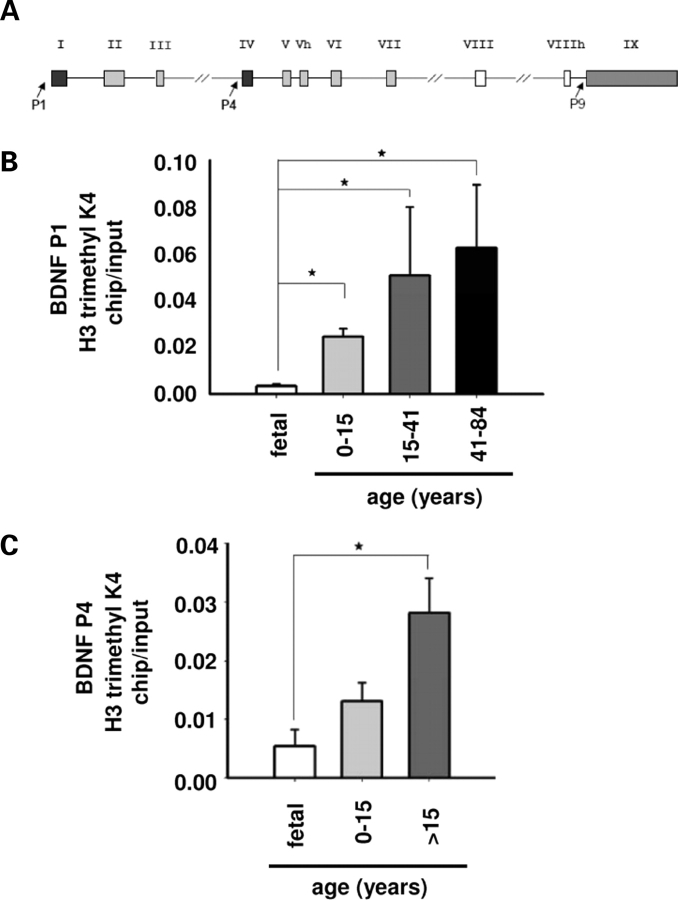
In order to address this issue, we measured levels of H3-trimethyl-lysine 4—an open chromatin mark related to transcriptional activity (37) that can be measured in postmortem brain (38)—at defined BDNF promoter sequences in PFC of fetal, child and adult samples. We assessed BDNF gene promoters I and IV (P1 and P4, Fig. 7A), as these are known to be epigenetically regulated in rodent cerebral cortex (39–42). As a control we also checked for changes of the same chromatin marker within the newly recognized promoter IX (P9, Fig. 7A), which reportedly shows only very low levels of activity in brain (43). Indeed, our results showed a significant increase in histone methylation at BDNF P1 occuring after birth (Fig. 7B), and at BDNF P4 after childhood (Fig. 7C). In contrast, histone methylation at BDNF P9 were very low and indistinguishable from background in all samples (data not shown). These results suggest that while miRNAs exert a robust effect on BDNF levels in mature and aging PFC, chromatin remodeling and transcriptional mechanisms might play a more prominent role at the earlier developmental stages.
Figure 7.
Chromatin remodeling at BDNF promoters during PFC development. (A) Illustration showing human BDNF gene, including its multiple exons and the three promoters (P1, P4 and P9, arrows) selected for chromatin immunoprecipitation studies (see text). (B and C) Developmental changes in tri-methylated histone H3-lysine 4 at BDNF P1 (B) and P4 (C). Bar graphs represent mean ± SEM of chip-to-input ratios (n = 3–6/age group). Notice significant increases in P1- and P4-associated histone methylation after birth or childhood [last two age groups merged in (C) due to limited number of samples], respectively. *P < 0.05, ANOVA post hoc Tukey.
DISCUSSION
Using multiple approaches, including microarray, LNA-ISH and qRT–PCR we identified a group of miRNAs that were abundantly expressed in different layers of human PFC and predicted to target a specific region within human BDNF 3′-UTR. Notably, albeit the sequence of these miRNAs is completely preserved in multiple mammalian species, there was lamina-specific expression in human but not in mouse neocortex. A subset of these miRNAs (miR-30a,b,c,d, miR-103/107, miR-191, miR-195) showed an inverse correlation with BDNF protein levels in the adult, but not in the immature human PFC. Among these miRNAs, miR-30a exerted a significant inhibitory interaction with BDNF 3′-UTR in functional assays and decreased BDNF protein levels in neuronal culture.
The significant inverse correlation between the group of the selected miRNAs and BDNF protein levels from late adolescence to old age suggests that these miRNAs could participate in post-transcriptional fine-tuning of BDNF expression in adult PFC, including the periods of late maturation and aging. Interestingly, BDNF mRNA levels in human PFC have been shown to increase from infancy to young adult age but subsequently are maintained at roughly the same levels during adulthood and old age (26). In contrast, BDNF protein levels are reportedly reduced during the aging of human PFC (44). Therefore, our studies could potentially explain these discrepancies in age-dependent changes of BDNF mRNA and protein, by showing that BDNF protein levels in mature and aging PFC could be driven in part by the post-transcriptional regulation mediated by BDNF-related miRNAs (Fig. 5 and Supplementary Material, Fig. S5).
In contrast, the absence of significant correlations between BDNF protein and the selected miRNAs in the younger PFC samples could be partly attributed to a more prominent transcriptional control of the BDNF gene in the immature PFC (defined here as the fetal and 0–15 years old samples). This hypothesis is further supported by dynamic increases in open chromatin-associated histone methylation at a subset of BDNF gene promoters during postnatal PFC development (Fig. 7). It has to be noted, though, that due to technical limitations we did not explore the expression levels of all BDNF alternative mRNA transcripts and the chromatin status of all BDNF gene promoters. In addition to the two layers of regulation outlined in the present study—miRNAs-mediated inhibition and chromatin remodeling—additional mechanisms that could affect expression and function of BDNF regulation may involve antisense non-coding transcripts originating from the BDNF locus (43).
The presence of miR-30a in large layer III pyramidal neurons of human PFC, as observed in the present study by qRT–PCR, LNA-ISH and laser capture (Fig. 2 and Supplementary Material, Fig. S2), is of particular interest given the fact that this neuronal population displays alterations in dendritic spine density (45) and soma size (46) in schizophrenia, a disease where deficits in BDNF levels have been reported in some postmortem cohorts (15,24). In this context it is intriguing that the developmental dynamics of miR-30a expression in human PFC include a pronounced decline in miR-30a levels during the late phase of PFC maturation (ages 15–41 years old), which coincides with the age of onset of the clinical symptomatology of psychiatric disease (29).
According to our present study, at least seven different miRNAs could contribute to the regulation of BDNF expression in human PFC (miR-30a-5p, miR-30b,c,d, miR-103/107, miR-191, miR-195). Interestingly, miR-107 was very recently shown to be significantly down-regulated in alzheimer's disease (13). Furthermore, in a previous study, miR-30b and miR-195 were shown to be reduced in schizophrenia PFC (11). Of note, miR-30a-3p was shown, by qRT–PCR, to be increased in cases of the same study; this miRNA is derived from the same precursor as miR-30a-5p, although its interactions with BDNF remain unclear. The potential role of these miRNAs for BDNF regulation and signaling in diseased brain remains to be clarified.
It is noteworthy that the BDNF-related miRNAs that were the focus of our study are also predicted to target numerous genes related to synaptogenesis, neuronal migration, neuronal growth and differentiation, according to multiple computational analysis tools (19–21). In this context, the miR-30 family of miRNAs is predicted to target multiples genes (19,47) implicated in the genetics or pathophysiology of schizophrenia other than BDNF; these include MAP6 (48), NR4A2 (49), GRM3 (50), GRM5 (51), CNR1 (52), NCAM1 (53,54) and NEUROG1 (55). Given their potential interaction with multiple schizophrenia risk genes and the reported interaction between BDNF and other miRNAs important for neuronal plasticity, additional studies are needed to elucidate the potential significance of this family of miRNAs in the context of psychiatric disease.
According to the present study, there is laminar specificity for several miRNAs expressed in human PFC, including miR-30a, miR-103/107, miR-495 (Fig. 2 and Supplementary Material, Fig. S2); for miR-103/107, this was also observed in (human) temporal neocortex (13). In striking contrast, the same miRNAs appeared to be either expressed evenly throughout layers II–VI of mouse neocortex (including frontal areas), or in the case of miR-495, below the detection limit (Supplementary Material, Fig. S2 and data not shown). It is possible that these species-related differences in cortical miRNA patterns could result in a more sophisticated lamina-specific regulation of BDNF expression in the human cortex.
Interestingly, a previous study has demonstrated that miR-134, a brain enriched miRNA that can inhibit the translation of the neurotrophin-related gene Lim-domain-containing protein kinase 1 (LIMK1) and regulate dendritic spine density, increases postnatally in mouse hippocampus, reaching its maximum levels at the age when synaptic maturation occurs (5). Therefore, one could hypothesize that maturation processes in the mammalian brain related to neurotrophin signaling could be influenced by the miRNA pathway. The findings presented here further support this scenario by showing that developmentally regulated miRNAs including members of the miR-30 family could modulate BDNF expression in human PFC.
Lastly, in addition to the miRNA-mediated inhibitory effects on BDNF levels as reported here, this neurotrophin might itself regulate the expression of neuronal miRNAs (56,57). Furthermore, BDNF could antagonize miRNA-mediated translational inhibition (5), possibly by activating the tropomyosin-related protein kinase B (TrK-B)/mammalian target of rapamycin (mToR) signaling pathway, which in turn interacts with subunits of the translation initiation complex (1,58). Interestingly, deficiency of protein kinase B (PKB or Akt1), a kinase that can activate the mTOR pathway alters neuronal morphology and leads to impaired PFC functions (59). It is therefore intriguing to speculate that miRNAs, including the ones discussed here, might participate in a molecular network involving multiple reciprocal nodes, that together orchestrate and fine-tune prefrontal BDNF expression and signaling in a developmental stage- and lamina-specific manner.
MATERIALS AND METHODS
Postmortem studies
Postmortem samples from 37 subjects, obtained from the dorso-rostral pole of the frontal lobe (Brodmann's area 10), were included in this study. All procedures were approved by the review boards of the participating institutions. All brains were fresh-frozen and stored at −80°C. The fetal, child and adolescent samples were obtained through the Brain and Tissue Banks for Development Disorders, University of Maryland (NICHD contract no. NO1-HD-8-3284). Adult samples were obtained from a brain bank located at the University of California at Davis (UCD), as described (38, 61). Demographics, medication status and postmortem confounds, including tissue pH and RNA integrity number (RIN) are provided in Supplementary Material, Table S1. For all experimental procedures, each assay included samples from all age groups. Adult brains were subjected to neuropathological examination to rule out neurodegenerative disease.
RNA isolation
Small RNAs (<200 nt) were isolated by using the mirVANA PARIS kit (Ambion), according to the manufacturer's instructions and treated with DNase I for 30 min at 37°C. Then, samples were incubated at RT (room temperature) for 2 min in DNase I inactivating buffer (Ambion—RNAqueous kit), followed by centrifugation (13 000g) for 1.5 min and supernatant was stored at −80°C. The mirVANA PARIS kit was also used for the extraction of large (>200nt) RNA that was used for measurement of BDNF mRNA. The mirVANA PARIS kit (Ambion) was used for total or small RNA isolation from rat neuronal cultures and the RNAqueous Micro kit (Ambion) was used for total RNA extraction in HEK-293 cells. All samples were treated with DNase I to avoid DNA contamination. For determination of RNA quality RNA RIN were calculated using the Agilent 2100 bioanalyzer and according to manufacturer's instructions.
RNA quantification
Amplicons were generated for 5S rRNA and the following miRNAs (see also Supplementary Material, Table S2): (i) mir-30a-5p, (ii) mir-128, (iii) miR-103, (iv) miR-30b (v) miR-30c, (vi) miR-30d, (vii) miR-191, (viii) miR-195, using mirVana qRT–PCR miRNA detection kit (Ambion). Applications were performed with an 7500 Applied Biosystems Real-Time PCR System and SDS software: Step 1, 95°C × 3 min; Step 2, 95°C × 15 s; Step 3, 60°C × 34 s, 40 cycles (Step 2, Step, 3), followed by dissociation step to obtain SYBR Green I-based melting curves. Specificity of the reactions was confirmed by melting curve analysis in conjunction with gel electrophoresis and, if necessary, subcloning and sequencing. For example, amplicons derived from miR-30a and miR-30d-specific PCR reactions (two miRNAs that differ in a single nucleotide in the middle portion of their mature sequence) yielded the correct sequence in >95% of clones (n = 37). For each sample and amplicon, cycle thresholds were averaged from triplicate reactions and normalized to 5S rRNA according to the following formula, E−CtmiRNA/E−Ct5srRNA, where E=(1+ln2/primer slope).
TaqMan One-Step RT–PCR (Applied Biosystems) was used according to manufacturer's instructions for the human BDNF 18S rRNA and rat BDNF and 18S rRNA with primers shown in Supplementary Material, Table S2.
miRNAs microarray
MiRNA expression profile was analyzed in pooled RNA samples isolated from parietal cortex of right hemisphere from 7 normal individuals (4 males with ages 41, 42, 52, 59 years and 3 females with ages 35, 44 and 57 years). Mixed RNA probes were labeled with Cy5 fluorescent dye and applied onto a µParaFlo™ Human miRNA chip (LcSciences). The chip contains seven redundant regions with miRNA probes corresponding to miRNA transcripts from miRNA Registry list (Human_V4E_050630 - Based on Sanger miRNA Registry Release 7.0), non-verified miRNA probes and multiple control probes. 5S rRNA was used as a housekeeping gene for normalization control.
The data were processed with background subtraction (regression-based background mapping method), Cy5 channel normalization (Locally-weighted regression method on the background-subtracted data) and detection determination (LcSciences data analysis). Transcripts were determined as detectable if their signal intensity was higher than 3× background standard deviation, spot CV (standard deviation/signal intensity) was <0.5 and transcripts had at least 50% of replicate probe signals registering above the detection level.
Solution hybridization
32P-UTP-labeled probes (mirVana miRNA Probe Construction Kit, Ambion) reverse antisense to the mature miRNAs and 5S rRNA were used in conjunction with the solution hybridization assay according to manufacturer's instructions (mirVana miRNA detection kit). Briefly, the small RNA sample was mixed with the probe and after hybridization in solution, samples were subjected to RNase digestion. The radiolabeled protected fragments of the probe after RNase inactivation and precipitation were separated in a denaturing polyacrylamide gel. Probe-specific sequences (without linker sequence) were as follows: 5′-CTTCCAGTCGAGGATGTTTACA-3′ (this is the reverse complement of the mature mir-30a-5p); 5′-ACTAGAGCCTTCGATT-3′ (this is the reverse complement of a conserved region within the 5S rRNA).
ISH with LNA-modified oligonucleotides
For LNA-ISH, 20 µm thick sections of immersion-fixed (human) or perfusion-fixed (mouse) cerebral cortex were mounted on SuperFrost-Plus slides (VWR), air-dried, then subjected to the following procedure with sterile solutions (DEPC-treated water): Washed with 1× PBS 3 × 5 min each, fixed with 4% paraformaldehyde in 0.1 M phosphate buffer for 15 min at RT, then washed again in 1× PBS 3 × 5 min each, then protein was denatured using 0.2 M HCL/1×PBS for 10 min at RT, then washed with 1×PBS 3 × 5 min, then treated with 0.25% acetic anhydride/0.1 M triethanolamine/1×PBS for 10 min at RT, washed again at 1× PBS 3 × 5 min, then prehybridized in 50–100 µl hybridization buffer/per section for 2 h at a specific temperature depending on probe based on the formula Tm probe −21°C, with Tm provided by probe vendor (Exiqon) and shown in Supplementary Material, Table S2. Each probe is a 5′-digoxigenin-labeled, 2′-O, 4′-C methylene bicyclonucleoside monomer-containing oligonucleotide (LNA, phosphoramidite). Sequences are reverse complement to the mature miRNAs (Supplementary Material, Table S2). The 20 ml of hybridization buffer was made of 50% deionized formamide/2×SSC/10% dextran sulfate/500 µg/ml sperm DNA/0.25 mg/ml yeast t-RNA/0.2 mg/ml BSA/50 µg/ml heparin/2.5 mm EDTA/0.1% Tween-20 in 2.3 ml DEPC-H20. After absorbing the prehybridization buffer with a kimwipe, 50–100 µl of hybridization buffer containing 0.17–0.25 µM of probe were added to each section and slides were covered with RNAse-free coverslips (HybriSlip, Molecular Probes) and incubated overnight at the specific temperatures (see above) in a humidified chamber. The following day, sections were washed twice in 2×SSC at RT for 15 min on a shaker, then washed in 1×SSC at 37°C for 15 min, then washed twice with 2×SSC/formamide, then with 0.1×SSC for 30 min at the probe-specific temperature (see above), then washed with 0.1×SSC for 15 min at RT, then incubated with buffer I (0.1% Tween-20/0.1 M Tris–HCL, pH 7.5/150 mm NaCl) for 10 min at RT, then with blocking solution [10% normal goat serum/1% blocking reagent (Roche) in buffer I] for 30 min at RT, then incubated with anti-digoxigenin-alkaline phosphate-conjugated antibody (goat, Roche) diluted 1:1000 in blocking solution (150 µl/slide) and parafilm-covered slides were incubated in a humidified chamber on shaker for 3 h at RT. Sections were then washed in buffer I 3 × 15 min each, then incubated in buffer III (0.1 M Tris–HCL, pH 9.5/0.1 M NaCl) at RT for 10 min, then 500 µl of color substrate solution (CSS) were added to each slide (CSS = nitroblue tetrazolium/BCIP stock solution (Roche) diluted 1:50 in buffer III) at RT under light-protected conditions overnight. Slides were then washed in TE buffer at RT for 10 min, then washed with 1×PBS at RT for 10 min, then with ddH20 at RT for 10 min. Finally the slides were coverslipped with 100 µl of VectaMount mounting medium (Vector Labs) for each slide, and were stored under light-protected conditions at RT for microscopic studies. Additional sections were first processed by LNA-ISH as described above, and then subjected to immunohistochemical labeling with the mouse monoclonal anti-NF-H (anti-SMI-32 antibody, Covance) and FITC-conjugated goat-anti mouse antibody, followed by diaminobenzidine (DAB)-based peroxidase detection with Vectastain ABC (Vector Labs).
BDNF immunoassay
Protein was extracted with the mirVANA PARIS kit according to manufacturer's instructions and after centrifugation the supernatants were used for estimation of total protein with BCA micro-kit (Pierce). BDNF levels were essayed with enzyme-linked immunosorbent assay (ELISA) and with the use of BDNF ELISA kit (Chemicon) according to manufacturer's instructions.
Immunohistochemistry, tissue dissection and laser capture microdissection procedures
Sections, 8–10 µm thick, were cut from frozen unfixed postmortem human tissue blocks (adult PFC—BA10) on a cryostat (Leica) on plain non-coated glass slides, stored at −80°C, than before staining they were dried for 2 min, fixed in 100% acetone for 2 min, air dried for 30 s, then washed in PBS and processed for immunohistochemistry with the mouse monoclonal anti-NF-H (anti-SMI-32 antibody, Covance) and FITC-conjugated goat-anti mouse antibody, with intermittent washing steps. This staining procedure was limited to altogether <100 min, and then sections were transferred to a Arcturus Veritas microdissection instrument (Molecular Devices) in order to collect somata of layer III NF-H immunoreactive pyramidal neurons, as defined by triangular shape and prominent, vertically oriented apical dendrite. As a control, tissue from deeper white matter was collected. Cells were collected in pools of 500–1000, using the CapSure MacroLCM Caps (Arcturus) collection caps and then transferred to RNase-free Eppendorf tubes and stored at −80°C until further processed. RNA was extracted with the mirVana miRNA isolation kit (Ambion). In particular, the plastic membrane containing harvested cells was removed from the CapSure cap and immersed into 300–400 µl of the kit's lysis-binding buffer, then incubated in the same solution at 42°C for 30 min with intermittent vortexing, in order to remove the laser-captured tissue from the membrane. The yield was ∼5 ng/µl small RNA/pool. For dissection of upper and deeper cortical layers, superficial cortical gray matter (approximately upper one-fifth of gray matter) and white matter from frozen unfixed postmortem tissue (n = 5, ages 30, 38, 56, 61, 68 years of age) was removed and the upper (roughly corresponding to parts of layers II and III) and lower one-third (roughly corresponding to parts of layers V and VI) of the remaining gray matter tissue was used for protein and RNA extraction.
Chromatin immunoprecipitation in postmortem tissue
Chromatin immunoprecipitation in postmortem tissue from human PFC of different age was done as described previously (38) by using 70–100 mg of tissue and with the primers shown in Supplementary Material, Table S2.
Luciferase assays
Ambion's pMIR-REPORT luciferase reporter plasmid was engineered to include a 551 bp fragment of human BDNF 3′-UTR (1500–2051 nt, Genbank ID NM_170735) at the 3′ end of the luciferase gene. Lipofectamine 2000 (Invitrogen) was used for transfection of HEK293 cells in 24-well plates. CMV-driven vectors containing chicken beta-actin promoter (CAG-RmiR plasmids) and expressing miRNA precursors (750 ng per well) were cotransfected with the luciferase reporter plasmid (150 ng per well) containing the 551 nt fragment of BDNF 3′-UTR and with Ambion's pMIR-REPORT β-galactosidase plasmid (100 ng per well) to control for transfection efficiency. Luciferase and β-galactosidase assays (Promega) were used to calculate the normalized luciferase expression. As controls 750 ng of vector expressing miR-NSC30 precursor with 3 bases difference in the seed sequence (see also below) or 750 ng of an EGFP expressing vector (control reference) were used. The overexpression of the mature miRNAs was measured with qRT–PCR and from at least two replicates.
Neuronal transduction
The pGIPZ self-inactivating lentiviral empty vector was purchased by Open Biosystems. Two sets of 111 bp oligos that encode the human miR-30a precursor or the miR-30a precursor with 3 bases difference in the seed sequence of the 5p mature miRNA (NSC30) and that contain XhoI and EcoRI restriction enzyme overhangs (purchased by IDT Integrated DNA Technologies) were annealed and initially ligated into a double digested (XhoI, EcoRI) self-inactivating retroviral vector pSM2c by Open Biosystems (Purchased by the shRNA Library Core Facility of UMass Medical School). After PCR and subsequent digestion this product was then ligated to the pGIPZ self-inactivating lentiviral empty vector. The final products (called pmiR30 and NSC30) are designed to drive expression of tGFP (turbo GFP) and the miRNA precursor molecule, through the same CMV RNA polymerase II promoter. The expected mature miRNA of the miR-NSC30 precursor molecule is not predicted to target BDNF mRNA at any region (RNA hybrid software). Standard methodologies were used for preparation of rat forebrain neuronal cultures from precursor cells (38), for viral production and infection (60). The production of the mature miR-30a was assayed with qRT–PCR. In addition, transduction efficiency was estimated by measuring GFP expression 4 days post-infection with epi-fluorescence microscopy (Nikon Eclipse E600). An average of 60% transfection efficiency was observed.
Statistical analysis
For the analysis of the Luciferase data, and after proper normalization of Luciferase activity to β-galactosidase activity and logarithmic transformation, data were evaluated using analysis of variance (ANOVA) for a mixed model by REML (restricted estimation by maximal likelihood). In the presence of significant main or interaction effects, pairwise comparisons were evaluated using Tukey Kramer adjustment for multiple comparisons. In the cases where no pairing was required, then ANOVA with post hoc Tukey was applied after ensuring normalized distribution of data. For presentation of data from age groups with different age representation and sample size and for allowing the comparison or combination of ‘relative’ changes in the values measured, weighted means were selected for data shown on Figs 5 and 6.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
FUNDING
This work was supported by National Institute of Mental Health (MH071476 to S.A.); National Institute of Child Health and Human Development (HD048489 to S.A.); NARSAD Distinguished Investigator Award (to E.R.); and Stanley Medical Research Institute (to E.R.).
AUTHOR CONTRIBUTIONS
N.M. conceived the hypothesis, designed and conducted the majority of the experiments. A.G. and E.R. conducted the microarray studies, H.-S.H. contributed to various experiments and S.A. designed experiments and wrote the manuscript together with N.M.
ACKNOWLEDGEMENTS
We would like to thank Dr Edward G. Jones (University of California Davis) and Dr William E. Bunney Jr. (University of California at Irvine) and the Brain and Tissue Bank for Developmental Disorders (NICHD contract number NO1-HD-8-3284) (Director: Dr Ron Zielke) for providing postmortem samples; Dr Zhuoshang Xu and Dr Chunxing Yang for sharing the CAG-RmiR vector design, Dr David Weaver for valuable feedback, Dr Stephen Baker for his assistance in statistical analysis, David Burns for help in live imaging and Yin Guo, Catheryne Whittle, Mathieu Guillaume, Katerina Ikonomu, Regina Bergmeier, Anouch Matevossian and Gulnaz Faskhutdinova for excellent technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Chang T.C., Mendell J.T. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 4.Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 5.Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 6.Bak M., Silahtaroglu A., Moller M., Christensen M., Rath M.F., Skryabin B., Tommerup N., Kauppinen S. MicroRNA expression in the adult mouse central nervous system. Rna. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miska E.A., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abelson J.F., Kwan K.Y., O'Roak B.J., Baek D.Y., Stillman A.A., Morgan T.M., Mathews C.A., Pauls D.L., Rasin M.R., Gunel M., et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 9.Beveridge N.J., Tooney P.A., Carroll A.P., Gardiner E., Bowden N., Scott R.J., Tran N., Dedova I., Cairns M.J. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum. Mol. Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 10.Kim J., Inoue K., Ishii J., Vanti W.B., Voronov S.V., Murchison E., Hannon G., Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins D.O., Jeffries C.D., Jarskog L.F., Thomson J.M., Woods K., Newman M.A., Parker J.S., Jin J., Hammond S.M. microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007;8:R27. doi: 10.1186/gb-2007-8-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang G., van der Walt J.M., Mayhew G., Li Y.J., Zuchner S., Scott W.K., Martin E.R., Vance J.M. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am. J. Hum. Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W.X., Rajeev B.W., Stromberg A.J., Ren N., Tang G., Huang Q., Rigoutsos I., Nelson P.T. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J. Neurosc.i. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertolino A., Callicott J.H., Elman I., Mattay V.S., Tedeschi G., Frank J.A., Breier A., Weinberger D.R. Regionally specific neuronal pathology in untreated patients with schizophrenia: a proton magnetic resonance spectroscopic imaging study. Biol. Psychiatry. 1998;43:641–648. doi: 10.1016/s0006-3223(97)00555-6. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto T., Bergen S.E., Nguyen Q.L., Xu B., Monteggia L.M., Pierri J.N., Sun Z., Sampson A.R., Lewis D.A. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J. Neurosci. 2005;25:372–383. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorski J.A., Zeiler S.R., Tamowski S., Jones K.R. Brain-derived neurotrophic factor is required for the maintenance of cortical dendrites. J. Neurosci. 2003;23:6856–6865. doi: 10.1523/JNEUROSCI.23-17-06856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angelucci F., Brene S., Mathe A.A. BDNF in schizophrenia, depression and corresponding animal models. Mol. Psychiatry. 2005;10:345–352. doi: 10.1038/sj.mp.4001637. [DOI] [PubMed] [Google Scholar]
- 18.Lugli G., Larson J., Martone M.E., Jones Y., Smalheiser N.R. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- 19.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 20.Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 21.Lall S., Grun D., Krek A., Chen K., Wang Y.L., Dewey C.N., Sood P., Colombo T., Bray N., Macmenamin P., et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 22.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q.R., Walther D., Drgon T., Polesskaya O., Lesnick T.G., Strain K.J., de Andrade M., Bower J.H., Maraganore D.M., Uhl G.R. Human brain derived neurotrophic factor (BDNF) genes, splicing patterns, and assessments of associations with substance abuse and Parkinson's Disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;134:93–103. doi: 10.1002/ajmg.b.30109. [DOI] [PubMed] [Google Scholar]
- 24.Weickert C.S., Hyde T.M., Lipska B.K., Herman M.M., Weinberger D.R., Kleinman J.E. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol. Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 25.Huntley G.W., Benson D.L., Jones E.G., Isackson P.J. Developmental expression of brain derived neurotrophic factor mRNA by neurons of fetal and adult monkey prefrontal cortex. Brain Res. Dev. Brain Res. 1992;70:53–63. doi: 10.1016/0165-3806(92)90103-4. [DOI] [PubMed] [Google Scholar]
- 26.Webster M.J., Weickert C.S., Herman M.M., Kleinman J.E. BDNF mRNA expression during postnatal development, maturation and aging of the human prefrontal cortex. Brain Res. Dev. Brain. Res. 2002;139:139–150. doi: 10.1016/s0165-3806(02)00540-0. [DOI] [PubMed] [Google Scholar]
- 27.Smirnova L., Grafe A., Seiler A., Schumacher S., Nitsch R., Wulczyn F.G. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 28.Nelson P.T., Baldwin D.A., Kloosterman W.P., Kauppinen S., Plasterk R.H., Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. Rna. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis D.A., Cruz D., Eggan S., Erickson S. Postnatal development of prefrontal inhibitory circuits and the pathophysiology of cognitive dysfunction in schizophrenia. Ann. N. Y. Acad. Sci. 2004;1021:64–76. doi: 10.1196/annals.1308.008. [DOI] [PubMed] [Google Scholar]
- 30.Campbell M.J., Morrison J.H. Monoclonal antibody to neurofilament protein (SMI-32) labels a subpopulation of pyramidal neurons in the human and monkey neocortex. J. Comp. Neurol. 1989;282:191–205. doi: 10.1002/cne.902820204. [DOI] [PubMed] [Google Scholar]
- 31.Rathjen T., Nicol C., McConkey G., Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 35.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greco S.J., Rameshwar P. MicroRNAs regulate synthesis of the neurotransmitter substance P in human mesenchymal stem cell-derived neuronal cells. Proc. Natl. Acad. Sci. USA. 2007;104:15484–15489. doi: 10.1073/pnas.0703037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eissenberg J.C., Shilatifard A. Leaving a mark: the many footprints of the elongating RNA polymerase II. Curr. Opin. Genet. Dev. 2006;16:184–190. doi: 10.1016/j.gde.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Huang H.S., Matevossian A., Whittle C., Kim S.Y., Schumacher A., Baker S.P., Akbarian S. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J. Neurosci. 2007;27:11254–11262. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W.G., Chang Q., Lin Y., Meissner A., West A.E., Griffith E.C., Jaenisch R., Greenberg M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 40.Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 41.Nelson E.D., Kavalali E.T., Monteggia L.M. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J. Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsankova N.M., Berton O., Renthal W., Kumar A., Neve R.L., Nestler E.J. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 43.Pruunsild P., Kazantseva A., Aid T., Palm K., Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durany N., Michel T., Zochling R., Boissl K.W., Cruz-Sanchez F.F., Riederer P., Thome J. Brain-derived neurotrophic factor and neurotrophin 3 in schizophrenic psychoses. Schizophr. Res. 2001;52:79–86. doi: 10.1016/s0920-9964(00)00084-0. [DOI] [PubMed] [Google Scholar]
- 45.Glantz L.A., Lewis D.A. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 46.Pierri J.N., Volk C.L., Auh S., Sampson A., Lewis D.A. Decreased somal size of deep layer 3 pyramidal neurons in the prefrontal cortex of subjects with schizophrenia. Arch. Gen. Psychiatry. 2001;58:466–473. doi: 10.1001/archpsyc.58.5.466. [DOI] [PubMed] [Google Scholar]
- 47.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu H., Iwayama Y., Yamada K., Toyota T., Minabe Y., Nakamura K., Nakajima M., Hattori E., Mori N., Osumi N., et al. Genetic and expression analyses of the STOP (MAP6) gene in schizophrenia. Schizophr. Res. 2006;84:244–252. doi: 10.1016/j.schres.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 49.Rojas P., Joodmardi E., Hong Y., Perlmann T., Ogren S.O. Adult mice with reduced Nurr1 expression: an animal model for schizophrenia. Mol. Psychiatry. 2007;12:756–766. doi: 10.1038/sj.mp.4001993. [DOI] [PubMed] [Google Scholar]
- 50.Egan M.F., Straub R.E., Goldberg T.E., Yakub I., Callicott J.H., Hariri A.R., Mattay V.S., Bertolino A., Hyde T.M., Shannon-Weickert C., et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc. Natl. Acad. Sci. USA. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Devon R.S., Anderson S., Teague P.W., Muir W.J., Murray V., Pelosi A.J., Blackwood D.H., Porteous D.J. The genomic organisation of the metabotropic glutamate receptor subtype 5 gene, and its association with schizophrenia. Mol. Psychiatry. 2001;6:311–314. doi: 10.1038/sj.mp.4000848. [DOI] [PubMed] [Google Scholar]
- 52.Ujike H., Takaki M., Nakata K., Tanaka Y., Takeda T., Kodama M., Fujiwara Y., Sakai A., Kuroda S. CNR1, central cannabinoid receptor gene, associated with susceptibility to hebephrenic schizophrenia. Mol. Psychiatry. 2002;7:515–518. doi: 10.1038/sj.mp.4001029. [DOI] [PubMed] [Google Scholar]
- 53.Barbeau D., Liang J.J., Robitalille Y., Quirion R., Srivastava L.K. Decreased expression of the embryonic form of the neural cell adhesion molecule in schizophrenic brains. Proc. Natl. Acad. Sci. USA. 1995;92:2785–2789. doi: 10.1073/pnas.92.7.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan P.F., Keefe R.S., Lange L.A., Lange E.M., Stroup T.S., Lieberman J., Maness P.F. NCAM1 and neurocognition in schizophrenia. Biol. Psychiatry. 2007;61:902–910. doi: 10.1016/j.biopsych.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 55.Fanous A.H., Chen X., Wang X., Amdur R.L., O'Neill F.A., Walsh D., Kendler K.S. Association between the 5q31.1 gene neurogenin1 and schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144:207–214. doi: 10.1002/ajmg.b.30423. [DOI] [PubMed] [Google Scholar]
- 56.Klein M.E., Lioy D.T., Ma L., Impey S., Mandel G., Goodman R.H. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 2007;10:1513–1514. doi: 10.1038/nn2010. [DOI] [PubMed] [Google Scholar]
- 57.Vo N., Klein M.E., Varlamova O., Keller D.M., Yamamoto T., Goodman R.H., Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gingras A.C., Kennedy S.G., O'Leary M.A., Sonenberg N., Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai W.S., Xu B., Westphal K.G., Paterlini M., Olivier B., Pavlidis P., Karayiorgou M., Gogos J.A. Akt1 deficiency affects neuronal morphology and predisposes to abnormalities in prefrontal cortex functioning. Proc. Natl. Acad. Sci. USA. 2006;103:16906–16911. doi: 10.1073/pnas.0604994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W., Qu Q., Smith F.I., Kilpatrick D.L. Self-inactivating lentiviruses: versatile vectors for quantitative transduction of cerebellar granule neurons and their progenitors. J. Neurosci. Methods. 2005;149:144–153. doi: 10.1016/j.jneumeth.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 61.Akbarian S., Kim J.J., Potkin S.G., Hagman J.O., Tafazzoli A., Bunney W.E., Jr, Jones E.G. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch. Gen. Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.