Abstract
The steroid 5-alpha reductase type II gene (SRD5A2) encodes the enzyme which converts testosterone (T) to the more active androgen dihydrotestosterone. A non-synonymous single-nucleotide polymorphism, A49T (rs9282858), in SRD5A2 has been implicated in prostate cancer risk; however, results have been inconsistent. In 1999, we reported a strong association between the A49T variant and prostate cancer risk among African-Americans and Latinos in the Hawaii–Los Angeles Multiethnic Cohort (MEC). We report here an updated analysis of MEC data including the five major ethnic groups of the MEC, an increased sample size, improved genotyping technology and a comprehensive meta-analysis of the published literature. We found a non-statistically significant positive association between prostate cancer risk and carrying either the AT or TT genotype [odds ratio (OR) = 1.16, 95% confidence interval (CI) 0.79–1.69] in the MEC. This finding is in contrast to our previous results of ORs of 3.28 and 2.50 for the association between prostate cancer risk and the variant in African-American and Latino men, respectively; this can be accounted for by genotyping error in our earlier study. Meta-analysis of the published literature, including the current MEC data, shows a summary OR of 1.13 (95% CI 0.95–1.34) for the A49T variant with prostate cancer risk among sporadic, unselected cases. After evaluating more than 6000 cases and 6000 controls, there is little evidence of a role for the SRD5A2 A49T variant in prostate cancer risk. Overall, this report highlights the importance of rigorous genotyping quality control measures and replication efforts in genetic association studies.
INTRODUCTION
The prostate is androgen dependent and it has been hypothesized that variation in genes involved in androgen biosynthesis and metabolism might be risk factors for prostate cancer. A key gene in this pathway is the steroid 5-alpha reductase type II gene (SRD5A2) which encodes the enzyme that converts testosterone (T) to its more potent form, dihydrotestosterone (DHT). In an early 1999 analysis from the Hawaii–Los Angeles Multiethnic Cohort (MEC) study that considered only African-Americans and Latinos, we reported that a single missense variant in exon 1, A49T (which replaces alanine at codon 49 with threonine; rs9282858) was associated with increased risk of prostate cancer (1). Higher enzymatic activity in the presence of this missense variant has been found in in vitro studies (1,2).
Since our original publication, there have been 19 additional publications across a variety of ethnic groups, with equivocal results (3–21). A meta-analysis of five studies published in 2003 (including our 1999 data) showed a fixed effects summary odds ratio (OR) of 1.39 (95% CI 0.98–1.96) under a dominant genetic model (22). This variant has also been examined in relation to benign prostatic hyperplasia (9,23,24), serum hormone levels (25) and prostate cancer prognosis (7,26), all without definitive results.
We report here on an updated analysis conducted with all five major ethnic groups of the MEC to further explore the association between the A49T variant and prostate cancer risk with substantially larger numbers and improved genotyping technology from that originally presented in 1999 as well as a comprehensive meta-analysis which includes 11 published studies.
RESULTS
The demographic and genotype characteristics of the MEC study participants are shown in Table 1. The age and education level distributions were comparable between cases and controls. Cases were more likely to have a first-degree relative with prostate cancer.
Table 1.
Descriptive characteristics of the study population by racial/ethnic groupa
| Characteristic | African-American |
Japanese-American |
Native-Hawaiian |
Latino |
White |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Co (n = 642) | Ca (n = 642) | Co (n = 470) | Ca (n = 430) | Co (n = 68) | Ca (n = 66) | Co (n = 564) | Ca (n = 585) | Co (n = 460) | Ca (n = 432) | |
| Age, mean (SD) | 67.3 (6.3) | 68.0 (6.7) | 69.3 (6.8) | 69.8 (6.3) | 67.9 (6.3) | 67.6 (5.9) | 67.3 (6.2) | 67.7 (6.1) | 67.9 (7.1) | 68.2 (7.0) |
| Education, n (%) | ||||||||||
| 0–10th grade | 96 (15.1) | 80 (12.7) | 26 (5.6) | 40 (9.3) | 7 (10.8) | 13 (19.7) | 185 (33.5) | 217 (37.6) | 22 (4.8) | 29 (6.7) |
| 11–12th grade | 151 (23.8) | 156 (24.7) | 155 (33.1) | 136 (31.7) | 29 (44.6) | 27 (40.9) | 113 (20.4) | 134 (23.2) | 69 (15.1) | 62 (14.4) |
| Vocational/some college | 234 (36.9) | 226 (35.8) | 141 (30.1) | 116 (27.0) | 17 (26.15) | 17 (25.8) | 171 (30.9) | 142 (24.6) | 117 (25.6) | 143 (33.2) |
| College graduate | 154 (24.3) | 169 (26.8) | 146 (31.2) | 137 (31.9) | 12 (18.5) | 9 (13.6) | 84 (15.2) | 84 (14.6) | 250 (54.6) | 197 (45.7) |
| Family history of PrCa, n (%) | ||||||||||
| 1 + first-degree relative | 71 (12.7) | 78 (13.6) | 24 (5.5) | 39 (9.6) | 6 (9.0) | 7 (11.7) | 36 (7.0) | 61 (11.8) | 35 (8.0) | 54 (13.2) |
| No first-degree degree relative | 490 (87.3) | 496 (86.4) | 415 (94.5) | 366 (90.4) | 61 (91.0) | 53 (88.3) | 480 (93.0) | 458 (88.2) | 404 (92.0) | 354 (86.8) |
| Genotype, n (%) | ||||||||||
| AA | 630 (98.1) | 628 (97.8) | 470 (100) | 430 (100) | 66 (97.1) | 66 (100) | 551 (97.7) | 566 (96.8) | 435 (94.6) | 406 (94.0) |
| AT | 12 (1.9) | 14 (2.2) | 0 | 0 | 2 (2.9) | 0 | 13 (2.3) | 19 (3.2) | 25 (5.4) | 25 (5.8) |
| TT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (0.02) |
| Stage | ||||||||||
| Localized, n | 413 | 235 | 39 | 343 | 270 | |||||
| AA, n (%) | 405 (98.1) | 235 (100) | 39 (100) | 329 (98.9) | 253 (93.7) | |||||
| AT, n (%) | 8 (1.9) | 0 | 0 | 14 (4.1) | 17 (6.3) | |||||
| TT, n (%) | 0 | 0 | 0 | 0 | 0 | |||||
| Advanced, n | 172 | 169 | 25 | 198 | 140 | |||||
| AA, n (%) | 168 (97.7) | 169 (100) | 25 (100) | 194 (98.0) | 132 (94.3) | |||||
| AT, n (%) | 4 (2.3) | 0 | 0 | 4 (2.0) | 7 (5.0) | |||||
| TT, n (%) | 0 | 0 | 0 | 0 | 1 (0.7) | |||||
Co, controls; Ca, cases; SD, standard deviation; PrCa, prostate cancer.
aNumbers in the tables do not always sum to total numbers in the study because of missing data.
No carriers of the T allele were identified among the Japanese-American cases and controls and only two native-Hawaiian controls carried a T allele. Only one man had the TT genotype, a white case. The variant allele was most common among the whites and was in Hardy–Weinberg equilibrium (HWE) among controls in all of the ethnic groups.
Risk analysis was carried out among the ethnic groups that were polymorphic at this site. We found no statistically significant association between the T allele and risk of prostate cancer among African-Americans, Latinos or whites. Native-Hawaiians were included in the ethnicity-adjusted analysis: the OR was 1.16 for all the four groups combined (95% CI 0.79–1.69; Table 2).
Table 2.
Association between A49T genotype and risk of prostate cancer by racial/ethnic group
| Race/ethnicitya | Genotype | OR (95% CI) |
||
|---|---|---|---|---|
| All cases | Localized cases | Advanced cases | ||
| African-Americanb | AA | 1.0 | 1.0 | 1.0 |
| AT/TT | 1.15 (0.53–2.51) | 1.01 (0.41–2.49) | 1.27 (0.40–3.99) | |
| Latinob | AA | 1.0 | 1.0 | 1.0 |
| AT/TT | 1.42 (0.69–2.90) | 1.76 (0.82–3.79) | 0.90 (0.29–2.79) | |
| Whiteb | AA | 1.0 | 1.0 | 1.0 |
| AT/TT | 1.11 (0.63–1.96) | 1.16 (0.62–2.20) | 1.06 (0.47–2.40) | |
| All groupsc | AA | 1.0 | 1.0 | 1.0 |
| AT/TT | 1.16 (0.79–1.69) | 1.23 (0.81–1.88) | 1.01 (0.57–1.79) | |
aJapanese were excluded from the analysis because there were no carriers of the T allele among cases or controls; native Hawaiians were included in the all groups analysis only because there were only T carriers among the controls.
bOR adjusted for age.
cAdjusted for age (<60/60+) and ethnicity.
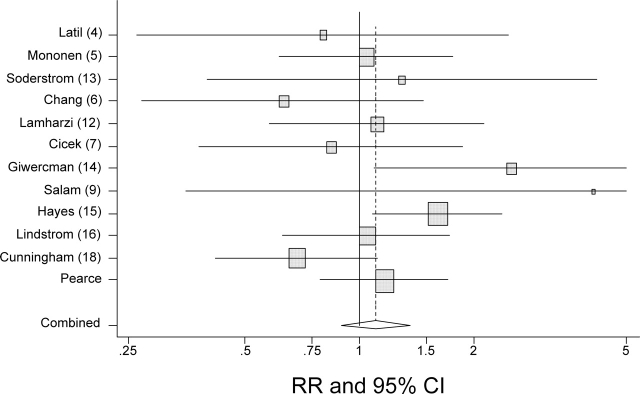
On meta-analysis of the published A49T prostate cancer literature (4–7,9,12–16,18) and excluding the present analysis, we find no association between this variant and risk of prostate cancer [OR = 1.11, 95% CI 0.91–1.34, P for heterogeneity 0.15; excludes Makridakis et al. (1) due to the genotyping error; data not shown]. When the results from the MEC Taqman genotyping are incorporated into the meta-analysis, the results are largely unchanged (OR = 1.13, 95% CI 0.95–1.34, P for heterogeneity 0.20; Fig. 1).
Figure 1.
Forest plot of relative risks (RR) and 95% CIs of the association between the AT/TT genotyping and risk of prostate cancer from all published studies and the current results. The study-specific (boxes) and summary ORs (diamond) and 95% CIs (lines) are shown. The size of each box is proportionate to the number of subjects genotyped. The combined RR is 1.13 (95% CI 0.95–1.34, P for heterogeneity 0.20).
DISCUSSION
We found no statistically significant association between the SRD5A2 A49T genotype and risk of prostate cancer in any of the four MEC ethnic groups in whom the polymorphism occurred (Table 2). This finding is in contrast to the original results from the MEC which showed a statistically significantly increased risk of prostate cancer among carriers of the SRD5A2 T variant for both African-Americans and Latinos which we now know to be based on genotyping error (1).
At the time our original results were published, single-strand conformation polymorphism (SSCP) was an accepted genotyping method; however, this screening method is now widely deemed unreliable with higher frequencies of both false-positive and false-negative results than occur with modern techniques such as Taqman. The genotype data from our original report were out of HWE in controls, and at that time it was believed this was because the minor allele was rare and thus tests for HWE were not reliable. We now know this was not the case and in fact this quality control measure had identified genotyping error (the most likely reason for deviations from HWE).
We observed bias away from the null in the presence of genotyping error in our original report (1). The explanation for this is likely due to the fact that African-American and Latino cases in Los Angeles were ascertained before controls to avoid losses due to death. Samples were sent to the laboratory and genotyped as they were collected, and cases were therefore genotyped earlier than controls. We speculate that laboratory technique may have changed over time, either due to the laboratory technicians becoming more experienced or to a change in technician such that, initially, more variant alleles were observed. Because cases were genotyped first, this would have led to a non-random error and spurious findings. This underscores the importance of rigorous quality-control measures in genetic association studies as described by Rebbeck et al. (27).
The literature surrounding the A49T variant has been inconsistent, demonstrating the complexity of interpreting genetic association study results and the importance of large sample sizes and replication of findings. On meta-analysis including more than 6000 cases and 6000 controls from 12 studies (including our current data), we find little or no association between this variant and risk of prostate cancer (OR = 1.13, 95% CI 0.95–1.34).
Further investigation into this variant with regard to sporadic prostate cancer is probably not warranted; even if this variant confers a 34% increased risk of prostate cancer (the upper bound of the 95% CI from the meta-analysis), the public health impact of such a modest effect and a low minor allele frequency would be minimal. We and others have shown that only one-third of genetic association studies replicate (28); the findings presented here highlight the importance of rigorous quality control and replication efforts in these types of studies.
MATERIALS AND METHODS
Study population
We completed a case–control study nested within the MEC study, including 2155 incident cases and 2204 male controls from the African-American, native-Hawaiian, Japanese-American, Latino and white subjects enrolled in the cohort. Details of the MEC study have been published previously (29). Briefly, more than 200 000 men and women between the ages of 45 and 75 and residing in Hawaii and California (mainly Los Angeles County) completed a questionnaire in the period 1993–96 which included data on demographic, lifestyle and health characteristics as well as a comprehensive dietary survey (29).
Participants in the MEC are followed for incident cancers by computer linkage of the cohort with the Surveillance, Epidemiology and End Results (SEER) cancer registries in Hawaii, Los Angeles and California. Both incident prostate cancer cases and a random sample of male controls in the MEC were contacted by telephone and asked to provide a blood specimen. The overall participation rate for blood collection was 72% for cases and 69% for controls.
The men who agreed to participate in the blood collection provided written informed consent following study approval by both the University of Hawaii and the University of Southern California Institutional Review Boards.
Sample preparation and genotyping
Blood components were separated and stored in 0.5 ml volumes in liquid nitrogen. The blood samples were processed within 4 h of collection. Individuals included in this study had their blood samples taken through April 2002. The African-American and Latino subjects included in this study overlap with those from Makridakis et al. (1). The only cases from the Makridakis et al. study who were excluded from the present study were those for whom DNA was no longer available.
DNA was purified from lymphocytes of peripheral blood samples for all cases and controls using the Qiagen Blood Kit (Qiagen, Chatsworth, CA, USA).
Genotyping by 5′ nuclease Taqman allelic discrimination assay (Taqman; Applied Biosystems, Foster City, CA, USA) was used for the A49T variant (rs9282858). The laboratory personnel were masked as to case–control status. Masked 5% replicate samples (n = 223) were also included: reproducibility was 100%.
In our original 1999 study, genotyping was done by SSCP (1). All samples for which SSCP genotyping had been conducted (both for samples included in our original paper and additional samples genotyped after the paper was published) and for which DNA was still available were genotyped by Taqman (n = 2498). Results obtained from the Taqman genotyping platform were compared with those originally obtained by SSCP and a total of 50 discordant results were observed (Table 3). All of the discordant results and a random sample of 46 concordant results were then sequenced using the following approach:
Table 3.
Concordance between SSCP and Taqman genotyping (n = 2498)
| SSCP genotype | Taqman genotype |
||
|---|---|---|---|
| AA (n = 2435) | AT (n = 63) | TT (n = 0) | |
| AA (n = 2426) | 2414 | 12 | 0 |
| AT (n = 53) | 19 | 34 | 0 |
| TT (n = 19) | 2 | 17 | 0 |
Amplification of genomic DNA by PCR for SRD5A_A49T was done using oligonucleotide primers (forward: 5′-gatgcaggttcagtgccag- 3′; reverse: 5′-cgctacctgtggaagtaatgtag-3′; fragment size: ∼300 bp) and the GeneAmp PCR system 9700 (Applied Biosystems). PCR reactions were carried out in a total volume of 25 μl. Reaction mix contained 20 ng of genomic DNA, 40 pmol of forward and reverse primers, 100 µm dNTPs, 2 U of AmpliTaq Gold (Applied Biosystems) and 1× reaction buffer. Thermal conditions consisted of one cycle at 95°C for 5 min, 35 cycles at 94°C for 30 s, 55°C for 30 s, 72°C for 1 min and one cycle at 72°C for 10 min. The amplified PCR products were purified using ExoSAP-IT (USB Corporation, Cleveland, OH, USA).
After PCR amplification and purification, the BigDye terminator Reaction Mix (Applied Biosystems) was utilized to labeling the extension reactions; the extension products were purified using DyeEx 96 Kit (Qiagen) and sequenced by capillary electrophoresis on an ABI3730xl DNA Analyzer (Applied Biosystems). PolyPhred was used to analyze the sequence traces to identify the presence of the single-nucleotide polymorphism.
Forty-eight of the 50 SSCP-Taqman discordant results and 43 of the 46 concordant results were successfully sequenced. Of the 91 sequenced samples, 97% of the Taqman genotypes were confirmed compared with 50% of the SSCP results. The SSCP genotyping results were considered unreliable on the basis of the lack of concordance, with the sequencing results and all analyses presented in this paper are based on the results obtained from the Taqman genotyping platform.
Data analysis
Prostate cancers were classified according to disease severity using a combination of stage and Gleason grade. Local disease was defined as tumors localized to the prostate with a Gleason grade of less than 8. Advanced disease was defined as tumors that had regional extension or distant metastases, regardless of grade, and tumors that were localized to the prostate, but had a Gleason grade of 8 or higher. The results were unchanged when advanced disease did not include localized cases with a Gleason grade of 8 or higher.
Unconditional logistic regression was used to model the association between risk of prostate cancer and A49T genotype. All analyses were adjusted for age (<60/60+) and ethnicity, as appropriate. Results were unchanged when age was not included in the model. Individuals with either an AT or TT genotypes were grouped together due to the rarity of the TT genotype.
Because the A49T variant has been examined by a number of investigators in relation to prostate cancer risk, a meta-analysis of published studies of sporadic prostate cancer was also undertaken. Published studies were identified by searching PubMed through the National Library of Medicine (www.pubmed.gov), using combinations of the search terms ‘SRD5A2’, ‘A49T’, ‘prostate cancer’, ‘prostate’, ‘5-alpha reductase’, ‘polymorphism’ and ‘SNP’. The identified papers were then reviewed and if additional references were found in these publications they were also reviewed. Papers or specific analyses within papers were excluded if they were not available in English (21), did not observe the variant allele in cases or controls (3,7,10,11,20), had overlapping subjects from another included report (17,19) or studied a selected set of cases, e.g. hereditary prostate cancer (6,18) or young cases (8). The reported ORs with associated 95% CIs or the reported genotype counts were extracted from the papers for the meta-analysis, and the software package Stata (Statacorp, College Station, TX, USA) was used to calculate the summary OR and associated P-values. AT and TT genotypes were combined for the meta-analysis.
FUNDING
This work was funded by the National Cancer Institute grants R01 CA68581, R01 CA63464, R01 CA54281 and 2 P30 CA14089-26.
ACKNOWLEDGEMENTS
We wish to thank Lucy Xia for her assistance with the sequencing conducted for this analysis. We also wish to thank the USC Genomics Core Facility for the genotyping conducted for this analysis.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Makridakis N.M., Ross R.K., Pike M.C., Crocitto L.E., Kolonel L.N., Pearce C.L., Henderson B.E., Reichardt J.K. Association of mis-sense substitution in SRD5A2 gene with prostate cancer in African-American and Hispanic men in Los Angeles, USA. Lancet. 1999;354:975–978. doi: 10.1016/S0140-6736(98)11282-5. [DOI] [PubMed] [Google Scholar]
- 2.Makridakis N.M., di Salle E., Reichardt J.K. Biochemical and pharmacogenetic dissection of human steroid 5 alpha-reductase type II. Pharmacogenetics. 2000;10:407–413. doi: 10.1097/00008571-200007000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Margiotti K., Sangiuolo F., De Luca A., Froio F., Pearce C.L., Ricci-Barbini V., Micali F., Bonafe M., Franceschi C., Dallapiccola B., et al. Evidence for an association between the SRD5A2 (type II steroid 5 alpha-reductase) locus and prostate cancer in Italian patients. Dis. Markers. 2000;16:147–150. doi: 10.1155/2000/683607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Latil A.G., Azzouzi R., Cancel G.S., Guillaume E.C., Cochan-Priollet B., Berthon P.L., Cussenot O. Prostate carcinoma risk and allelic variants of genes involved in androgen biosynthesis and metabolism pathways. Cancer. 2001;92:1130–1137. doi: 10.1002/1097-0142(20010901)92:5<1130::aid-cncr1430>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 5.Mononen N., Ikonen T., Syrjakoski K., Matikainen M., Schleutker J., Tammela T.L., Koivisto P.A., Kallioniemi O.P. A missense substitution A49T in the steroid 5-alpha-reductase gene (SRD5A2) is not associated with prostate cancer in Finland. Br. J. Cancer. 2001;84:1344–1347. doi: 10.1054/bjoc.2001.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang B.L., Zheng S.L., Isaacs S.D., Turner A.R., Bleecker E.R., Walsh P.C., Meyers D.A., Isaacs W.B., Xu J. Evaluation of SRD5A2 sequence variants in susceptibility to hereditary and sporadic prostate cancer. Prostate. 2003;56:37–44. doi: 10.1002/pros.10225. [DOI] [PubMed] [Google Scholar]
- 7.Cicek M.S., Conti D.V., Curran A., Neville P.J., Paris P.L., Casey G., Witte J.S. Association of prostate cancer risk and aggressiveness to androgen pathway genes: SRD5A2, CYP17, and the AR. Prostate. 2004;59:69–76. doi: 10.1002/pros.10358. [DOI] [PubMed] [Google Scholar]
- 8.Forrest M.S., Edwards S.M., Houlston R., Kote-Jarai Z., Key T., Allen N., Knowles M.A., Turner F., Ardern-Jones A., Murkin A., et al. Association between hormonal genetic polymorphisms and early-onset prostate cancer. Prostate Cancer Prostatic Dis. 2005;8:95–102. doi: 10.1038/sj.pcan.4500785. [DOI] [PubMed] [Google Scholar]
- 9.Salam M.T., Ursin G., Skinner E.C., Dessissa T., Reichardt J.K. Associations between polymorphisms in the steroid 5-alpha reductase type II (SRD5A2) gene and benign prostatic hyperplasia and prostate cancer. Urol. Oncol. 2005;23:246–253. doi: 10.1016/j.urolonc.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Yamada Y., Watanabe M., Murata M., Yamanaka M., Kubota Y., Ito H., Katoh T., Kawamura J., Yatani R., Shiraishi T. Impact of genetic polymorphisms of 17-hydroxylase cytochrome P-450 (CYP17) and steroid 5alpha-reductase type II (SRD5A2) genes on prostate-cancer risk among the Japanese population. Int. J. Cancer. 2001;92:683–686. doi: 10.1002/1097-0215(20010601)92:5<683::aid-ijc1255>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Hsing A.W., Chen C., Chokkalingam A.P., Gao Y.T., Dightman D.A., Nguyen H.T., Deng J., Cheng J., Sesterhenn I.A., Mostofi F.K., et al. Polymorphic markers in the SRD5A2 gene and prostate cancer risk: a population-based case–control study. Cancer Epidemiol. Biomarkers Prev. 2001;10:1077–1082. [PubMed] [Google Scholar]
- 12.Lamharzi N., Johnson M.M., Goodman G., Etzioni R., Weiss N.S., Dightman D.A., Barnett M., DiTommaso D., Chen C. Polymorphic markers in the 5alpha-reductase type II gene and the incidence of prostate cancer. Int. J. Cancer. 2003;105:480–483. doi: 10.1002/ijc.11126. [DOI] [PubMed] [Google Scholar]
- 13.Soderstrom T., Wadelius M., Andersson S.O., Johansson J.E., Johansson S., Granath F., Rane A. 5alpha-reductase 2 polymorphisms as risk factors in prostate cancer. Pharmacogenetics. 2002;12:307–312. doi: 10.1097/00008571-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Giwercman Y.L., Abrahamsson P.A., Giwercman A., Gadaleanu V., Ahlgren G. The 5alpha-reductase type II A49T and V89L high-activity allelic variants are more common in men with prostate cancer compared with the general population. Eur. Urol. 2005;48:679–685. doi: 10.1016/j.eururo.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Hayes V.M., Severi G., Padilla E.J., Morris H.A., Tilley W.D., Southey M.C., English D.R., Sutherland R.L., Hopper J.L., Boyle P., et al. 5alpha-reductase type 2 gene variant associations with prostate cancer risk, circulating hormone levels and androgenetic alopecia. Int. J. Cancer. 2007;120:776–780. doi: 10.1002/ijc.22408. [DOI] [PubMed] [Google Scholar]
- 16.Lindstrom S., Zheng S.L., Wiklund F., Jonsson B.A., Adami H.O., Balter K.A., Brookes A.J., Sun J., Chang B.L., Liu W., et al. Systematic replication study of reported genetic associations in prostate cancer: strong support for genetic variation in the androgen pathway. Prostate. 2006;66:1729–1743. doi: 10.1002/pros.20489. [DOI] [PubMed] [Google Scholar]
- 17.Loukola A., Chadha M., Penn S.G., Rank D., Conti D.V., Thompson D., Cicek M., Love B., Bivolarevic V., Yang Q., et al. Comprehensive evaluation of the association between prostate cancer and genotypes/haplotypes in CYP17A1, CYP3A4, and SRD5A2. Eur. J. Hum. Genet. 2004;12:321–332. doi: 10.1038/sj.ejhg.5201101. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham J.M., Hebbring S.J., McDonnell S.K., Cicek M.S., Christensen G.B., Wang L., Jacobsen S.J., Cerhan J.R., Blute M.L., Schaid D.J., et al. Evaluation of genetic variations in the androgen and estrogen metabolic pathways as risk factors for sporadic and familial prostate cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:969–978. doi: 10.1158/1055-9965.EPI-06-0767. [DOI] [PubMed] [Google Scholar]
- 19.Giwercman C., Giwercman A., Pedersen H.S., Toft G., Lundin K., Bonde J.P., Giwercman Y.L. Polymorphisms in genes regulating androgen activity among prostate cancer low-risk Inuit men and high-risk Scandinavians. Int. J. Androl. 2008;31:25–30. doi: 10.1111/j.1365-2605.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- 20.Onen I.H., Ekmekci A., Eroglu M., Polat F., Biri H. The association of 5alpha-reductase II (SRD5A2) and 17 hydroxylase (CYP17) gene polymorphisms with prostate cancer patients in the Turkish population. DNA Cell Biol. 2007;26:100–107. doi: 10.1089/dna.2006.0534. [DOI] [PubMed] [Google Scholar]
- 21.Tong M., Xu Z., Ai J.K., Yuan Y.M., Yin Y., Wang J.Q., Li H.W., Liu J.H., Xin D.Q., Zhou L.Q., et al. Association of polymorphisms in testosterone 5-alpha-reductase II genotype and prognosis factors of prostate cancer. Zhonghua Wai Ke Za Zhi. 2004;42:1493–1496. [PubMed] [Google Scholar]
- 22.Ntais C., Polycarpou A., Ioannidis J.P. SRD5A2 gene polymorphisms and the risk of prostate cancer: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 2003;12:618–624. [PubMed] [Google Scholar]
- 23.Roberts R.O., Bergstralh E.J., Farmer S.A., Jacobson D.J., McGree M.E., Hebbring S.J., Cunningham J.M., Anderson S.A., Thibodeau S.N., Lieber M.M., et al. Polymorphisms in the 5alpha reductase type 2 gene and urologic measures of BPH. Prostate. 2005;62:380–387. doi: 10.1002/pros.20142. [DOI] [PubMed] [Google Scholar]
- 24.Schatzl G., Madersbacher S., Gsur A., Preyer M., Haidinger G., Haitel A., Vutuc C., Micksche M., Marberger M. Association of polymorphisms within androgen receptor, 5alpha-reductase, and PSA genes with prostate volume, clinical parameters, and endocrine status in elderly men. Prostate. 2002;52:130–138. doi: 10.1002/pros.10101. [DOI] [PubMed] [Google Scholar]
- 25.Allen N.E., Reichardt J.K., Nguyen H., Key T.J. Association between two polymorphisms in the SRD5A2 gene and serum androgen concentrations in British men. Cancer Epidemiol. Biomarkers Prev. 2003;12:578–581. [PubMed] [Google Scholar]
- 26.Jaffe J.M., Malkowicz S.B., Walker A.H., MacBride S., Peschel R., Tomaszewski J., Van Arsdalen K., Wein A.J., Rebbeck T.R. Association of SRD5A2 genotype and pathological characteristics of prostate tumors. Cancer Res. 2000;60:1626–1630. [PubMed] [Google Scholar]
- 27.Rebbeck T.R., Martinez M.E., Sellers T.A., Shields P.G., Wild C.P., Potter J.D. Genetic variation and cancer: improving the environment for publication of association studies. Cancer Epidemiol. Biomarkers Prev. 2004;13:1985–1986. [PubMed] [Google Scholar]
- 28.Lohmueller K.E., Pearce C.L., Pike M., Lander E.S., Hirschhorn J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 29.Kolonel L., Henderson B., Hankin J., Nomura A., Wilkens L., Pike M., Stram D., Monroe K., Earle M.E., Nagamine F. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]