Abstract
Aggresomes are juxtanuclear inclusion bodies that have been proposed to act as staging grounds for the disposal of protein aggregates via the autophagic route. To examine whether the composition of an aggresome influences its clearance by autophagy, we ectopically expressed a variety of aggregation-prone proteins in cultured cells to generate aggresomes that differ in their protein content. We found that whereas aggresomes generated in cells expressing mutant huntingtin or mutant tau, or co-expressing synphilin-1 and α-synuclein, are amenable to clearance by autophagy, those produced in AIMP2 (p38)- or mutant desmin-expressing cells are apparently resistant to autophagic clearance. Notably, AIMP2 (p38)- and desmin-positive inclusions fail to recruit key components of the autophagic/lysosomal system. However, by altering the composition of inclusions, ‘autophagy-resistant’ aggresomes could be rendered ‘autophagy-susceptible’. Taken together, our results demonstrate that not all aggresomes are efficiently primed for autophagic clearance and highlight a certain degree of selectivity for the supposedly non-discriminative pathway.
INTRODUCTION
The presence of intracellular protein inclusions is a common hallmark of a wide variety of human disorders. These include neurofibrillary tangles in Alzheimer's disease, Lewy bodies (LBs) in Parkinson's disease (PD), polyglutamine-enriched inclusions in Huntington's disease (HD), Mallory bodies in alcoholic and non-alcoholic hepatitis, as well as intermediate filament inclusions in specific myopathies (1–3). Notably, a diverse array of pathogenic proteins bearing little structural and functional similarities with each other has been identified as key contributors to the formation of inclusion bodies (IBs) in these varied diseases. Despite their differences, the various disease-associated proteins share the tendency to become misfolded and thereby aggregation-prone, suggesting that intracellular pathways responsible for the management of misfolded proteins, such as the ubiquitin-proteasome system (UPS) that identifies and degrades unwanted proteins, are linked to the generation of IBs. Consistent with this, aggregation-prone proteins often generate peri-nucleus-localized IBs when ectopically expressed in cultured cells in the presence of proteasome inhibition. These peri-nuclear protein deposits, commonly known as aggresomes (4), are formed via a microtubule-driven process by which non-degradable protein aggregates are trafficked along microtubules to the microtubule organizing center juxtaposed to the nucleus (4,5). Aggresomes are seemingly inert structures that are often enriched with molecular chaperones, components of the UPS and intermediate filaments such as vimentin (4–6). Their formation could be considered a proactive way by which the cell deals with its non-disposable proteins. Interestingly, protein inclusions associated with several of the diseases mentioned above bear striking resemblance to aggresomes (7–9).
Although a subject of intense research, the role of IBs in disease pathogenesis remains a vigorously debated topic. However, several reports, including an elegant live cell-imaging study conducted by Arrasate et al. (10), suggest a neuroprotective function for protein inclusions formation (11–14). Notwithstanding this, IBs are space-filling entities that could physically disrupt cellular function if their sizes were to exceed a certain threshold. Conceivably, the growth of an IB is a regulated process. Emerging evidence implicates macroautophagy (hereafter referred to as autophagy), a lysosome-mediated bulk degradation system, as a key regulator of inclusion dynamics (5,15). For example, two recent independent reports clearly indicated the importance of autophagy in neurodegeneration and inclusion formation (16,17). Further, several other studies demonstrated that pharmacological activation of autophagy promotes the degradation of aggregation-prone proteins, including huntingtin (Htt) and tau mutants, and concomitantly improves cellular survivability (18–21). Together, these findings suggest that harnessing the autophagic pathway may offer innovative approaches in the treatment of diseases linked to altered protein conformation. Relevant to this is the proposal that aggresome formation facilitates the delivery of dispersed protein aggregates to the autophagic pathway (4). Support for such a mechanism came from the study by Fortun et al. (22), who demonstrated that pharmacological inhibition of autophagy retards the clearance of aggresomes formed by mutant peripheral myelin protein PMP22, a Schwann cell protein linked to demyelinating neuropathies, as well as a study by Iwata et al. (23), who showed that aggresomes generated by mutant Htt stain positively for several autophagy-linked proteins and are amenable to clearance by autophagy. However, whether aggresomes formed by other disease-related proteins are amenable to autophagic clearance remains to be established.
Here, we investigated the predisposition of a variety of disease-associated proteins toward aggresome formation and clearance by autophagy. These include HD-linked Htt mutant (24), frontal temporal dementia-linked tau mutant (25), myopathy-linked desmin mutant (1) as well as PD-linked AIMP2 (p38) (26), α-synuclein and synphilin-1 (27). We found that all of these disease-associated proteins mediate the formation of ubiquitinated juxtanuclear inclusions resembling aggresomes when they are ectopically expressed in cultured cells in the presence of proteasome inhibition. However, the various aggresome-like inclusions show different susceptibility to clearance by autophagy. Whereas autophagy induction significantly reduces the number of aggresome-like inclusions generated in cells expressing mutant Htt, mutant tau and α-synuclein/synphilin-1, it has no apparent effects on the number of inclusions generated in cells expressing AIMP2 (p38) and mutant desmin. Consistent with this, we found that AIMP2 (p38)- and desmin-positive inclusions fail to engage key components of the lysosomal machinery. Interestingly, these ‘autophagy-resistant’ aggresome-like structures could be rendered ‘autophagy-susceptible’, albeit in a context-specific manner, by altering the inclusions composition. For example, the presence of α-synuclein and synphilin-1 in AIMP2 (p38)-positive inclusions facilitates the latter's engagement with lysosomal components and thereby its clearance by autophagy. Taken together, our results suggest that autophagy-mediated clearance of aggresome-like inclusions is a selective phenomenon that is dependent on the nature of the inclusions.
RESULTS
Aggregation-prone proteins associated with various degenerative diseases form morphologically similar, aggresome-like structures in cultured cells
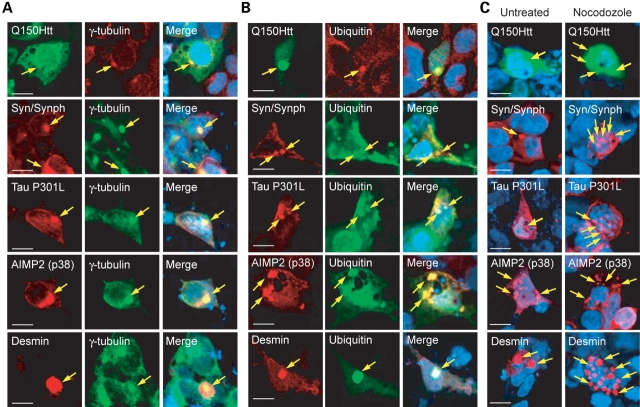
To address whether the clearance of aggresomes by autophagy is a universal phenomenon, we first ascertain whether aggregation-prone proteins linked to various degenerative diseases are capable of forming aggresome-like structures when ectopically expressed in cultured cells. For this purpose, we transfected SH-SY5Y cells with cDNA encoding an N-terminal fragment of mutant Htt carrying 150 CAG repeats (Q150Htt), tau P301L mutant, AIMP2 (p38), α-synuclein, synphilin-1 or desmin mutant. Transfected cells were treated with lactacystin, a selective proteasome inhibitor, to facilitate inclusions formation. Consistent with previous reports (9,24), we found that Q150Htt-expressing cells exhibit a tendency to generate ubiquitin-positive, peri-nuclear inclusions that co-localize with the centrosome marker, γ-tubulin (Fig. 1A and B), as well as with vimentin, which forms a cage-like structure surrounding the inclusions (Supplementary Material, Fig. S1A). Upon treatment of Q150Htt-expressing cells with nocodozole, a microtubule depolymerizing agent, the juxtanucleus-localized inclusions disperse into multiple smaller aggregates that scatter throughout the cytoplasm (Fig. 1C). Q150Htt-positive inclusions thus exhibit the classical hallmarks of aggresomes (4). Similarly, inclusions generated by ectopically expressed mutant tau, mutant desmin, AIMP2 (p38) and α-synuclein/synphilin-1 in the presence of proteasome inhibition all show characteristics of aggresomes and resemble Q150Htt-positive inclusions, at least on the light microscopic level, except that AIMP2 (p38)- and desmin-positive inclusions frequently appear to be comparatively larger in size (Fig. 1A–C). We also noted a higher tendency for cells expressing AIMP2 (p38) and desmin to generate IBs, especially in comparison to cells expressing α-synuclein/synphilin-1 or mutant tau (Supplementary Material, Fig. S1B). Together, our results demonstrate that expression of aggregation-prone proteins associated with various degenerative diseases in SH-SY5Y cells in the presence of proteasome inhibition generates intracellular IBs that bear striking resemblance to aggresomes.
Figure 1.
Aggregation-prone proteins associated with various degenerative diseases form morphologically similar, aggresome-like inclusions in SH-SY5Y cells. (A and B) Representative confocal images of SH-SY5Y cells ectopically expressing green fluorescent protein-tagged huntingtin mutant (Q150Htt), α-synuclein and myc-tagged synphilin-1, myc-tagged tau P301L mutant, AIMP2 (p38) or desmin mutant immunostained either with relevant primary protein-directed antibodies or with (A) anti-γ-tubulin or (B) anti-ubiquitin, as indicated. Arrows in merge pictures show the co-localization of inclusions generated by the various aggregation-prone proteins with γ-tubulin or ubiquitin (yellow). (C) All the inclusions examined dispersed into micro-aggregates when cells were treated with 10 µg/ml nocodozole for 16 h. Each of these experiments was repeated at least three times (scale bar, 10 µm).
Clearance of aggresome-like inclusions is not a universal phenomenon
Next, autophagic clearance of aggresome-like inclusions generated by various disease-associated proteins was investigated using a method described by Fortun et al. (22). SH-SY5Y cells ectopically expressing mutant Q150Htt, mutant tau, mutant desmin, and AIMP2 (p38) or co-expressing α-synuclein and synphilin-1 (α-synuclein/synphilin-1) were initially treated with lactacystin for 16 h to facilitate the formation of aggresome-like inclusions before a recovery period (24 h) in culture medium containing normal 10% serum or without serum. Serum withdrawal is known to stimulate autophagy (28). Supporting this, we observed an apparent expansion of the lysosomal compartment in cells that recover in starvation medium compared with those that recover in normal serum-supplemented medium, as visualized with the acidotropic lysosomal stain, lysotracker, as well as with the lysosomal-specific marker, LAMP1 (Supplementary Material, Fig. S2A). Quantification of inclusions in Q150Htt-expressing cells reveals a significant reduction in the number of inclusion-positive cells when they recover in serum-free medium compared with those that recover in normal serum-containing medium, suggesting the removal of Q150Htt-positive inclusions by starvation-induced autophagy (Fig. 2A). This is in consistent with previous demonstration by others that Htt-positive aggregates could be cleared by autophagy (20,29). Similarly, inclusions formed in cells expressing either mutant tau or α-synuclein/synphilin-1 are amenable to clearance by autophagy, as can be seen from the dramatic decrease in the number of tau-positive or α-synuclein/synphilin-1-positive inclusions following serum starvation (Fig. 2A). Furthermore, quantification of number of inclusions in these cells at different time points following serum starvation reveals a rapid decline in the first 6 h, followed by a more gradual decline thereafter (Supplementary Material, Fig. S2B). This rate of inclusions clearance correlates well with the reported activity of macroautophagy, which peaks at about 6 h following starvation (30–32). Direct stimulation of autophagy in these cells via rapamycin treatment essentially reproduces the phenomenon brought about by serum starvation (Fig. 2A). On the other hand, no reduction in the number of inclusions is observed in these cells when they recover in serum-free medium in the presence of the widely used inhibitor of autophagy, 3-methyladenine (3-MA) (Fig. 2A), suggesting that the observed reduction in Q150Htt-, tau- and α-synuclein/synphilin-1-positive inclusions in cells undergoing stimulated autophagy is attributable to an increase in their clearance rather than a decrease in their formation. Consistent with this, we did not observe an appreciable change in the number of inclusions in tau- and α-synuclein/synphilin-1 or even AIMP2 (p38)-expressing cells during their entire recovery period in normal serum-containing medium (Supplementary Material, Fig. S2B). Surprisingly, despite being morphologically similar to inclusions containing Q150Htt-, mutant tau- or α-synuclein/synphilin-1, both AIMP2 (p38)- and mutant desmin-positive inclusions appear to be resistant to clearance by starvation- or rapamycin-induced autophagy (Fig. 2A). In these cases, the number of inclusions remains constant throughout the 24 h recovery period regardless of culture conditions (dynamics of AIMP2 (p38)-positive inclusions are shown in Supplementary Material, Fig. S2B).
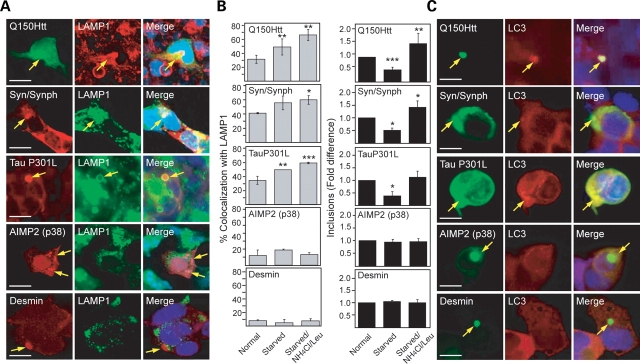
Figure 2.
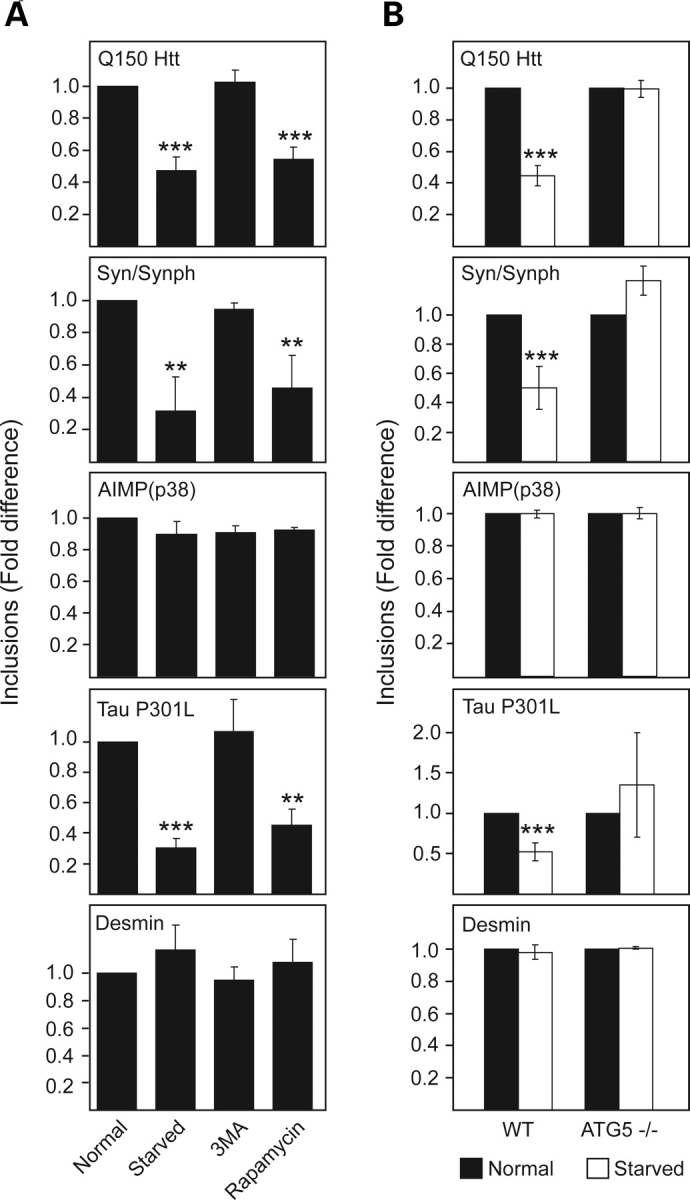
Clearance of aggresome-like inclusions by autophagy is not a universal phenomenon. (A) Bar graph showing the relative fold difference in the number of inclusions observed in SH-SY5Y cells ectopically expressing various aggregation-prone proteins (as indicated) under different recovery conditions. (B) Bar graph showing the relative fold difference in the number of inclusions observed in wild-type (WT) or autophagy-deficient ATG5−/− mouse embryonic fibroblasts ectopically expressing various aggregation-prone proteins (as indicated) recovering under normal or starved conditions. Each of these experiments was repeated at least three times (*P < 0.05, **P < 0.01, ***P < 0.001 versus control group).
To further confirm the clearance of aggresome-like inclusions by autophagy, we repeated the above experiments in mouse embryonic fibroblasts (MEFs) derived from wild-type mice or autophagy-deficient ATG5 knockout (ATG5−/−) mice. When ectopically expressed in these cells, all the aggregation-prone proteins examined produce aggresome-like inclusions that are morphologically similar to those generated in SH-SY5Y cells (not shown). As with SH-SY5Y cells, aggresome-like inclusions generated in wild-type MEFs by expression of Q150Htt, tau and α-synuclein/synphilin-1, but not of AIMP2 (p38) or mutant desmin, are apparently amenable to clearance by starvation-induced autophagy (Fig. 2B). However, this phenomenon is not observed in autophagy-deficient ATG 5−/− MEFs expressing the same compendium of proteins (Fig. 2B). Taken together, our results suggest that autophagy induction facilitates the clearance of aggresome-like inclusions formed by Q150Htt, mutant tau and α-synuclein/synphilin-1, but not those generated by AIMP2 (p38) and mutant desmin.
Autophagic response is not impaired in cells containing various disease-associated inclusions, including those containing AIMP2 (p38)- and desmin-positive inclusions
To rule out the possibility that the autophagic system may be defective in cells containing AIMP2 (p38)- and desmin-positive inclusions, we examined the dynamics of autophagic vacuole (AV) formation and clearance in these cells following starvation-induced autophagy. Since the cellular distribution of the autophagosome marker LC3 changes from a diffuse cytosolic staining to a punctuate appearance upon stimulation of autophagy, and the LC3 protein is degraded following the fusion between the autophagosome and the lysosome, quantifying the number of LC3 puncta in the absence and presence of lysosomal protease inhibitors will thus provide an indication of AV formation and clearance, respectively, in the cell (33). Indeed, we observed an apparent increase in the number of LC3 puncta in control SH-SY5Y cells in the presence of starvation-induced autophagy, which increased further when these cells were treated with the lysosomal protease inhibitors, NH4Cl and leupeptin (Fig. 3A and B). Accordingly, we then measured the number of LC3 puncta in cells expressing either AIMP2 (p38)- or desmin, as well as in cells expressing the other disease-associated proteins under different conditions. Upon induction of autophagy by starvation, we recorded an apparent increase in the number of LC3 puncta in all the transfected cells which is especially significant for those containing tau- and α-synuclein/synphilin-1-positive inclusions compared with their corresponding controls (Fig. 3A and B). Among these, the ones containing tau- and α-synuclein/synphilin-1-positive inclusions also appear to elicit a greater than normal autophagic response, as suggested by their more abundant LC3 puncta compared with control starved cells (Fig. 3B). Upon inhibition of lysosomal degradation in various cells by NH4Cl and leupeptin treatment, we observed a further accumulation of LC3 puncta (Fig. 3B). Again, the accumulation of LC3 puncta in tau- and α-synuclein/synphilin-1-inclusion positive cells significantly exceeds that of control cells cultured under the same conditions (Fig. 3B). The formation and clearance of AVs thus appears to be enhanced in cells containing tau- and α-synuclein/synphilin-1-inclusions. Taken together, our results demonstrate that AV formation and clearance are not impaired in cells expressing various disease-associated proteins, including those containing AIMP2 (p38)-positive inclusions, suggesting that the autophagic response is not compromised in these cells. However, the level of autophagic response seems to be dependent on the composition of the IBs. Although cells containing AIMP2 (p38)- and desmin-positive inclusions appear to mount a comparatively weaker autophagic response, the number of LC3 puncta in these cells is not significantly different from their corresponding untransfected control cells or those containing Q150Htt-positive inclusions in the absence or presence of autophagy stimulation (Fig. 3B). Hence, it is unlikely that the resistance of AIMP2 (p38)- and desmin-positive inclusions to clearance is due to a defective autophagic system.
Figure 3.
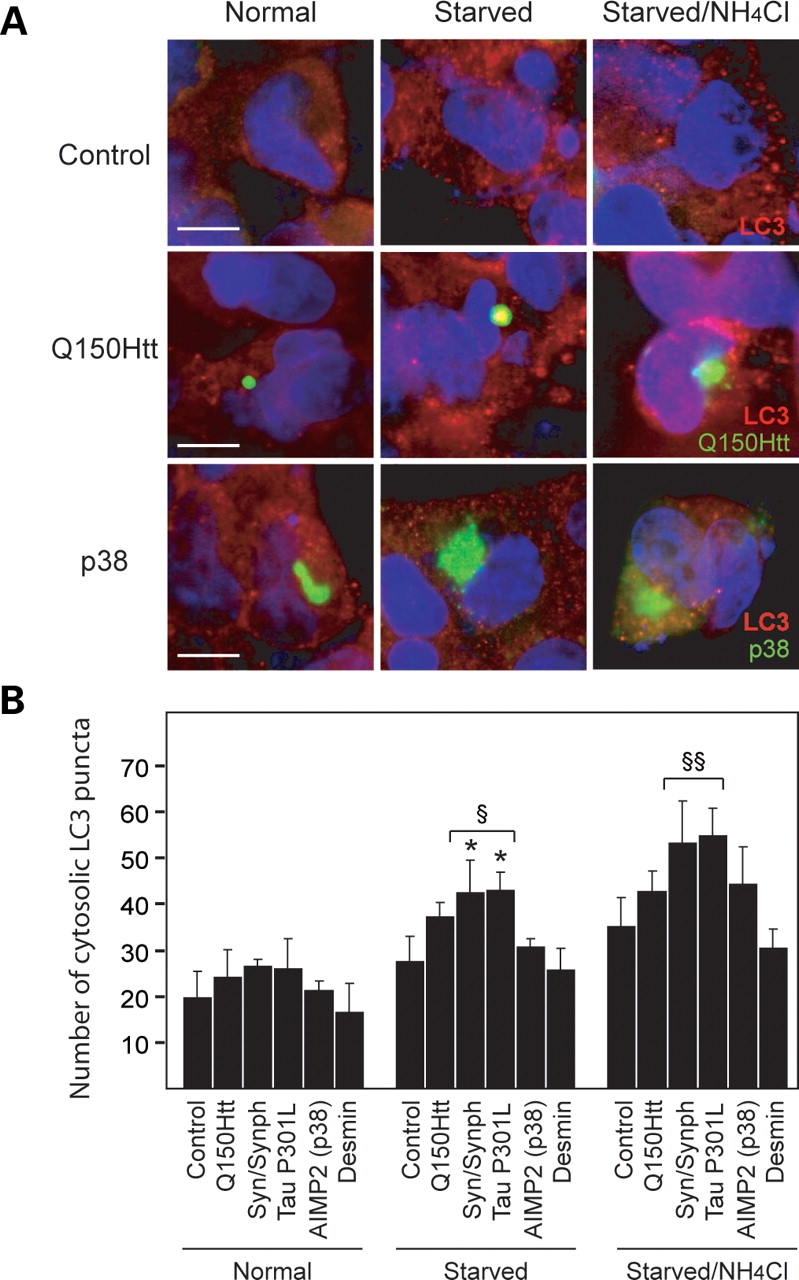
Formation of AIMP2 (p38)- and desmin-positive inclusions does not impair the autophagic system. (A) Representative confocal images showing LC3 staining in untransfected SH-SY5Y cells (control) (top panels), or cells containing either Q150Htt-positive inclusions (middle panels) or AIMP2 (p38)-positive inclusions (bottom panels) cultured under different conditions (as indicated) following proteasome inhibition. (B) Bar graph showing the average number of cytosolic LC3 puncta per cell in control cells or those containing various types of inclusions (as indicated) cultured under different conditions following proteasome inhibition. Each of these experiments was repeated at least three times (*P < 0.05 versus respective value under normal conditions, §P < 0.01, §§P < 0.001 versus control cells under same conditions).
Inclusions formed by various disease-associated proteins exhibit different competencies in recruiting key components involved in autophagy
Given the apparent inertness of AIMP2 (p38)- and desmin-positive inclusions under conditions of stimulated autophagy, it is likely that these ‘autophagy-resistant’ structures fail to engage the lysosomal machinery. To address this, we examined the ability of various aggresome-like inclusions to recruit the lysosomal marker, LAMP1 (34). Under normal culture conditions, we found that a significant percentage (30–40%) of Q150Htt-, tau- and α-synuclein/synphilin-1-positive inclusions co-localizes with LAMP1 (Fig. 4B). However, both AIMP2 (p38)- and desmin-positive inclusions show significantly less (∼12%) co-localization with the lysosomal marker (Fig. 4B). During starvation-induced autophagy, there is a general increase in the average percentage of inclusions that co-stains with LAMP1 which is particularly significant in Q150Htt- and tau-expressing cells (Fig. 4A and B, left panels). Following treatment of the cells with the lysosomal protease inhibitors, NH4Cl and leupeptin, a further significant increase in co-localization between inclusions and LAMP1 is also observed in Q150Htt- and tau-expressing cells, as well as in α-synuclein/synphilin-1 expressing cells (Fig. 4B, left panels). As expected, the inhibition of lysosomal activity also blocks the clearance of aggresome-like structures in these cells which otherwise occur in serum-free media (Fig. 4B, right panels). On the other hand, NH4Cl and leupeptin treatment of AIMP2 (p38)- and desmin-expressing cells has no effects on the co-localization of inclusions formed in these cells with LAMP1 (Fig. 4B, left panels). That LAMP1 is recruited by a significant population of Q150Htt-, tau- and α-synuclein/synphilin-1-positive inclusions even in the absence of starvation-induced autophagy, and that the recruitment increases further upon autophagy induction, suggesting the interesting possibility that ‘autophagy-susceptible’ aggresomes may already be primed for clearance during their formation. We also examined the recruitment of LC3 by various disease-associated inclusions in the presence of starvation-induced autophagy. Consistent with our above observations with LAMP1, we found that LC3 is frequently recruited to Q150Htt-, tau- and α-synuclein/synphilin-1-positive inclusions but not to AIMP2 (p38)- and desmin-positive inclusions (Fig. 4C). Both AIMP2 (p38)- and desmin-positive inclusions thus appear to be impaired in recruiting the lysosomal machinery, a phenomenon that could explain their failed clearance under conditions of stimulated autophagy. This impairment is unlikely a result of microtubule disruption, as anti-α-tubulin immunostaining of these cells reveals an intact microtubule network that is similar to those found in tau- and α-synuclein/synphilin-1-expressing cells (Supplementary Material, Fig. S3).
Figure 4.
Differential recruitment of autophagy-related components by various aggresome-like inclusions. (A) Representative confocal images of SH-SY5Y cells ectopically expressing various aggregation-prone proteins under serum-starved conditions immunostained with relevant primary protein-directed antibodies or anti-LAMP1, as indicated. Arrows in merge pictures show the co-localization of inclusions generated by the various aggregation-prone proteins with LAMP1 (yellow). (B) Left panels: bar graph showing the percentage of LAMP-1-positive inclusions in these cells. Note that the inactivation of lysosomal proteases with NH4Cl and leupeptin promotes the co-localization of LAMP1 to Q150Htt-, tau- and syn/synph-positive, but not AIMP2 (p38)- or desmin-positive inclusions. Right panels: bar graph showing the relative fold difference in the number of inclusions observed in SH-SY5Y cells ectopically expressing various aggregation-prone proteins (as indicated) under different recovery conditions (*P < 0.05, **P < 0.01, ***P < 0.001 versus control group). (C) Representative confocal images of transfected SH-SY5Y cells showing the co-localization of the autophagosome marker, LC3, to mutant Q150Htt, tau- and syn/synph-positive, but not AIMP2 (p38)- or desmin-positive inclusions under serum-starved conditions. Each of these experiments was repeated at least three times (scale bar, 10 µm).
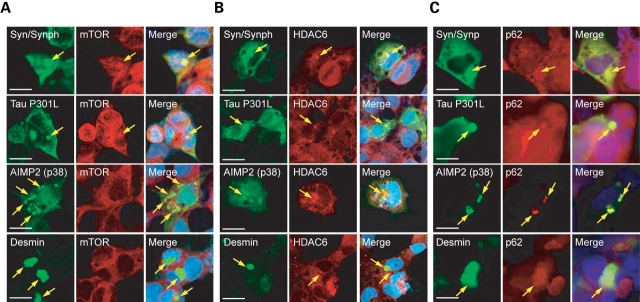
Next, we examined the competency of various aggresome-like inclusions in recruiting key components known to be involved in the aggresome–lysosome pathway, including the mammalian target of rapamycin (mTOR), an inhibitor of autophagy (35); histone deacetylase 6 (HDAC6), a cytoplasmic deacetylase that is apparently important for the transport of lysosomes to aggresomes (29); and p62, an ubiquitin-associated (UBA) protein that is involved in the formation of protein aggregates and in linking polyubiquitinated protein aggregates to the autophagy machinery (36,37). Aggregates containing polyglutamine proteins have recently been shown to sequester mTOR, the immobilization of which inhibits mTOR activity and induces autophagy (35). Consistent with this, we observed the recruitment of mTOR into Q150Htt-, mutant tau- or α-synuclein/synphilin-1-positive inclusions (Fig. 5A, data not shown for Q150Htt). On the other hand, no such co-localization was observed between mTOR and AIMP2 (p38)- or desmin-positive inclusions (Fig. 5A), providing thus a possible explanation for the comparatively more discrete upregulation of autophagy in these cells in response to these two pathogenic proteins (Fig. 3B). However, mTOR activity as measured by the phosphorylation of its downstream mediator, p70 S6 kinase, does not appear to be overtly affected in the presence of the various disease-associated inclusions, regardless of their ability to recruit mTOR (not shown). Similarly, tau-positive and α-synuclein/synphilin-1-positive inclusions, but not desmin-positive inclusions, are competent in recruiting HDAC6 (Fig. 5B). Interestingly, AIMP2 (p38)-positive inclusions also co-localize with HDAC6 (Fig. 5B). All the inclusions examined are, however, enriched with p62 (Fig. 5C). Taken together, these results demonstrate that inclusions formed by different pathogenic proteins exhibit differential abilities in recruiting key components involved in the lysosomal pathway, a property that could, in part, account for their respective amenability to autophagic clearance. A summary of our above findings is provided in Table 1.
Figure 5.
Differential recruitment of mTOR and HDAC6 by various aggresome-like inclusions. (A) Representative confocal images of SH-SY5Y cells ectopically expressing various aggregation-prone proteins (as indicated) immunostained with relevant primary protein-directed antibodies (green) or with (A) anti-mTOR (red), (B) anti-HDAC6 (red) or (C) anti-p62 (red). Arrows in merge pictures show the presence or absence of co-localization between aggresome-like inclusions and mTOR, HDAC6 or p62. Each of these experiments was carried out in cells cultured under normal serum conditions but in the presence of proteasome inhibition and was repeated at least three times (scale bar, 10 µm).
Table 1.
Recruitment of autophagy-associated components to aggresomes formed by different aggregation-prone proteins
| Aggresome | LAMP1 | LC3 | mTOR | HDAC6 | p62 |
|---|---|---|---|---|---|
| Q150Htt | + | + | + | + | + |
| Syn/synph | + | + | + | + | + |
| Tau P301L | + | + | + | + | + |
| AIMP2 (p38) | − | − | − | + | + |
| Desmin | − | − | − | − | + |
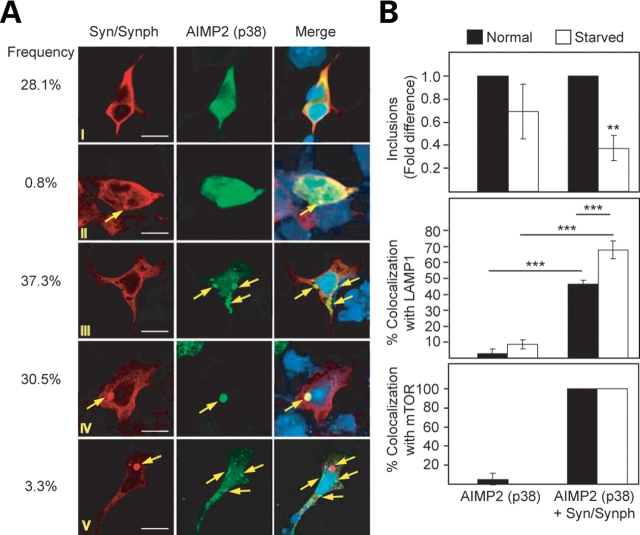
Presence of α-synuclein/synphilin-1 in AIMP2 (p38)-positive inclusion influences its clearance by autophagy
It is noteworthy that α-synuclein, synphilin-1 and AIMP2 (p38) are all components of the PD-associated LBs (38,39). Yet, inclusions formed by these proteins in cultured cells show remarkably different susceptibility toward autophagy-mediated clearance (Fig. 2). Accordingly, we co-expressed α-synuclein, synphilin-1 and AIMP2 (p38) in SH-SY5Y cells to examine whether the presence of α-synuclein/synphilin-1 in AIMP2 (p38)-positive inclusions would influence inclusions clearance by autophagy. Quantitative analysis of cells expressing α-synuclein, synphilin-1 and AIMP2 (p38) reveals that 30.5% of the inclusions formed contain all three proteins, while ∼37% of the inclusions formed are positive for AIMP2 (p38) only (Fig. 6A). Consistent with our above results, the number of inclusions that are positive for AIMP2 (p38) only remains relatively unaffected under conditions of stimulated autophagy (Fig. 6B). However, under the same conditions, we observed a significant reduction in the number of AIMP2 (p38)-positive inclusions that also contains α-synuclein and synphilin-1, suggesting their clearance by starvation-induced autophagy. Further, the binding of AIMP2 (p38)/α-synuclein/synphilin-1-positive inclusions with LAMP1 increases by about 16- and 8-fold under normal and stimulated autophagy, respectively, compared with those positive for LAMP1 and AIMP2 (p38) alone (Fig. 6B; Supplementary Material, Fig. S4). Remarkably, inclusions positive for all three proteins examined [i.e. AIMP2 (p38), α-synuclein and synphilin-1] are extremely efficient in sequestering mTOR. Whereas the population of AIMP2 (p38)-positive inclusions that co-localize with mTOR is almost negligible, the recruitment of mTOR into inclusions that contain α-synuclein, synphilin-1 and AIMP2 (p38) is a robust 100% (Fig. 6B; Supplementary Material, Fig. S4). Clearly, the co-aggregation of α-synuclein and synphilin-1 with AIMP2 (p38) could augment the interaction between AIMP2 (p38)-positive inclusions and mTOR or LAMP1.
Figure 6.
Co-aggregation of α-synuclein/synphilin-1 and AIMP2 (p38) promotes clearance of AIMP2 (p38)-positive inclusions by autophagy. (A) Representative confocal images of SH-SY5Y cells cultured under normal serum conditions but in the presence of proteasome inhibition and expressing α-synuclein, synphilin-1 and AIMP2 (p38) immunostained with anti-α-synuclein (leftmost panels) or AIMP2 (p38) (middle panels). Arrows in merge pictures (rightmost panels) show the presence or absence of co-localization between AIMP2 (p38)-positive inclusions and α-synuclein, as indicated. (B) Top panel: bar graph showing the relative fold difference in the number of AIMP2 (p38)-positive inclusions in the absence [AIMP2 (p38) only] or presence of α-synuclein/synphilin-1 co-localization [AIMP2 (p38)+Syn/Synph] under normal or serum-starved conditions. The percentage of these inclusion types that co-localized with LAMP1 and mTOR under different conditions are shown in the middle and bottom panels, respectively. Each of these experiments was repeated at least three times (*P < 0.05, **P < 0.01, ***P < 0.001).
We also examined whether co-aggregation of AIMP2 (p38) with either α-synuclein or synphilin-1 alone is sufficient to promote its clearance and found that AIMP2 (p38)-positive inclusions could be cleared in both cases (Supplementary Material, Fig. S5A). Notably, inclusions formed by α-synuclein or synphilin-1 alone are themselves amenable to clearance (data not shown), although we have previously demonstrated that their co-expression in cultured cells enhances the formation of inclusions and that these α-synuclein/synphilin-1-positive inclusions more closely resemble PD-associated LBs (27). It may therefore seem intriguing that AIMP2 (p38)-positive inclusions generated in SH-SY5Y cells are otherwise non-amenable to autophagic clearance, given that α-synuclein is also present endogenously in these cells. However, when compared with over-expressed protein, endogenous α-synuclein is present in SH-SY5Y cells at much lower levels even in the presence of proteasome inhibition (Supplementary Material, Fig. S5B). This low level of endogenous α-synuclein is expected to reduce its tendency to aggregate and thereby its tendency to co-aggregate with AIMP2 (p38)-positive inclusions, which appears to be the case (Supplementary Material, Fig. S5C and D). Nonetheless, although the small number of AIMP2 (p38)/endogenous α-synuclein precluded reliable quantification to be performed, they appear to be amenable to clearance by autophagy (Supplementary Material, Fig. S5D). Interestingly, when we repeated our co-localization studies with mutant tau in place of ectopically expressed α-synuclein and synphilin-1, we observed a comparatively much less remarkable tendency (∼13%) for AIMP2 (p38)-positive inclusions and tau-positive inclusions to co-aggregate (Supplementary Material, Fig. S6A). On the other hand, we observed the co-localization of a population of desmin-positive inclusions with those generated by α-synuclein and synphilin-1 co-expression in a related experiment. However, unlike AIMP2 (p38)/α-synuclein/synphilin-1-positive inclusions, inclusions containing desmin, α-synuclein and synphilin-1 remain inert to clearance by autophagy (Supplementary Material, Fig. S6B). Finally, we also looked at the effects of mutant tau expression on the dynamics of desmin-positive inclusions. Although co-aggregation between inclusions mediated by these two proteins did occur, albeit infrequently, these structures also appear non-amenable to autophagic clearance (Supplementary Material, Fig. S6C). Taken together, these results demonstrate that the clearance of AIMP2 (p38)/α-synuclein/synphilin-1-positive inclusions by autophagy is a rather specific phenomenon and, at the same time, suggests that the composition of inclusions influences their ability to recruit key components involved in the lysosomal pathway and thereby its clearance by autophagy.
DISCUSSION
The major finding of this study is that autophagy-mediated clearance of aggresome-like inclusions is a selective phenomenon that is dependent on the nature of the inclusions. We have demonstrated in this report that whereas aggresomes generated in cells expressing mutant Htt or mutant tau, or co-expressing synphilin-1 and α-synuclein are amenable to clearance by autophagy, those produced in AIMP2 (p38)- or mutant desmin-expressing cells are apparently resistant to autophagic clearance. Although our principal finding does not contradict the proposal that aggresomes may act as staging areas for the disposal of protein aggregates resistant to proteasomal degradation via the autophagic route (4), it posits that hitherto unknown factor(s) that is/are important in recruiting the lysosomal machinery must be present within aggresomes for these structures to be amenable to clearance by autophagy. Clearly, the elucidation of such factors would be of importance in view of recent suggestions that autophagy upregulation may represent a therapeutic strategy in mitigating diseases caused by alterations in protein conformation (40).
It is noteworthy that several studies supporting a neuroprotective role for inclusions formation have emerged recently (10–14). These studies collectively suggest that the accumulation of disease-associated proteins in a diffuse non-aggregated form is more toxic than when they are sequestered into IBs. Unquestionably, exquisite protein homeostasis is important for cellular survival. The cell has evolved several complex systems, including the chaperone, ubiquitin–proteasome and autophagic systems to maintain the quality of its proteins. The dynamic interaction among these different systems is well exemplified by the cellular management of α-synuclein, a natively unfolded protein whose accumulation is associated with neurotoxicity in a spectrum of brain diseases collectively known as synucleinopathies. Chaperones such as Hsp70 could interact with α-synuclein and suppress its toxic effects (41). Chaperones also mediate the binding of α-synuclein to LAMP-2A on lysosomal membrane, which facilitates the protein clearance via chaperone-mediated autophagy (CMA) (42). Disease-associated mutations and modifications of α-synuclein by dopamine are known to impair CMA-mediated clearance of α-synuclein (42,43). However, α-synuclein degradation could also occur via the UPS or macro-autophagy route, and the co-chaperone carboxyl terminus of Hsp70-interacting protein has been suggested to act as a molecular switch mediating α-synuclein degradation decisions between proteasomal and lysosomal pathways (44). Not surprisingly, the steady-state levels of α-synuclein are affected by inhibitors of proteasome, CMA or macroautophagy. Indeed, simultaneous inhibition of these systems promotes a synergic formation of α-synuclein inclusions (45). These findings strengthen the importance of the connection between the different proteolytic systems in the formation and clearance of inclusions relevant to neurodegenerative diseases. Clearly, all these elaborate systems act in synergy to prevent the build up of α-synuclein, which could otherwise be detrimental to cellular survival, especially when the protein is present as an oligomeric, protofibrillar form. Consistent with this, we have previously speculated that when the proteasome system becomes overwhelmed under conditions of proteolytic stress, IBs formation provides a defense mechanism against a build up of a soluble toxic load by channeling this load to an inert location for subsequent handling by autophagy (46). In this way, a susceptible neuron may prolong its survival and limit neurodegeneration. However, since a growing IB potentially could affect cellular functions if its size was not regulated, it is conceivable that the amenability of an IB to clearance by autophagy may also influence its role in disease pathogenesis, although this possibility remains to be proven. Interestingly, Arrasate et al. (10) has previously demonstrated that IB formation by mutant Htt mitigates its toxicity in neurons. Similarly, aggresomes formed by α-synuclein and synphilin-1 are apparently also cytoprotective (47,48). On the other hand, desmin aggregation appears to promote cytotoxicity (49). These findings thus correlate well with our results regarding the relative amenability of Htt-, α-synuclein/synphilin-1 and desmin-positive inclusions to be cleared by autophagy. However, the enhanced formation of AIMP2 (p38) aggregates in the presence of its cognate ubiquitin ligase, parkin, apparently reduces, and not promotes, its toxicity (26), although it is possible that parkin, being a broad-spectrum neuroprotectant, could mitigate the toxicity associated with AIMP2 (p38) aggregates. The relationship between protein aggregation and cell death is therefore by no means straightforward and requires further clarifications. Nonetheless, their impaired clearance in the absence of autophagy certainly appears to be intimately associated with neurodegeneration, as elegantly demonstrated recently by two independent groups who analyzed the consequence of ablating autophagy function specifically in the neural cells of mice (50,51). Thus, while the aggregation of toxic diffuse proteins into IBs may confer a protective role, it would seem that the protective effect of IBs is dependent on the continuous clearance of these structures from the cell. This concept is consistent with the aggresome theory (4), where IBs are thought to represent mere transit stations for protein cargoes destined for degradation to make their final exit from the cell.
Notwithstanding the aggresome theory, it is important to mention that it remains controversial whether autophagy is clearing monomeric and oligomeric precursors of aggregates, or inclusions themselves. For example, work from Rubinstzein's laboratory suggests that autophagy can efficiently target mutant species of α-synuclein that do not form aggregates large enough to be seen with light microscopy (52). On the other hand, a very recent live cell imaging study by a German group suggests that the sequestration of α-synuclein into aggresomes facilitates its clearance from the cell (53). Similarly, Fortun et al.’s demonstration that autophagy inhibition leads to a slowdown of aggresome clearance would support a role for autophagy-mediated clearance at the level of IBs (22). Notably, the experimental paradigm used in our current study is similar to the one described by Fortun et al. (22); our analysis of inclusions in the presence or absence of stimulated autophagy is performed after, and not before, they are formed. Hence, our results showing that autophagy induction promotes a significant reduction of inclusions number in cells expressing Q150Htt, mutant tau and α-synuclein/synphilin-1 would favor a role for autophagy in clearing inclusions. However, it is tempting to think that autophagy could clear proteins of all conformations, particularly under conditions of proteasome impairment. Supporting this, a previous study by Kopito's group (23) and a recent report by Pandey et al. (54) demonstrated that autophagy act as a compensatory degradation system when the UPS is impaired. The principal lysosomal pathway involved in the clearance of aggresomes is likely macro-autophagy, since the clearance of these structures could be induced and prevented by rapamycin and 3-MA treatment, respectively, which primarily influence macroautophagy (18,22). Further, aggresome-like inclusions are resistant to clearance when they are produced in ATG5−/− MEFs, a cell line deficient in macroautophagy (55). Harnessing the macroautophagy pathway may therefore offer innovative approaches in the treatment of several human disorders caused by altered protein conformation. However, our current study cautions the universal application of this strategy as not all IBs are amenable to autophagic degradation.
Given that the various disease-associated inclusions examined in this study all form ubiquitinated juxtanuclear inclusions, it is intriguing to note their differential susceptibility to clearance by autophagy. Furthermore, the apparent resistance of p38- and desmin-positive inclusions to clearance by autophagy does not appear to be a result of global defects on autophagy activation as the autophagic response in these cells is comparable with that in untransfected control cells as indicated by their similar increase in number of LC3 puncta following starvation-induced autophagy and their comparable clearance rates of the LC3-positive compartments. In our search, for protein factors that might account for the observed variability among the different types of inclusions toward clearance by autophagy (Table 1), we found that ‘autophagy-susceptible’ inclusions mediated by ectopically expressed mutant Htt, tau or α-synuclein/synphilin-1 are capable of recruiting mTOR while ‘autophagy-resistant’ inclusions mediated by ectopically expressed AIMP2 (p38) or mutant desmin fail to recruit mTOR. Since mTOR normally functions as an inhibitor of autophagy, the sequestration of mTOR into inclusions might influence the kinetics of autophagy stimulation by serum starvation or rapamycin treatment, which might in turn influence the clearance of inclusions. However, the number of AIMP2 (p38)-positive inclusions persists throughout the 24 h experimental period of stimulated autophagy, by which time the cellular levels of autophagy would have long peaked. Further, AIMP2 (p38)-positive inclusions are apparently impaired in engaging the lysosomal (LAMP-1) and autophagy (LC3) machinery under conditions of activated autophagy. Together, these results suggest that factors other than mTOR are responsible for the apparent inertness of AIMP2 (p38)-positive inclusions toward autophagic clearance. One such factor could be the microtubule-associated deacetylase HDAC6, which has been identified to be an important component of the aggresome (56). HDAC6 interacts with both polyubiquitinated misfolded proteins and dynein motors, and this interaction appears to be essential for aggresome formation (56), as well as for subsequent degradation of mutant Htt-positive aggresomes by autophagy (29). Consistent with a pivotal role of HDAC6 in aggresome formation, we found that HDAC6 is a component of all the different types of inclusions examined, except desmin-positive inclusions. The lack of binding between desmin-positive inclusions and HDAC6 is intriguing, given that the former exhibits the classical hallmarks of aggresomes including being ubiquitin-positive and that HDAC6 is apparently important for the formation of ubiquitin-positive aggresomes (56). It would be interesting to examine whether mutant desmin expression could produce aggresome-like inclusions in cells deficient in HDAC6. Nonetheless, our results suggest that despite association of HDAC6 to AIMP2 (p38)-positive inclusion, their clearance by autophagy is impaired. Another candidate factor that could influence the lysosomal clearance of protein inclusions is p62, a UBA-containing protein commonly found in IBs that has been demonstrated recently by Johansen's group to facilitate the formation and autophagic clearance of inclusions via its direct interaction with LC3 (36,57). However, we found that p62 is recruited to all the inclusions examined irrespective of their amenability to clearance by autophagy. Further, AIMP2 (p38)-positive inclusions are apparently impaired in engaging LC3 despite being competent in recruiting p62 (Table 1). Given the demonstrated dual role of p62 in promoting the formation and autophagic clearance of protein aggregates (36,37,57), it would appear that the balance between the two functions of p62 is dependent on the type of IBs recruiting it.
Although the missing factor(s) responsible for the inertness of AIMP2 (p38)- and desmin-positive inclusions toward autophagy remain(s) to be elucidated, its role in aggresomes that are amenable to autophagic clearance can be appreciated by the efficient clearance of AIMP2 (p38)-positive inclusions in the presence of α-synuclein/synphilin-1 co-localization. This alteration in inclusion composition not only facilitates the recruitment of mTOR into AIMP2 (p38)-positive inclusions, but also significantly increases the engagement of AIMP2 (p38)-positive inclusions with LAMP1. However, an important caveat is that the participation of this unknown factor appears to be context specific. Thus, unlike AIMP2 (p38)/α-synuclein/synphilin-1-positive inclusions, a population of desmin-positive inclusions that are also capable of forming co-aggregates with synuclein/synphilin-1-positive inclusions (or otherwise, tau-positive inclusions) remains inert to clearance by autophagy. This specificity may not be too surprising, given that α-synuclein, synphilin-1 and AIMP2 (p38) are all components of PD-associated LBs and their co-aggregation would presumably be physiologically more relevant compared with those generated by mutant desmin and synuclein/synphilin-1 (or tau). At the same time, our results also suggest that LBs are probably amenable to clearance by autophagy. Accordingly, therapeutic strategies directed at clearing LBs from PD patients via autophagy induction may produce beneficial outcomes. On the other hand, such strategies might not work in patients suffering from myopathies related to desmin mutations, as our results demonstrate that desmin-positive inclusions fail to engage the lysosomal apparatus efficiently and are thereby resistant to autophagic clearance. Desmin is a type III intermediate filament protein. Ultrastructurally, desmin-positive inclusions resulting from an S13F desmin mutation (same one used in this study) do not appear remarkably different from LBs, which could otherwise explain their inertness to clearance by autophagy (39,58). Interestingly, the expression of Alexander disease-associated mutant form of glial fibrillary acidic protein, another type III intermediate filament protein, in astrocytes similarly produces aggresomes that do not co-localize with LAMP1 (59). It thus appears that inclusions formed by this class of proteins may not be amenable to clearance by autophagy, suggesting that therapeutic strategies alternative to autophagy induction are probably required for diseases associated with these proteins.
In view of emerging evidence suggesting that enhancement of macroautophagy with drugs such as rapamycin could offer a viable therapeutic strategy for diseases caused by cellular mismanagement of aggregation-prone proteins, it is clearly important to distinguish between disease-associated proteins that could and could not present themselves as substrates to the lysosome. We have attempted in this study toward such a classification. Although the non-amenability of some disease-associated protein inclusions to clearance by autophagy may limit the utility of autophagy-based therapy, we anticipate that future elucidation of factors essential for lysosomal cargo selection would allow us to better exploit the therapeutic potential of autophagy.
MATERIALS AND METHODS
Plasmids and cell lines
Plasmids expressing α-synuclein, myc-tagged synphilin-1, HA-tagged AIMP2 (p38) and green fluorescent protein (GFP)-tagged mutant Htt exon 1 containing 150 polyglutamine repeats have been described previously (24,60). Expression constructs for myc-tagged mutant TauP301L and mutant S13F desmin were kind gifts from L. Petrucelli (Mayo Clinic, USA) and W.C. Yee (National Neuroscience Institute, Singapore), respectively. SH-SY5Y neuroblastoma cell line was from American Type Culture Collection. Wild-type and ATG5−/− MEFs were kind gifts from N. Mizushima (Tokyo Medical and Dental University, Japan).
Antibodies and reagents
The following antibodies were used: monoclonal anti-myc (clone 9E10) and anti-HA (both from Roche Diagnostics); monoclonal anti-desmin, rabbit anti-HDAC6 and rabbit anti-myc (all from Santa Cruz Biotechnology); monoclonal anti-vimentin, rabbit anti-γ-tubulin, rabbit anti-α-tubulin and rabbit anti-desmin (clone V9) (all from Sigma); monoclonal anti-LAMP1 (clone H4A3, Developmental Studies Hybridoma Bank, Iowa City, IA, USA); rabbit anti-LC3 (Novus Biologicals); rabbit anti-mTOR (Cell Signaling); rabbit anti-ubiquitin (Dako); rabbit anti-HA (Zymed); rabbit anti-p62 (BD Transduction); sheep anti-α-synuclein (Calbiochem); rhodamine-conjugated anti-mouse IgG (Molecular Probes); FITC-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories Inc.); FITC-conjugated anti-rabbit IgG (BD Pharmingen); Rhodamine-conjugated anti-rabbit IgG (Molecular Probes); Texas Red-conjugated anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc.); LysoTracker® Red DND-99, Alexa555-labeled anti-mouse IgG, Alexa488-labeled anti-rabbit IgG and Alexa647 anti-sheep IgG (all from Molecular Probes). Stock solutions of the following chemicals were prepared and stored at −20°C: rapamycin (0.2 mg/ml in DMSO) and nocodozole (2 mg/ml in DMSO) (both from Sigma), leupeptin (10 mm in dH2O) and NH4Cl (2 mm in dH2O) (both from Fisher Scientific), clasto-lactacystin-β-lactone (Affiniti Research) (5 mm in dH2O). 3-Methyladenine (Sigma) was dissolved directly in serum-free Dulbecco's modified Eagle's medium (DMEM) medium to 10 mm final concentration prior to treatment.
Cell culture and transfections
SH-SY5Y cells and MEFs were grown in DMEM containing 10% fetal bovine serum (FBS) in a 5% CO2 atmosphere. A total of 2.5 × 104 SH-SY5Y cells and MEFs were seeded on coverslips for transient transfection with various expression vectors using the LipofectAMINE 2000 reagent (Invitrogen) according to manufacturer's instructions. An amount of 0.25 µg of DNA was used for all transfection reactions except for coexpression studies whereby 0.25 µg of AIMP2 (p38) plasmid DNA was coexpressed with either 0.25 µg pCDNA3 vector or 0.25 µg of α-synuclein/synphilin-1 plasmid DNA (in equal ratio).
Immunocytochemistry and confocal microscopy
To characterize the properties of various inclusions, transfected cells were treated with 5 µm lactacystin for the last 16 of the 48 h transfection period. Thereafter, the cells were fixed with 3% paraformaldehyde (Sigma) and stained as described previously (61). Methanol fixation was performed for the staining of endogenous γ- and α-tubulin. To determine whether the formation of these inclusions is dependent on microtubule polymerization, the various transfected cells were treated with 10 µg/ml nocodozole and lactacystin simultaneously. Negative controls omitting each primary antibody were performed in each case, and no staining was seen (data not shown). Lysosome tracking was carried out by incubating cells with 75 nm LysoTracker for 2 h at 37°C followed by three washes in PBS before fixing in 3% paraformaldehyde and mounting with FluorSave reagent (Calbiochem) before viewing. Nuclei were counterstained with Hoechst (Sigma) or 4′-6-Diamidino-2-phenylindole (DAPI) (Molecular Probes) dyes. Images were collected using either an Axiovert 200 fluorescence microscope (Carl Zeiss MicroImaging, Inc) equipped with ×63 and ×100 objectives and an Axiovision 4.2 software (Carl Zeiss MicroImaging, Inc.) or a laser scanning confocal microscope (Leica) equipped with ×40 or ×60 objectives and analyzed using Leica Confocal software.
Inclusion formation and autophagic removal
The autophagic clearance of inclusions formed under conditions of proteasome impairment was investigated using a method recently described by Fortun et al. (22). Twenty-four hours post-transfection, cells were first treated with 5 µm lactacystin to enhance the inclusion formation. After 16 h incubation, the treated cells were allowed to recover in normal serum-containing media for 24 h. Concurrently, a parallel set of treated cells were washed extensively before incubating in serum-free media to stimulate autophagy either in the presence or in the absence of autophagy inhibitors, 10 mm 3-MA or 20 mm NH4Cl/100 µm leupeptin. Alternatively, autophagy in the treated cells was also activated by treatment with 200 µg/ml rapamycin in serum-containing media. Thereafter, cells were processed for immunocytochemical staining for evaluation of inclusions. For certain experiments, the co-localization of various inclusions with LAMP1 under different conditions were also examined. For time-course experiments, lactacystin-treated cells recovering in normal and serum-free conditions were fixed after 6, 9 and 24 h incubation periods. A minimum of 50 transfected cells were counted for each experiment. Cells were counted in a blinded manner and quantitative results reported are an average of at least three experiments.
LC3 puncta quantification
Cytosolic LC3 puncta were visualized by means of anti-LC3 immunofluorescence-based microscopy, and quantified using the ‘analyze particle’ function of the NIH IMAGE J program after thresholding. For this purpose, lactacystin-treated untransfected or transfected SH-SY5Y cells were washed out either in normal serum-containing media or in serum-free media as described above. In a parallel set, the recovery media was supplemented with 20 mm NH4Cl/100 µm leupeptin to inhibit lysosomal proteases. Five randomly selected inclusion-positive cells and five untransfected control cells were analyzed for each condition. The experiment was repeated at least three times to demonstrate the reproducibility.
Statistical significance
Statistical significance for all the quantitative data obtained was analyzed using ANOVA with post hoc Tukey's test (InStat; GraphPad software; *P < 0.05, **P < 0.01, ***P < 0.001).
Conflict of Interest statement. None declared.
FUNDING
This work was supported by grants from the Singapore Biomedical Research Council 0613319483, National Medical Research Council NMRC/0776/2003, and SingHealth Group (L.K.L.), Parkinson's Disease Foundation (E.W.), Singapore Millennium Foundation (J.T.), NIH/NIA AG021904 (A.M.C.) and NINDS Grants NS38377 and NS48206 (T.M.D.) and NS038370 (A.M.C.). T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Supplementary Material
REFERENCES
- 1.Costa M.L., Escaleira R., Cataldo A., Oliveira F., Mermelstein C.S. Desmin: molecular interactions and putative functions of the muscle intermediate filament protein. Braz. J. Med. Biol. Res. 2004;37:1819–1830. doi: 10.1590/s0100-879x2004001200007. [DOI] [PubMed] [Google Scholar]
- 2.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(suppl.):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 3.Zatloukal K., French S.W., Stumptner C., Strnad P., Harada M., Toivola D.M., Cadrin M., Omary M.B. From mallory to Mallory-Denk bodies: what, how and why? Exp. Cell Res. 2007;313:2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 4.Kopito R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Mata R., Gao Y.S., Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 6.Wigley W.C., Fabunmi R.P., Lee M.G., Marino C.R., Muallem S., DeMartino G.N., Thomas P.J. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olanow C.W., Perl D.P., DeMartino G.N., McNaught K.S. Lewy-body formation is an aggresome-related process: a hypothesis. Lancet Neurol. 2004;3:496–503. doi: 10.1016/S1474-4422(04)00827-0. [DOI] [PubMed] [Google Scholar]
- 8.Mishra R.S., Bose S., Gu Y., Li R., Singh N. Aggresome formation by mutant prion proteins: the unfolding role of proteasomes in familial prion disorders. J. Alzheimers Dis. 2003;5:15–23. doi: 10.3233/jad-2003-5103. [DOI] [PubMed] [Google Scholar]
- 9.Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., Wanker E.E. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 11.Bowman A.B., Yoo S.Y., Dantuma N.P., Zoghbi H.Y. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 2005;14:679–691. doi: 10.1093/hmg/ddi064. [DOI] [PubMed] [Google Scholar]
- 12.Cummings C.J., Reinstein E., Sun Y., Antalffy B., Jiang Y., Ciechanover A., Orr H.T., Beaudet A.L., Zoghbi H.Y. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24:879–892. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 13.Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 14.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 15.Cuervo A.M. Autophagy in neurons: it is not all about food. Trends Mol. Med. 2006;12:461–464. doi: 10.1016/j.molmed.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J.I., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 18.Berger Z., Ravikumar B., Menzies F.M., Oroz L.G., Underwood B.R., Pangalos M.N., Schmitt I., Wullner U., Evert B.O., O'Kane C.J., et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 19.Ravikumar B., Berger Z., Vacher C., O'Kane C.J., Rubinsztein D.C. Rapamycin pre-treatment protects against apoptosis. Hum. Mol. Genet. 2006;15:1209–1216. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 20.Ravikumar B., Duden R., Rubinsztein D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 21.Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 22.Fortun J., Dunn W.A., Jr, Joy S., Li J., Notterpek L. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 2003;23:10672–10680. doi: 10.1523/JNEUROSCI.23-33-10672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwata A., Christianson J.C., Bucci M., Ellerby L.M., Nukina N., Forno L.S., Kopito R.R. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl. Acad. Sci. USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaoka U., Kim K., Jana N.R., Doi H., Maruyama M., Mitsui K., Oyama F., Nukina N. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J. Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- 25.Gotz J., Chen F., Barmettler R., Nitsch R.M. Tau filament formation in transgenic mice expressing P301L tau. J. Biol. Chem. 2001;276:529–534. doi: 10.1074/jbc.M006531200. [DOI] [PubMed] [Google Scholar]
- 26.Corti O., Hampe C., Koutnikova H., Darios F., Jacquier S., Prigent A., Robinson J.C., Pradier L., Ruberg M., Mirande M., et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum. Mol. Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- 27.Chung K.K., Zhang Y., Lim K.L., Tanaka Y., Huang H., Gao J., Ross C.A., Dawson V.L., Dawson T.M. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat. Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 28.Mortimore G.E., Poso A.R. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- 29.Iwata A., Riley B.E., Johnston J.A., Kopito R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 30.Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn P.F., Mesires N.T., Vine M., Dice J.F. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005;1:141–145. doi: 10.4161/auto.1.3.2000. [DOI] [PubMed] [Google Scholar]
- 32.Fuertes G., Martin De Llano J.J., Villarroya A., Rivett A.J., Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2007;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravikumar B., Vacher C., Berger Z., Davies J.E., Luo S., Oroz L.G., Scaravilli F., Easton D.F., Duden R., O'Kane C.J., et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 36.Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 38.Wakabayashi K., Engelender S., Yoshimoto M., Tsuji S., Ross C.A., Takahashi H. Synphilin-1 is present in Lewy bodies in Parkinson's disease. Ann. Neurol. 2000;47:521–523. [PubMed] [Google Scholar]
- 39.Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 40.Rubinsztein D.C., Gestwicki J.E., Murphy L.O., Klionsky D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 41.Auluck P.K., Chan H.Y., Trojanowski J.Q., Lee V.M., Bonini N.M. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 42.Cuervo A.M., Stefanis L., Fredenburg R., Lansbury P.T., Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Vicente M., Talloczy Z., Kaushik S., Massey A.C., Mazzulli J., Mosharov E.V., Hodara R., Fredenburg R., Wu D.C., Follenzi A., et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J. Clin. Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin Y., Klucken J., Patterson C., Hyman B.T., McLean P.J. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J. Biol. Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 45.Rott R., Szargel R., Haskin J., Shani V., Shainskaya A., Manov I., Liani E., Avraham E., Engelender S. Monoubiquitylation of alpha-synuclein by seven in absentia homolog (SIAH) promotes its aggregation in dopaminergic cells. J. Biol. Chem. 2008;283:3316–3328. doi: 10.1074/jbc.M704809200. [DOI] [PubMed] [Google Scholar]
- 46.Lim K.L., Dawson V.L., Dawson T.M. Parkin-mediated lysine 63-linked polyubiquitination: a link to protein inclusions formation in Parkinson's and other conformational diseases? Neurobiol. Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 47.Liani E., Eyal A., Avraham E., Shemer R., Szargel R., Berg D., Bornemann A., Riess O., Ross C.A., Rott R., et al. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2004;101:5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka M., Kim Y.M., Lee G., Junn E., Iwatsubo T., Mouradian M.M. Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. J. Biol. Chem. 2004;279:4625–4631. doi: 10.1074/jbc.M310994200. [DOI] [PubMed] [Google Scholar]
- 49.Sugawara M., Kato K., Komatsu M., Wada C., Kawamura K., Shindo P.S., Yoshioka P.N., Tanaka K., Watanabe S., Toyoshima I. A novel de novo mutation in the desmin gene causes desmin myopathy with toxic aggregates. Neurology. 2000;55:986–990. doi: 10.1212/wnl.55.7.986. [DOI] [PubMed] [Google Scholar]
- 50.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 51.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 52.Webb J.L., Ravikumar B., Atkins J., Skepper J.N., Rubinsztein D.C. Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 53.Opazo F., Krenz A., Heermann S., Schulz J.B., Falkenburger B.H. Accumulation and clearance of alpha-synuclein aggregates demonstrated by time-lapse imaging. J. Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05407.x. May 6, Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 54.Pandey U.B., Nie Z., Batlevi Y., McCray B.A., Ritson G.P., Nedelsky N.B., Schwartz S.L., DiProspero N.A., Knight M.A., Schuldiner O., et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 55.Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 57.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 Binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 58.Pica E.C., Kathirvel P., Pramono Z.A., Lai P.S., Yee W.C. Characterization of a novel S13F desmin mutation associated with desmin myopathy and heart block in a Chinese family. Neuromuscul. Disord. 2008;18:178–182. doi: 10.1016/j.nmd.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 59.Mignot C., Delarasse C., Escaich S., Della Gaspera B., Noe E., Colucci-Guyon E., Babinet C., Pekny M., Vicart P., Boespflug-Tanguy O., et al. Dynamics of mutated GFAP aggregates revealed by real-time imaging of an astrocyte model of Alexander disease. Exp. Cell Res. 2007;313:2766–2779. doi: 10.1016/j.yexcr.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 60.Wang C., Tan J.M., Ho M.W., Zaiden N., Wong S.H., Chew C.L., Eng P.W., Lim T.M., Dawson T.M., Lim K.L. Alterations in the solubility and intracellular localization of parkin by several familial Parkinson's disease-linked point mutations. J. Neurochem. 2005;93:422–431. doi: 10.1111/j.1471-4159.2005.03023.x. [DOI] [PubMed] [Google Scholar]
- 61.Lim K.L., Chew K.C., Tan J.M., Wang C., Chung K.K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C.A., et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.