Abstract
Somatic microindels (microdeletions with microinsertions) have been studied in normal mouse tissues using the Big Blue lacI transgenic mutation detection system. Here we analyze microindels in human cancers using an endogenous and transcribed gene, the TP53 gene. Microindel frequency, the enhancement of 1–2 microindels and other features are generally similar to that observed in the non-transcribed lacI gene in normal mouse tissues. The current larger sample of somatic microindels reveals recurroids: mutations in which deletions are identical and the co-localized insertion is similar. The data reveal that the inserted sequences derive from nearby but not adjacent sequences in contrast to the slippage that characterizes the great majority of pure microinsertions. The microindel inserted sequences derive from a template on the sense or antisense strand with similar frequency. The estimated error rate of the insertion process of 13% per bp is by far the largest reported in vivo, with the possible exception of somatic hypermutation in the immunoglobulin gene. The data constrain possible mechanisms of microindels and raise the question of whether microindels are ‘scars’ from the bypass of large DNA adducts by a translesional polymerase, e.g. the ‘Tarzan model’ presented herein.
INTRODUCTION
The term ‘indel’ has different definitions in different fields. In evolutionary studies, indel is used to mean an insertion or a deletion (1,2) and ‘indels’ simply refers to the mutation class that includes both insertions, deletions and the combination thereof (3–5) including insertion and deletion events that may be separated by many years (6). In germline and somatic mutation studies, however, indel describes a special mutation class, defined as a co-localized insertion and deletion (7), and sometimes defined (8) to include tandem-base mutations (TBMs), mutations in which the insertion and deletion are the same size (9). TBMs, however, may result from fundamentally different mechanisms (9,10). Herein, the term indel is defined as a mutation resulting in a co-localized insertion and deletion and a net gain or loss in nucleotides, and ‘microindel’ is defined as an indel which has a deletion and/or insertion size of 1–50 nucleotides and that results in a net gain or loss of 1–50 nucleotides. The notation N-M indicates an indel with M nucleotides deleted and N nucleotides inserted. An example of a microindel that is common in a genetic disorder is the ‘blmAsh’ mutation, a 7–6 microindel common in Askenazi Jews with Bloom Syndrome (11).
A two-step mechanism has been suggested for microindels: a deletion followed by an insertion, or vice-versa (8). Chuzhanova et al. (8) concluded that ‘the majority of indels (>90%) are explicable in terms of a two-step process involving established mutational mechanisms’. Microindels were generally attributed to combinations of the mutational mechanisms of insertion and deletion (e.g. strand switching, single strand loops, slipped strand mispairing) (8).
Initial Astrogenetic analyses (see Terminology) of mouse somatic microindels, however, suggest a more complicated story which includes mechanisms that are microindel specific (12,13). A sample of 30 spontaneous somatic microindels in normal mouse tissues was previously analyzed in the context of 5562 independent spontaneous somatic mutations using the Big Blue transgenic mouse mutation detection system (12,13). The Big Blue system utilizes the non-transcribed bacterial lacI gene as the mutation target. The mostly young mice were fed a standard diet and housed under controlled conditions with care taken to avoid mutagen exposure. 1–2 microindels (2 bp deleted and 1 bp inserted) are by far the most frequent class (12). When compared with pure microinsertions and pure microdeletions, the microindels in mice were characterized by an absence of hotspots, inserted sequences that rarely repeat the adjacent base (compared with 97% of pure microinsertions that do so) (7), generally larger and more varied sizes of inserted and deleted sequences, and different sequence contexts. For the six microindels with the longest insertions, the nature of the insertions seemed heterogeneous and without a clear pattern, not like the mechanisms that cause pure microinsertions, pure microdeletions and single-base substitutions. Are the somatic mutations seen in the non-transcribed bacterial lacI gene reflective of microindels in endogenous transcribed mammalian genes? Are the results in mice extendable to microindels in a transcribed endogenous human gene in cancers in typically older individuals on varied diets and having varied mutagen exposures?
To address these questions, we created a database of TP53 microindels by reviewing 126 primary articles ascertained by reviewing version 10 of the IARC TP53 Mutation Database (14) for mutations described as ‘complex’ and for multiple mutations reported in the same individual. Sixty-six TP53 somatic microindels were identified and analyzed in the context of the other mutations in the IARC database. To compare the inserted sequences in microindels with those in pure microinsertions, we also created a database of TP53 pure microinsertions by reviewing over 150 primary publications.
The somatic TP53 microindels in human cancer analyzed herein illuminate the unique features of microindels in humans relative to pure microinsertions and pure microdeletions and extend the previous results in mice to an endogenous transcribed human gene in human cancer. TP53 microindels in cancer are remarkably similar to spontaneous microindels in the non-transcribed lacI transgene in normal Big Blue mouse tissues, suggesting that the selective pressures associated with human oncogenesis as well as any mutagens associated with cancers have minor effects relative to endogenous mechanisms. In addition, the data reveal (i) the presence of recurroids: deletions with similar but not identical insertions; (ii) inserted sequences derived from unexpectedly close templates, as demonstrated by statistical analyses; (iii) nearly equal numbers of insertions deriving from the sense and antisense strands and (iv) an insertion process that is highly error-prone with an estimated error rate of 13% per bp. The data are consistent with a model in which microindels are the ‘scars’ of error-prone repair of large, potentially lethal DNA adducts (‘Tarzan model’ of indelogenesis).
RESULTS
Microindels are uncommon and exhibit sequence context effects
Sixty-six somatic microindels distributed widely over TP53 were identified (Supplementary Material, Table S5, Fig. S4) by analysis of the primary literature. These microindels constitute a subset of the ‘complex mutations’ and the closely spaced double mutations within version 10 of the IARC TP53 Mutation Database (14) and should account for all microindel events in that database. These 66 microindels account for 0.3% of the mutations in the IARC database. The microindel frequency among individual cancers appears similar (data not shown).
Recurroids occur
At one nucleotide position, the same microindel recurred once, and at three other positions, microindel ‘recurroids’ occurred: microindels with an identical deletion and a similar but not identical insertion (Fig. 1). In these recurroids, the insertion differs by only one nucleotide. These identical or recurroid microindels involve 6% of the observed microindels. In only one instance was the deleted sequence of the recurroid microindel also seen as a pure microdeletion (14 555del24). To our knowledge, this is the first description of the microindel recurroid phenomenon. A search of other databases revealed multiple recurroids in the EGFR gene in lung cancer including sites with deletion identical recurroids as found in TP53, sites with insertion identical recurroids and hybrid sites with both deletion and insertion identical recurroids (Supplementary Material, Table S8).
Figure 1.
Sequence context of identical or recurroid microindels. Sequence context of the four sites where two TP53 somatic microindels occurred and deleted the same sequence. Deleted sequence is shown in square brackets and inserted sequence is shown above. Base numbering below the sequence indicates the number of the base that is aligned with the first digit according to GenBank genomic sequence accession no. X54156.1.
Microindels usually shorten sequence and shift the reading frame
Most TP53 somatic microindels (83%) result in a net loss of sequence (Supplementary Material, Table S2, Fig. S5). The 1–2 microindel (2 bp deleted and 1 bp inserted) is the most common type, comprising 14% (9/66) of TP53 somatic microindels (Supplementary Material, Table S2, Fig. S6A, Supplementary text on 1−M and N−1 microindels). Most TP53 somatic microindels (79%) shift the reading frame and all the in-frame microindels result in a net deletion (Supplementary Material, Table S5, Fig. S5).
Insertions and deletions in microindels tend to be larger than in pure microinsertions and microdeletions
A database of pure microinsertions was constructed from the primary literature and pure microdeletions were extracted from the IARC database. One nucleotide is the most common size of somatic TP53 pure microinsertions (63%) and pure microdeletions (45%). In microindels, the sizes of the inserted sequence and the sizes of the deleted sequence are more dispersed and larger overall; the distributions are significantly different from the distributions of the sizes of pure microinsertions and pure microdeletions (P = 0.03 and P = 0.00000002, respectively, by the Kolmogorov–Smirnov test; P = 0.001 and P < 0.0000005, respectively, by Fisher’s exact test on the sizes binned as shown in Supplementary Material, Fig. S7). The distributions in Supplementary Material, Figure S7 are significantly different even if the common single-base pure microinsertions and pure microdeletions are excluded from the analysis (P = 0.01 and P = 0.02, respectively).
The larger insertions in microindels suggest a highly error-prone mechanism
The longer inserted sequences in microindels derive from nearby sequence and may involve error-prone processes (Fig. 2). In the sample of TP53 microindels, the inserted sequence is at least six nucleotides for four somatic microindels (Supplementary Material, Table S5A) and for one germline microindel (Supplementary Material, Table S5B), and for all these, a putative sense or antisense template sequence exists significantly closer than expected by chance (Fig. 2). The putative template was nearby but not adjacent in three instances and overlapping the deletion in two instances. In two instances, error-prone duplication is suggested by a putative template that is not a perfect match but is significantly closer than expected by chance even after accounting for the mismatches.
Figure 2.
Sequence context of microindels with larger insertions. Sequence context of TP53 microindels (Supplementary Material, Table S5) plus microindels in CFTR (Human Gene Mutation Database, public version as of 4 April 2007) (17), microindels in EGFR (Epidermal Growth Factor Receptor Mutation Database, http://www.cityofhope.org/cmdl/egfr_db) (16), microindels in HPRT (Human HPRT Mutation Database, http://www.ibiblio.org/dnam/mainpage.html, Supplementary Material, Table S9) and microindels in PTEN (Catalogue of Somatic Mutations in Cancer, as of 8 June 2007) (15) in which the inserted sequence is at least six nucleotides. Deleted sequence is shown in square brackets, inserted sequence is shown above and the putative sense or antisense template for the inserted sequence is shown underlined. Bold font indicates sense or antisense matching bases in the inserted sequence and template (regular font indicates a mismatch). The first number shown in parentheses above the putative insertion template is the expected distance to the first instance of the inserted sequence with the same number of mismatches as the putative template. The P-value is the chance probability of the putative template or its reverse complement occurring at the observed distance or closer. Both values are calculated by simulation (see Materials and Methods). Base numbering below the sequence indicates the number of the reference sequence nucleotide that is aligned with the first digit (TP53, X54156.1; CFTR, NC_000007.12 region 116907253–117095955; EGFR, NC_000007.12 region 55054219–55242525; HPRT, NC_000023.9 region 133421923–133462362; PTEN, NC_000010.9 region 89613175–89716382).
Large mutation databases were searched for genes having more than one reported microindel with at least six nucleotides inserted. The Catalogue of Somatic Mutations in Cancer (COSMIC) (15) contains two somatic PTEN microindels in which the inserted sequence is at least six nucleotides and has a putative template nearby. The Epidermal Growth Factor Receptor (EGFR) Mutation Database (http://www.cityofhope.org/cmdl/egfr_db) contains two such somatic EGFR microindels (16). The Human HPRT Mutation Database described in Supplementary Material contains two such microindels (one somatic and one germline), and three such germline microindels are available from CFTR mutation data in the Human Gene Mutation Database (17). Combined with the TP53 microindels (Fig. 2), these data indicate that the sequence contexts of microindels are dispersed among three binary characteristics: (i) insertion template nearby but not adjacent to the deleted sequence versus overlapping the deleted sequence, (ii) sense versus antisense duplication and (iii) error-prone versus error-free duplication. Seven of the eight possible categories are represented by the 14 microindels in Figure 2.
The five cases in Figure 2 that are interpreted as a microindel in which the insertion is a sense duplication of a template overlapping the deleted sequence (one TP53 case, two CFTR cases and two EGFR cases) can also be interpreted as doublet mutations since, in all five cases, the duplication errors occur at the ends of the putative insertion template. The TP53 case could be interpreted as a doublet consisting of an insertion of T separated by 6 bp from a 1–5 microindel (delCGCGCinsG). The CFTR cases can be interpreted, respectively, as a doublet consisting of an insertion of C separated by 4 bp from a G > A substitution, and a doublet consisting of an A > T substitution separated by 10 bp from an insertion of T. The EGFR cases can be interpreted, respectively, as a doublet consisting of an insertion of nine nucleotides separated by 1 bp from a C > T substitution, and a doublet consisting of an insertion of five nucleotides separated by 1 bp from an insertion of T. The microindels in the complementary class of antisense duplication of a template overlapping the deleted sequence cannot be interpreted as doublet mutations.
The microindels in Figure 2 can be used to estimate the overall error rate for the mechanisms of microindels with larger insertions. Among these microindels in which at least six nucleotides were inserted, 13% (16/120) of the nucleotides inserted do not match the putative insertion template (Supplementary Material, Table S3) of all the nucleotides shown inserted in Figure 2 or 12% (12/103) excluding the two microindels with putative insertion templates at an observed distance with a P-value > 0.05.
DISCUSSION
Astronomic analyses utilize electromagnetic frequency, pattern and spectrum to make inferences about the universe. ‘Astrogenetic’ analyses utilize mutation frequency spectrum and pattern to make inferences about in vivo mutagenesis, which cannot be observed directly (18). We present the first comprehensive ‘Astrogenetic’ analysis of human somatic microindels in (i) an endogenous gene and (ii) in human cancer. The data herein describe the ‘anatomy’ of microindels and constrain hypotheses of their nature and origin. We conclude that (i) microindels are uncommon with remarkably similar frequencies in human cancers and in the human germline (0.3–0.4%; Supplementary Material, Table S2), (ii) microindels tend to shorten the nucleotide sequence with the majority resulting in a net deletion that also shifts the reading frame, (iii) ‘recurroids’ occur (Fig. 1), (iv) novel sequence contexts occur (Fig. 2) as well as sequence contexts previously observed for mouse somatic microindels, (v) the alternative model of a base substitution with a deletion or insertion for 1−M and N−1 microindels, respectively, is not consistent with the signatures of single-base substitutions, pure deletions and pure insertions in TP53 (analysis in Supplementary Material), (vi) microindels do not result predominantly from simple combinations of the mechanisms that cause pure microinsertions and pure microdeletions, (vii) the mechanisms of microindels, at least those with larger insertions, are highly error-prone overall with an estimated error rate of 13% per bp, consistent with the error rates of certain Y-family translesion polymerases and (viii) the existence of similar but not identical short insertions in recurroids and the preferential production of 1–2 microindels by at least one error-prone polymerase (see in what follows) are consistent with microindels with short insertions also deriving from these highly error-prone mechanisms. A possible mechanism of indelogenesis is presented in what follows.
Overall, somatic microindels in cancer are similar to germline microindels (Supplementary Material, Table S2) in the F8 (Sommer, unpublished data) and F9 genes (19) and in the Human Gene Mutation Database (HGMD) (8,17). Although the sizes of the inserted sequence in human germline microindels from HGMD are skewed slightly to larger sizes than for somatic TP53 microindels (P = 0.04, Supplementary Material, Fig. S7A), there is no significant difference between the distributions of deletion sizes (P = 0.1; Supplementary Material, Fig. S7B). Human somatic microindels in cancer are also similar to somatic microindels in normal mouse tissues (12,13) (Table 1), although recurroids were not observed in the mouse data. Note that the TP53 recurroids were deletion identical recurroids while both deletion and insertion identical recurroids are found in EGFR (Supplementary Material, Table S8) (16). The similarity of TP53 microindels in cancer to germline microindels and to spontaneous somatic microindels in normal mouse tissues is consistent with microindels in human cancer generally deriving from spontaneous endogenous processes.
Table 1.
Features of somatic microindels in a transcribed endogenous human gene (TP53) in cancers versus in a non-transcribed transgenic reporter gene (lacI) in mouse normal tissues
| TP53 | lacI | |
|---|---|---|
| Tissue | Cancer | Normal |
| Transcribed gene | + | − |
| Endogenous gene | + | − |
| Number of events | 66 | 30 |
| Microindel frequency, % | 0.3 | 0.5 |
| Microindels resulting in net loss of sequence, % | 83 | 70 |
| Microindels causing frameshift, % | 79 | 87 |
| 1–2 type microindels, % | 14 | 23 |
| Recurroidsa | + | − |
aIdentical deletion with a similar insertion (see text).
Microindels are uncommon and exhibit sequence context effects
The TP53 somatic microindel frequency is 0.3%, similar to the germline microindel frequency [0.4% in F8 (Sommer, unpublished data) and F9 (19), and 0.4% in HGMD (8)] and similar to the somatic microindel frequency in the mouse (0.5%) (12,13).
The previous analyses of the HGMD human germline microindel data (8) and the mouse somatic microindel data (12,13) did not reveal positional hotspots. In the TP53 microindel data analyzed herein, however, there are five microindels that start at the same nucleotide as another (Supplementary Material, Fig. S4). One of these is an identical recurrence and three are ‘recurroids’ with an identical deletion and a similar insertion (Fig. 1), suggesting the effects of sequence context and error-prone processes.
The two identical 1–4 microindels at bp 14 042 occur at a site that is a hotspot of pure microinsertions (17 of 596 pure microinsertions in the IARC database) and pure microdeletions (14 of 1782 pure microdeletions), suggesting shared sequence context effects. Indeed, bp 14 042 is the most frequent hotspot of pure microinsertions in the IARC database (Supplementary Material, Fig. S8). Reviewing the primary publications for a subset of the single base microinsertions at bp 14 042, however, revealed that only 25% (2/8) are insertions of T, as observed in the pair of 1–4 microindels. In addition, none of the pure microdeletions at bp 14 042 are deletions of 4 bp, as observed in the pair of 1–4 microindels. The three recurroid sites are not at hotspots of either pure microinsertions or pure microdeletions. These differences are not consistent with the identical and recurroid microindels being due to the same mechanisms as pure microinsertions and pure microdeletions.
Microindels usually shorten sequence and shift the reading frame
The majority (83%) of TP53 somatic microindels result in a net loss of sequence (Supplementary Material, Table S2, Fig. S5). The 1–2 microindel (2 bp deleted and 1 bp inserted) is the most common type, comprising 14% of TP53 somatic microindels, similar to the 20% of HGMD germline microindels and to the 23% of mouse microindels (12).
The majority (79%) of TP53 somatic microindels result in a reading frame shift. Microindels can also be in-frame and thereby alter the protein by adding or subtracting a few amino acids (‘protein tinkering’). The minority (21%) of TP53 somatic microindels that are in-frame result in the deletion of one or more amino acids that are presumably critical to TP53 function. None of the in-frame TP53 somatic microindels result in a net insertion. In other genes, such as EGFR (http://www.cityofhope.org/cmdl/egfr_db/), protein tinkering does result in gain of function (16,20–22).
Microindels are not TBMs
TBMs might also be called ‘N-N’ microindels (e.g. a mutation that changes two adjacent bases (2 bp TBM) might be called a 2–2 microindel). The former notation implies a mechanism involving adjacent base substitutions, whereas the latter notation implies a mechanism involving a coordinate deletion and insertion. The overwhelming majority of somatic TBMs are those that change two adjacent bases (2 bp TBM) and these are 2.4-fold (160/66) more frequent than all types of somatic microindels combined. The majority of 2 bp TBMs are known mutagen signatures. CC > TT TBMs in skin cancer are a signature of UV exposure (23) and account for 51% (169/329) of the 2 bp TBMs in the IARC TP53 Mutation Database (version 10). Of the remaining 2 bp TBMs, 8% (13/160) are GG > TT TBMs, a reported signature of peroxyacetyl nitrate (24), of acetaldehyde in the mouse (9) and of lipid peroxidation (25). In the mouse, the frequency of 2 bp TBMs varies dramatically with tissue type (9,10) while no such tissue specificity was observed for microindels (13).
Somatic TBMs of three or more base pairs are rare. In two Big Blue mouse studies (9,10), only one 3 bp and no larger TBMs were observed; a frequency that is four orders of magnitude lower than that for single-base substitutions. In the IARC database, 10 3 bp and zero larger TBMs are reported (as ‘complex’ mutations). Of these 3 bp TBMs, three contain a CC > TT change and can be viewed as a UV-associated 2 bp TBM with an adjacent substitution. Thus, unambiguous TBMs larger than 2 bp are extremely rare.
We conclude that 2 bp TBMs are fundamentally different from microindels and that 3 bp or larger TBMs are rare with a substantial fraction of those reported deriving from the same mechanisms that commonly generate 2 bp TBMs. While future work may demonstrate that a minority of TBMs arise from the same mechanisms producing microindels, it currently seems reasonable to classify TBMs as distinct and separate from microindels.
Towards a descriptive nomenclature for uncommon mutations
Towards discarding the catchall category of ‘complex’ mutations, the term ‘microindels’ is meant as a descriptive term for a class of mutations with characteristics that differ from those of other common mutation classes. A distinct practical advantage of descriptive classes of uncommon mutation is that they serve as a nomenclature to facilitate further study. It would be much easier to collect microindels for analysis if they were described as such in the literature and in databases.
Microindel ≠ pure microinsertion + pure microdeletion
Several lines of evidence support the conclusion that microindels are not caused predominantly by simple combinations of the same mechanisms that cause pure microinsertions and pure microdeletions: (i) the sizes of the insertions and deletions in microindels are larger and more varied than in pure microinsertions and pure microdeletions reflecting significant differences in the size distributions (P = 0.001 and P < 0.0000005, respectively), (ii) microinsertions overwhelmingly repeat the adjacent sequence (83% of TP53 microinsertions, 151/183) while the insertions in microindels do so only infrequently (30%, 20/66, P < 0.000000005) and (iii) microindels do not occur preferentially at mononucleotide and dinucleotide repeats which are hotspots of pure microinsertion and pure microdeletion (7,26).
In vivo microindels arise by error-prone mechanisms
The microindels with inserted sequences of at least six nucleotides showed insertions of sequences nearby or overlapping with the deletion (Fig. 2). Of TP53 microindels with insertions of at least six nucleotides, 40% (2/5) involve putative error-prone duplications, raising the intriguing possibility of mechanisms that involve error-prone polymerases such as the Y-family polymerases (27–31).
Using an in vitro forward mutation assay that measures errors made during synthesis from an undamaged DNA template (a 407 nuclotide single-stranded gap), Matsuda et al. (32) measured an average base substitution error rate of 3% for human Y-family polymerase eta. This error rate is comparable with the 13% error rate estimated herein for errors made during putative duplication of a nearby template sequence. Among the mutations detected by Matsuda et al. (32) were 27 microindels of type 1–2 and 7 of type 2–1, a ratio of 1–2 to 2–1 type microindels similar to that observed in vivo for TP53 and Big Blue lacI (16 versus 5; P = 1.0). Mammalian Y-family polymerase kappa can also produce microindels and these most frequently are type 1–2 (30,33). Polymerase iota, another highly error-prone Y-family polymerase, exhibits a dramatic variation in error rate depending on the template, ranging from 0.01 to 13% for A, C and G templates, but up to 300% for T templates (34). The errors indicated in Figure 2 do not exhibit this dramatic base specificity in error rate (data not shown). The hypothesis that error-prone polymerases are involved in microindels might be tested by over production or knockout in a model organism such as yeast.
Tarzan model of indelogenesis
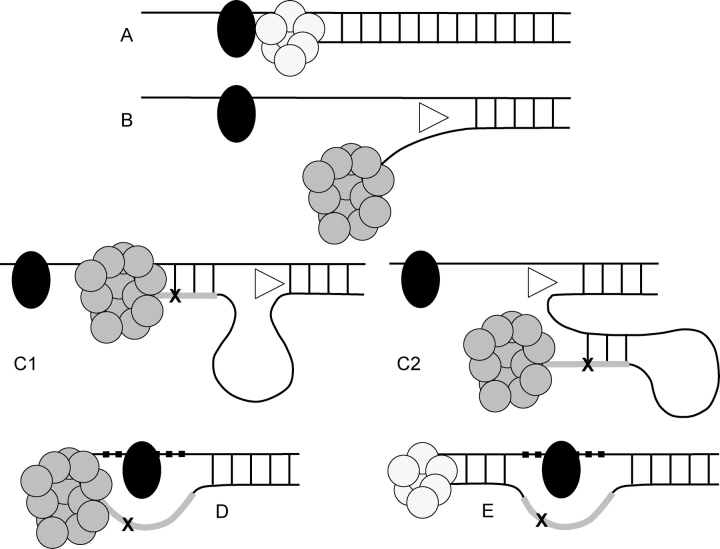
We present the Tarzan Model (Fig. 3) of indelogenesis, as a model consistent with our Astrogenetic in vivo data. The Tarzan model is reminiscent of the old TV series in which Tarzan swings on vines to cross obstacles and save the day. A large DNA adduct that blocks replication by the normal polymerase is not easily bypassed, even by translesion polymerases (29,35,36). To save the cell, an error-prone translesion polymerase complex may be recruited along with a helicase. In the Tarzan model, the helicase unwinds the nearby nucleotides of the nascent strand from the template so that the translesion polymerase, its forward progress blocked by the adduct, can back up on the template strand or loop back on itself. Some additional length is synthesized on the nascent strand constituting the inserted sequence of the microindel and acting like a vine that the translesion polymerase uses to swing across the adduct. A few nucleotides on the template are bypassed, constituting the deleted sequence of the microindel. The normal polymerase can then continue with replication. The Tarzan model makes predictions that are testable including: intrastrand and interstrand cross-linkers like cisplatin or mitomycin C will increase the frequency of microindels, a mouse or yeast knockout of a relevant DNA polymerase or helicase will reduce the frequency of microindels and an in vitro system in which large DNA adducts are synthesized will result in microindels when incubated with an appropriate cellular extract.
Figure 3.
Tarzan Model: proposed mechanism of indelogenesis. Replication by the normal DNA polymerase (small cluster of circles) is blocked by a large DNA adduct (black oval) on the template DNA (A). To bypass the adduct, an error-prone translesion polymerase (large cluster of circles) and a helicase (triangle) are recruited. The translesion polymerase is also blocked by the adduct but the helicase disassociates the nascent strand from the template (B), allowing the translesion polymerase to synthesize a few nucleotides from the template (C1) or the nascent strand (C2). A few additional nucleotides are synthesized, sometimes with errors (X), generating the inserted sequence of the microindel (thick line). Thus, the inserted sequence is either a sense (in the case of C1) or antisense (in the case of C2) copy, sometimes with errors, of nearby sequence. The helicase then disassociates the segment synthesized by the translesion polymerase from the template (in the case of C1) or the nascent strand (in the case of C2). With the additional length of synthesized DNA, the translesion polymerase is able to swing across the adduct and save the cell, as Tarzan would use a vine to ‘swing’ across an obstacle and save the day. This process results in the skipping of some nucleotides on the template (D, thick dashes) resulting in the microdeletion part of the microindel. Some of the skipped bases (or their complements on the nascent strand in the case of C2) may have been part of the template for the inserted sequence in which case there is overlap of the inserted and deleted sequences of the microindel. After the adduct is bypassed, the normal polymerase can proceed with replication (E).
Serial replication slippage models offer a possible alternative mechanism for the microindels with larger insertion sizes (37,38). However, these models require the presence of repeat sequences and would not explain the high error-rate observed herein during synthesis from the putative insertion template (Supplementary Material, Fig. S9). Non-homologous end joining (NHEJ) after DNA double-strand breaks is another possible mechanism for microindels (discussed in Supplementary Material). We present a new model that does not require repeat sequences and that explains the observed high error-rate during template duplication.
In conclusion, human TP53 somatic microindels in cancer are uncommon, exhibit recurroids and can derive from unique error-prone mechanisms.
MATERIALS AND METHODS
Identification of microindels
Somatic microindels in the human TP53 gene were identified by analyzing 126 publications in the primary literature. Version 10 of the IARC TP53 Mutation Database (14), which contains 21,587 somatic mutations (21,073 excluding complex mutations, TBM, and insertions and deletions larger than 50 bp), was initially examined as a starting point for the identification of microindels in the mutation literature. Eighteen TP53 microindels were identified directly from the IARC database; additional microindels were identified by analyzing the original publications for mutations labeled ‘complex’ and for cases with multiple mutations.
For comparisons to microindels in the human germline, ‘HGMD’ (as used herein) refers to the 155 microindels (as defined herein) extracted by Chuzhanova et al. (8) from the Human Gene Mutation Database (HGMD) (17). Among the 211 mutations examined in that meta-analysis, 56 are excluded from the analyses herein since they are TBMs, not microindels, as defined herein.
Analysis of sequence context
Genomic sequence was extracted from GenBank accession no. X54156.1 containing the TP53 gene. For the inserted sequence in pure microinsertions or microindels, the IARC database records the size of the inserted sequence but not the actual base sequence inserted. Therefore, the inserted sequence in the microindels was extracted from the primary publications (reference PubMed ID provided in Supplementary Material, Table S5). In addition, since the IARC database does not generally show the sequence of insertions, the inserted sequences of 183 pure microinsertions were identified by reviewing the literature (data not shown). This sample of pure microinsertions was used to estimate the fraction of pure microinsertions that repeat adjacent sequence and for comparison with the inserted sequences in microindels.
Statistics
The patterns of mutation counts and the distributions of insertion or deletion sizes (counts in size distribution bins) were tested for significant differences between two groups by analysis as unordered R × C contingency tables using the ‘Fisher-Freeman-Halton’ test implemented by StatXact (CYTEL Software Corporation, Cambridge, MA). When the reference set is too large for practical computation of the exact P-value, StatXact estimates the P-value and the corresponding 99% confidence interval by a Monte Carlo sampling of the tables in the reference set, and the P-value shown herein is the most conservative bound of the 99% confidence interval.
Insertion and deletion size distributions were compared by two methods. In the first method, the distributions are compared without binning using the Kolmogorov–Smirnov test implemented by StatXact (CYTEL Software Corporation, Cambridge, MA). In the second method, the sizes are binned as shown in Supplementary Material, Figure S7 and the bin counts are compared using the Fisher–Freeman–Halton test as described earlier.
A Monte Carlo simulation was used to test the significance of the observed recurrent sites of microindels. Target sites are drawn randomly from a 1003 nt uniform likelihood target representing the coding sequence of exons 3 through 9 of TP53 (NM_000546.3) plus the splice site regions (six nucleotides on each side of each exon). The P-value is calculated by counting the number of iterations of 66 target sites in which the number of recurrent sites is the same or higher than observed and dividing by the total number of iterations (100 000 000). To assess the reliability of the P-value estimate, the 99% confidence interval is calculated. The most conservative bound of the 99% confidence interval is shown.
A random sequence simulation was used to quantify the likelihood that the putative insertion templates indicated in Figure 2 would occur at the observed distance or closer by chance. Random sequences are generated by randomly selecting (with replacement) nucleotides from the genomic sequence (defined in Fig. 2 legend). During each iteration of the simulation, the random sequence is extended downstream and upstream simultaneously until a match to the observed putative template or its reverse complement is found. If the putative template contains mismatches compared with the inserted sequence (i.e. the inserted sequence is said to result from an error-prone duplication), then the same number of mismatches are allowed when searching for a match. The mean distance to a match is calculated over all iterations. The P-value is calculated by dividing the number of iterations having a match as close as or closer than the putative template by the total number of iterations (1 000 000). To assess the reliability of the P-value estimate, the 99% confidence interval is calculated. The most conservative bound of the 99% confidence interval is shown.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
FUNDING
This work was partially supported by NIH R01AG19784.
TERMINOLOGY
Astrogenetics: Study of in vivo mutagenesis by quantitative analysis of mutation frequency, pattern and spectrum, analogous to the study of the universe by astronomers by quantitative analysis of electromagnetic frequency, pattern and spectrum.
Indel: A mutation resulting in a co-localized insertion and deletion and a net gain or loss of nucleotides.
Microindel: An indel which has a deletion and/or insertion size of 1–50 nucleotides and results in a net gain or loss of 1–50 nucleotides.
Tandem-base mutation (TBM): Substitution at two or more adjacent nucleotides (not an indel as defined herein since it results in no net gain or loss of nucleotides). Cases of substitutions separated by one or more unchanged bases (doublet substitutions) are not defined as TBMs.
Protein tinkering: Lengthening or shortening of a protein by a few amino acids resulting from an in-frame microinsertion, microdeletion or microindel.
Recurroid: A microindel with an identical deletion and a similar but not identical insertion (deletion identical recurroid), or conversely, a microindel with an identical insertion and a similar but not identical deletion (insertion identical recurroid).
ACKNOWLEDGEMENTS
We thank Lin-Ling Chen for helping to find microindels in the literature.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kondrashov A.S., Rogozin I.B. Context of deletions and insertions in human coding sequences. Hum. Mutat. 2004;23:177–185. doi: 10.1002/humu.10312. [DOI] [PubMed] [Google Scholar]
- 2.Ogurtsov A.Y., Sunyaev S., Kondrashov A.S. Indel-based evolutionary distance and mouse-human divergence. Genome Res. 2004;14:1610–1616. doi: 10.1101/gr.2450504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregory T.R. Insertion-deletion biases and the evolution of genome size. Gene. 2004;324:15–34. doi: 10.1016/j.gene.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths A.J.F., Gelbart W.M., Lewontin R.C., Miller J.H. Modern Genetic Analysis. New York: W.H. Freeman & Company; 2002. p. 736. [Google Scholar]
- 5.Ball E.V., Stenson P.D., Abeysinghe S.S., Krawczak M., Cooper D.N., Chuzhanova N.A. Microdeletions and microinsertions causing human genetic disease: common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum. Mutat. 2005;26:205–213. doi: 10.1002/humu.20212. [DOI] [PubMed] [Google Scholar]
- 6.Mank R., Wilson M.D., Rubio J.M., Post R.J. A molecular marker for the identification of Simulium squamosum (Diptera: Simuliidae) Ann. Trop. Med. Parasitol. 2004;98:197–208. doi: 10.1179/000349804225003118. [DOI] [PubMed] [Google Scholar]
- 7.Halangoda A., Still J.G., Hill K.A., Sommer S.S. Spontaneous microdeletions and microinsertions in a transgenic mouse mutation detection system: analysis of age, tissue, and sequence specificity. Environ. Mol. Mutagen. 2001;37:311–323. doi: 10.1002/em.1038. [DOI] [PubMed] [Google Scholar]
- 8.Chuzhanova N.A., Anassis E.J., Ball E.V., Krawczak M., Cooper D.N. Meta-analysis of indels causing human genetic disease: mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum. Mutat. 2003;21:28–44. doi: 10.1002/humu.10146. [DOI] [PubMed] [Google Scholar]
- 9.Hill K.A., Wang J., Farwell K.D., Sommer S.S. Spontaneous tandem-base mutations (TBM) show dramatic tissue, age, pattern and spectrum specificity. Mutat. Res. 2003;534:173–186. doi: 10.1016/s1383-5718(02)00277-2. [DOI] [PubMed] [Google Scholar]
- 10.Buettner V.L., Hill K.A., Halangoda A., Sommer S.S. Tandem-base mutations occur in mouse liver and adipose tissue preferentially as G:C to T:A transversions and accumulate with age. Environ. Mol. Mutagen. 1999;33:320–324. doi: 10.1002/(sici)1098-2280(1999)33:4<320::aid-em9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Eng C., Desnick R.J., German J., Ellis N.A. Carrier frequency of the Bloom syndrome blmAsh mutation in the Ashkenazi Jewish population. Mol. Genet. Metab. 1998;64:286–290. doi: 10.1006/mgme.1998.2733. [DOI] [PubMed] [Google Scholar]
- 12.Hill K.A., Gonzalez K.D., Scaringe W.A., Wang J.C., Sommer S.S. Preferential occurrence of 1-2 microindels. Hum. Mutat. 2006;27:55–61. doi: 10.1002/humu.20260. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez K.D., Hill K.A., Li K., Li W., Scaringe W.A., Wang J.C., Gu D., Sommer S.S. Somatic microindels: analysis in mouse soma and comparison with the human germline. Hum. Mutat. 2007;28:69–80. doi: 10.1002/humu.20416. [DOI] [PubMed] [Google Scholar]
- 14.Olivier M., Eeles R., Hollstein M., Khan M.A., Harris C.C., Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 15.Forbes S., Clements J., Dawson E., Bamford S., Webb T., Dogan A., Flanagan A., Teague J., Wooster R., Futreal P.A., et al. COSMIC 2005. Br.J Cancer. 2006;94:318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu D., Scaringe W.A., Li K., Saldivar J.S., Hill K.A., Chen Z., Gonzalez K.D., Sommer S.S. Database of somatic mutations in EGFR with analyses revealing indel hotspots but no smoking-associated signature. Hum. Mutat. 2007;28:760–770. doi: 10.1002/humu.20512. [DOI] [PubMed] [Google Scholar]
- 17.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z., Feng J., Saldivar J.S., Gu D., Bockholt A., Sommer S.S. EGFR somatic doublets in lung cancer are frequent and generally arise from a pair of driver mutations uncommonly seen as singlet mutations: one-third of doublets occur at five pairs of amino acids. Oncogene. 2008 doi: 10.1038/onc.2008.71. [DOI] [PubMed] [Google Scholar]
- 19.Sommer S.S., Scaringe W.A., Hill K.A. Human germline mutation in the factor IX gene. Mutat. Res. 2001;487:1–17. doi: 10.1016/s0921-8777(01)00108-2. [DOI] [PubMed] [Google Scholar]
- 20.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 21.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 22.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brash D.E., Rudolph J.A., Simon J.A., Lin A., McKenna G.J., Baden H.P., Halperin A.J., Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeMarini D.M., Shelton M.L., Kohan M.J., Hudgens E.E., Kleindienst T.E., Ball L.M., Walsh D., de Boer J.G., Lewis-Bevan L., Rabinowitz J.R., et al. Mutagenicity in lung of big Blue((R)) mice and induction of tandem-base substitutions in Salmonella by the air pollutant peroxyacetyl nitrate (PAN): predicted formation of intrastrand cross-links. Mutat. Res. 2000;457:41–55. doi: 10.1016/s0027-5107(00)00121-4. [DOI] [PubMed] [Google Scholar]
- 25.Nath R.G., Ocando J.E., Chung F.L. Detection of 1, N2-propanodeoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 1996;56:452–456. [PubMed] [Google Scholar]
- 26.Cooper D.N., Krawczak M. Human Gene Mutation. Oxford, England: Bios Scientific Publishers; 1993. [Google Scholar]
- 27.Friedberg E.C. Why do cells have multiple error-prone DNA polymerases? Environ. Mol. Mutagen. 2001;38:105–110. doi: 10.1002/em.1059. [DOI] [PubMed] [Google Scholar]
- 28.Friedberg E.C., Fischhaber P.L., Kisker C. Error-prone DNA polymerases: novel structures and the benefits of infidelity. Cell. 2001;107:9–12. doi: 10.1016/s0092-8674(01)00509-8. [DOI] [PubMed] [Google Scholar]
- 29.Rattray A.J., Strathern J.N. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- 30.Goodman M.F. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 31.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda T., Bebenek K., Masutani C., Rogozin I.B., Hanaoka F., Kunkel T.A. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 33.Goodman M.F., Tippin B. The expanding polymerase universe. Nat. Rev. Mol. Cell Biol. 2000;1:101–109. doi: 10.1038/35040051. [DOI] [PubMed] [Google Scholar]
- 34.Tissier A., McDonald J.P., Frank E.G., Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 35.Dumstorf C.A., Clark A.B., Lin Q., Kissling G.E., Yuan T., Kucherlapati R., McGregor W.G., Kunkel T.A. Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl Acad. Sci. USA. 2006;103:18083–18088. doi: 10.1073/pnas.0605247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCulloch S.D., Kunkel T.A. Multiple solutions to inefficient lesion bypass by T7 DNA polymerase. DNA Repair (Amst) 2006;5:1373–1383. doi: 10.1016/j.dnarep.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J.M., Chuzhanova N., Stenson P.D., Ferec C., Cooper D.N. Complex gene rearrangements caused by serial replication slippage. Hum. Mutat. 2005;26:125–134. doi: 10.1002/humu.20202. [DOI] [PubMed] [Google Scholar]
- 38.Chen J.M., Chuzhanova N., Stenson P.D., Ferec C., Cooper D.N. Intrachromosomal serial replication slippage in trans gives rise to diverse genomic rearrangements involving inversions. Hum. Mutat. 2005;26:362–373. doi: 10.1002/humu.20230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.