Abstract
Juvenile neuronal ceroid lipofuscinoses (JNCL), commonly known as Batten disease, is a progressive neurodegenerative disorder of childhood characterized by blindness, seizures, motor and cognitive decline, leading to death in early adulthood. Mutations within the CLN3 gene, which encodes a putative lysosomal protein of unknown function, are the underlying cause of JNCL. Over 85% of JNCL patients harbor a 1 kb deletion that is predicted to result in a truncated CLN3 protein and is presumed to be a null mutation. A recent study by Kitzmuller et al. ( 1) suggested that the 1 kb deletion-associated truncated protein may have partial function, and proposed that JNCL is a mutation-specific disease. In addition, the validity of the original and most widely utilized JNCL mouse model, the Cln3Δex1-6 mouse, as a true null mutant was questioned. We report a substantial decrease in the transcript level of the truncated CLN3 gene product in cells from 1 kb deletion patients. We contend that the truncated CLN3 protein is unlikely to be expressed in JNCL patients since cellular quality control mechanisms at the RNA and protein levels are likely to degrade the mutant transcript and polypeptides. Moreover, we present analysis identifying the expressed transcripts present in Cln3Δex1-6 mouse brain. From the analysis of expressed Cln3Δex1-6 mouse transcripts, combined with in silico prediction of the expected consequences of the Cln3Δex1-6 mutation on these transcripts, we argue that aberrant Cln3 proteins are unlikely to be expressed in this disease model. Taken together our results indicate that the most common mutation associated with JNCL results in a loss of functional CLN3, that the Cln3Δex1-6 mouse harbors a null Cln3 allele, and that it therefore represents a valid model for this disease.
INTRODUCTION
Juvenile neuronal ceroid lipofuscinosis (JNCL), or Batten disease, is a recessively inherited lysosomal storage disorder characterized by progressive neurodegeneration. Typical symptoms include blindness, seizures, motor and cognitive decline, culminating in premature death in early adulthood. The underlying genetic defect in JNCL has been identified as arising from mutations in the CLN3 gene, which encodes a putative lysosomal protein of unknown function. Thus far, over 40 different mutations have been identified (http://www.ucl.ac.uk/ncl/mutation.shtml), the most common of which is a 1 kb deletion that eliminates exons 7 and 8 and represents over 85% of reported cases, with the remainder comprising deletions, insertions, missense and splicing mutations (2–4). There is extensive heterogeneity in the clinical phenotype even in patients carrying the same CLN3 mutations (5). However, some evidence exists to support genotype–phenotype correlations since certain mutations are associated with a late-onset or protracted disease course, such as the E295K missense mutation (3,6). Nevertheless, the rarity of this disorder and lack of sufficient numbers of patients harboring mutations other than the common deletion has hindered substantial genotype–phenotype studies. The clinical variability and a lack of antisera that is able to detect the native endogenous CLN3 protein makes it somewhat unclear whether there are robust correlations between the type of mutation and the clinical progression of disease. Recent work by Kitzmuller et al. (1) has suggested that some JNCL patients and mouse models may express a mutant transcript that encodes an aberrant CLN3 protein retaining biological function.
To further our understanding of the function of CLN3 and to gain an insight into the disease process, several laboratories have developed murine models of disease. The original, and most widely utilized model, known as the Cln3Δex1-6 mouse was created by Mitchison and colleagues and was intended to provide a complete null mutation of Cln3 by replacing the start codon and first six exons with a neo cassette (7,8). The Cln3Δex7/8 Katz mouse was created at around the same time using a different strategy that replaced the majority of exon 7 and all of exon 8 with a neo cassette in an attempt to create a genetically similar mutation to the 1 kb deletion mutation found in the majority of human patients (9). Cotman et al. (10) utilized a ‘knock-in’ approach to create the Cln3Δex7/8 Cotman mouse in which exons 7 and 8 and the neo cassette were removed in their entirety, resulting in a more genetically accurate model of the 1 kb deletion. Finally, Eliason et al. (11) recently generated a fourth mouse model by replacing exons 1–8 of Cln3 with the LacZ reporter gene in order to create a reporter mouse expressing β-galactosidase under the native Cln3 promoter (Cln3LacZ/LacZ).
Characterization of these murine models has shown that each exhibits classical hallmarks of JNCL, including the accumulation of autofluorescent storage material, neurodegenerative changes and loss of specific neuronal populations in accordance with the human disease process (7,9–11). The onset and severity of these pathological changes appear to vary somewhat among the mouse models but direct comparisons are confounded by differences in strain backgrounds of these mice. This is illustrated in the report of the initial characterization of the Cln3Δex7/8 Cotman mouse in which the authors noted that disease phenotype exhibited variability of onset, which was attributed to the mixed CD1 albino strain background of these mice (10). Strain-specific effects have been reported in another NCL mouse model, the motor neuron degeneration (mnd) model of CLN8, which has an earlier onset when placed on an AKR background rather than the original C57BL/6 background in which the mutation was first identified (12). Thus, there is controversy surrounding how comparable the different Cln3 mutant mice are, either the null models (Cln3Δex1-6 and Cln3LacZ/LacZ), or the mice designed to mimic the human 1 kb deletion mutation (Cln3Δex7/8 Cotman or Cln3Δex7/8 Katz).
In this study, we examined the expression of CLN3 mRNA transcripts in JNCL lymphoblasts and two mouse models of JNCL using quantitative reverse transcriptase-polymerase chain reaction (qRT–PCR) and demonstrate a significant decrease in mutant CLN3/Cln3 message levels. In addition, we performed in silico transcript analysis in the Cln3Δex1-6 mouse model to ascertain whether mutant Cln3 transcripts are present that could encode aberrant CLN3 proteins. In the Cln3Δex1-6 mouse brain, we demonstrate that although residual transcripts comprising the 3′ portion of Cln3 are detectable, the expression of these transcripts is decreased approximately two-fold in comparison with wild-type. The expression of partial mouse CLN3 protein is unlikely, due to a lack of appropriate translation initiation signals. Therefore, we propose that the expression of partial CLN3 protein products in the Cln3Δex1-6 mouse is improbable and that it is doubtful whether these truncated transcripts could exert an effect on the mouse phenotype. Thus, the transcript analysis of mRNA in patients indicates decreased mRNA that is likely targeted for degradation by the nonsense-mediated decay pathway, and the Cln3Δex1-6 mouse is suggestive of a similar null mutation. Taken together, this evidence indicates that 1 kb deletion patients lack a CLN3 protein and that the Cln3Δex1-6 mouse does mimic the human disease, providing a valid model of juvenile Batten disease.
MATERIALS AND METHODS
Experimental animals
Homozygous Cln3Δex1-6 mice were maintained on a 129/SvEv background and Cln3Δex7/8 Cotman mice were maintained on a C57BL/6J background. Age- and sex-matched mice were used in this study along with appropriate wild-type control mice for each strain. All procedures were carried out according to the Guidelines of the Animal Welfare Act, NIH policies and the University of Rochester Animal Care and Use Committee.
Lymphoblast cell culture
Lymphoblast cell lines derived from JNCL patients, designated DT8, DT25 and DT27, were prepared and maintained as described previously (13). Age- and sex-matched control cell lines 7535 and 15792 were obtained from Coriell Cell Repositories (Camden, NJ, USA).
Total RNA isolation
Total RNA was isolated from P14 mouse cerebellum tissue (n = 4 for each genotype) and human lymphoblast cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. To reduce genomic DNA contamination, the isolated RNA was incubated with 3 U of Turbo DNase (Ambion, Austin, TX, USA) for 1 h at 37°C. The Turbo DNase was inactivated by brief incubation with DNase inactivation reagent (Ambion) and the resulting total RNA was stored at −80°C until use.
Quantitative reverse transcriptase-polymerase chain reaction
For comparative gene expression studies, 1 µg of total RNA was reverse-transcribed using the High-Capacity cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's protocol. Quantitative PCR was performed using exon-specific primers as listed in Table 1 and normalized to actin expression. Amplification was carried out using Power SYBR Green supermix (Applied Biosystems) containing appropriate primer pairs and 2 µl of cDNA in a 96-well plate. Thermal cycling and fluorescence data collection were performed on a Mx3005p real-time PCR instrument (Stratagene, La Jolla, CA, USA) using the following cycling parameters: 95°C for 10 min, followed by 40 cycles of 95°C for 20 s, 60°C for 1 min. Specificity of the amplified product was determined by melt–curve analysis immediately following completion of the final amplification cycle. A total of three independent experiments were performed for each comparative expression study. Relative gene expression and statistical analysis was calculated using the REST-XL program (14) and expressed as fold-change versus wild-type.
Table 1.
Primer sequences used for qualitative reverse transcriptase-polymerase chain reaction
| Primer | Sequence 5′ to 3′ |
|---|---|
| hCln3ex3F | TAG CCA CAA GAG GAC ATC GGG AAA |
| hCln3ex4/5R | ACA GCA GCC GTA GAG ACA GAG TT |
| h18sF | TCA ACT TTC GAT GGT AGT CGC CGT |
| h18sR | TCC TTG GAT GTG GTA GCC GTT TCT |
| mCln3ex11F | TAT CAA CCA GGG ACT TTT CGA GCT C |
| mCln3ex13R | ACA CAC CAG CCT GGT ATA GCA TCT |
| mActB F | CTG TCG AGT CGC GTC CAC CC |
| mActB R | CGT CAT CCA TGG CGA ACT GG |
Reverse-transcriptase polymerase chain reaction
For transcript analysis, 3 µg of total RNA was reverse-transcribed using the AffinityScript cDNA synthesis kit and oligo dT primers (Stratagene) according to the manufacturer's protocol. Amplification was performed using EconoTaq Plus Mastermix (Lucigen, Middleton, WI, USA) or HotMasterMix (5′ Prime, Gaithersburg, MD, USA) with the primer sets listed in Table 2 under standard cycling conditions (95°C for 3 min, followed by 35 cycles of 95°C for 20 s, 60°C for 20 s, 72°C for 2 min). PCR products were separated by electrophoresis on 1% or 2% agarose gels, imaged and analyzed using Visionworks LS software (UVP, Upland, CA, USA).
Table 2.
Primer sequences used for reverse transcriptase-polymerase chain reaction
| Primer | Sequence 5′ to 3′ | Product size | |
|---|---|---|---|
| Ex6F | TCA GTG GAG TTT GTT CTG CTG GGA | 207 bp | |
| Ex8R2 | TAA GAC AGC GAT CCA AGA AGC CCT | ||
| Ex7F | TTC CTC TCA CTG ACT GCC TT | 831 bp | |
| Ex15R | TGT GTC GAG CAA CTC CTG | ||
| In6F | CAC TGT TTG CTG TTT GTC CTC CCA | 406 bp | |
| Ex10/11R | AGA CCC TTG AAC ACT GTC CAC CTT | ||
| neoF5 | ATG CTC CAG ACT GCC TTG GGA AA | 168 bp | |
| Ex8R1 | AGA CCA CCA TGA GAT CAC AGC ACT |
Cloning and sequencing
DNA fragments generated by PCR using the neoF5 and Ex8R1 primers were cloned into pCR®2.1-TOPO® vector using the TOPO-TA cloning system (Invitrogen). Sequencing of the cloned insert was performed using the Big Dye Terminator 3.1 kit on an ABI 3730 DNA sequencer (Applied Biosystems).
In silico analysis
Genbank sequences for mouse Cln3 (NM_009907 and NC_000073.5) and for human CLN3 (NM_000086 and NC_000016.8) were used for computer analysis. The genomic sequence of the Cln3Δex1-6 mouse was assembled using Vector NTI software (Invitrogen) from the available sequences of the targeting vector and NC_000073.5 (genomic Cln3). Sequence manipulations, translational initiation codon analysis, in silico translations and sequence alignments were performed using Vector NTI.
RESULTS
Juvenile neuronal ceroid lipofuscinosis arises from mutations in the CLN3 gene that encodes a putative lysosomal protein of unknown function. Although more than 40 distinct mutations have been described, the vast majority (over 85%) of patients are homozygous for the 1 kb deletion of exons 7 and 8 (2,3). In addition, a number of compound heterozygotes harboring the 1 kb deletion in combination with a second mutation have also been reported. The 1 kb deletion causes a frameshift and premature termination, resulting in a truncated protein comprising the first 153 amino acids of CLN3 and a novel 28 residue C-terminal tail (15). Thus, the fact that the most common mutation predicts that a truncated protein could be made has led some researchers to question the fate of this putative polypeptide. In an effort to deduce whether a truncated protein could even exist or impart an impact on the pathogenicity of JNCL, we have looked at both 1 kb deletion patient cell lines and two Cln3 mouse models to establish the possible fate of any transcript resulting from this mutation.
Transcripts resulting from the 1 kb deletion are less abundant
Whether mutant CLN3 proteins have a significant biological activity may depend upon their abundance in the cell. The abundance of any putative mutant CLN3 proteins would likely be regulated at the most basic level by the relative abundance of mRNA transcripts in the cell. Therefore, we used qRT–PCR to investigate the expression level of CLN3 transcripts in lymphoblasts from JNCL patients homozygous for the 1 kb deletion. Using a primer set designed to amplify the 5′ region of Cln3 mRNA upstream of the mutation and therefore present in both wild-type and patient cell lines (hCln3ex3F and hCln3ex4/5R amplifying exons 3–5), we observed a 2.9- to 5.3-fold decrease in the relative abundance of the transcript in comparison with normal control cells (Table 3). No transcripts were detectable using primers against the deleted region in JNCL patient cells (data not shown).
Table 3.
Relative expression of Cln3 in juvenile neuronal ceroid lipofuscinoses (JNCL) lymphoblasts
| JNCL cell line | Mutation | Fold change | P-value |
|---|---|---|---|
| DT8 | 1 kb deletion | 4.7 | 0.04 |
| DT25 | 1 kb deletion | 2.9 | 0.02 |
| DT27 | 1 kb deletion | 5.3 | 0.03 |
Since the Cln3Δex7/8 Cotman model was engineered to mimic the common JNCL 1 kb deletion mutation, we sought to determine whether a similar decrease in transcript abundance could be observed in these mice. Using primers directed to the 3′ region of Cln3 mRNA downstream of the mutation but predicted to be expressed in both wild-type and Cln3Δex7/8 Cotman mice (mCln3ex11F and mCln3ex13R amplifying exons 11–13), we demonstrate a 2.6-fold decrease (P < 0.001) in the expression of transcripts lacking exons 7 and 8, when compared with levels in wild-type mice (Table 4). Similar results were observed using primers to the 5′ region upstream of the deletion and no transcripts were detected using a primer set directed to the deleted region (exons 6 and 7) (data not shown).
Table 4.
Relative expression of Cln3 in mouse models
| Mouse model | Fold change | P-value |
|---|---|---|
| Cln3Δex1-6 | 2.5 | 0.001 |
| Cln3Δex7/8 Cotman | 2.6 | 0.001 |
In silico analysis of Cln3Δex1-6 allele demonstrates a lack of alternative translational initiation sequences
We performed an extensive in silico analysis of the published mouse Cln3 mRNA sequence (accession no. NM_009907) to identify alternate in-frame start codons in the non-deleted portions of the transcript arising from the Cln3Δex1-6 allele mutation. As shown in Figure 1, a total of three in-frame AUG codons are present in the undeleted exons 7–15 region of Cln3, located in exon 8 (residue M216), exon 12 (M324) and exon 15 (M409). If translated, these would result in protein products of 223 amino acid, 115 amino acid or 30 amino acid residues, respectively. We next compared these putative polypeptides with those recently reported by Kitzmuller et al. (1) as retaining partial biological function. In that study, the authors utilized Btn1p, the yeast homolog of CLN3, in the Schizosaccharomyces pombe fission yeast model of JNCL (1,16). The absence of Btn1p in Sz. pombe has previously been shown to result in a vacuolar phenotype that was complemented by human CLN3 protein (16). The Btn1p103–396 construct described by Kitzmuller et al. as retaining significant biological function is equivalent to a transcript containing the last 284 amino acid residues of human CLN3 (CLN3154–438) and this is significantly longer than those in Figure 1 predicted by sequence analysis using in-frame start codons. In addition, expression of this longer protein product would also require that initiation begins at the +3 position of exon 7 in order to preserve the correct reading frame of CLN3. It is therefore doubtful that the purported CLN3154–438 would actually be expressed.
Figure 1.
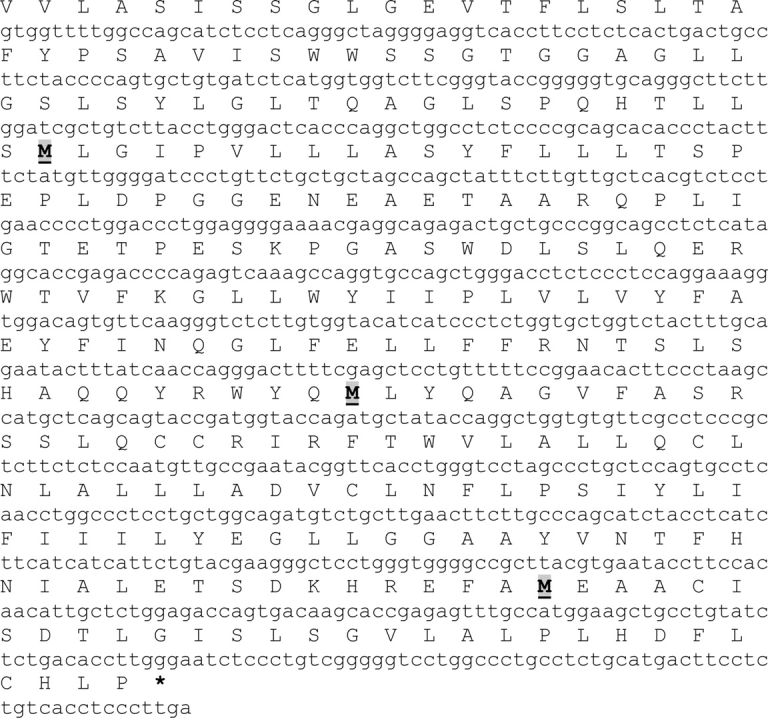
Putative in-frame polypepetides encoded by the non-deleted sequence in the Cln3Δex1-6 mouse (cDNA sequence shown here is base 711–1492 of NM_009907). Alternative start codons (M) are located at M216, M324 and M409. Note the absence of Kozak sequences at the M216 and M324 locations.
The presence of an AUG start codon alone does not constitute an initiation site since other sequence elements are required for efficient translation. The most important of these elements is the Kozak sequence—a short consensus sequence present on eukaryotic mRNA that plays a major role in the initiation of translation. The consensus sequence has the general structure (gcc)gccRccAUGG where R is generally a purine (adenine or guanine) located three bases upstream of the AUG codon, and is then followed by a guanine residue (17). It has been shown that point mutations in the sequence surrounding the AUG codon can have adverse effects on translational efficiency, especially if the −3 purine or +4 guanine residues are substituted (18,19). We therefore analyzed the RNA sequence surrounding the three predicted alternate AUG start sites for the presence of a Kozak consensus sequence (Fig. 1). Our analysis shows that only the third alternate AUG codon (M408) has the pre-requisite Kozak sequence required for efficient translational initiation, and if expressed, would result in a short peptide comprising the final 30 amino acid residues of CLN3. The other two alternate AUG codons, which would encode longer protein products, do not possess either element of the Kozak sequence that is generally required for start codon recognition.
Cln3Δex1-6 knockout mice express partial Cln3 mRNA transcripts
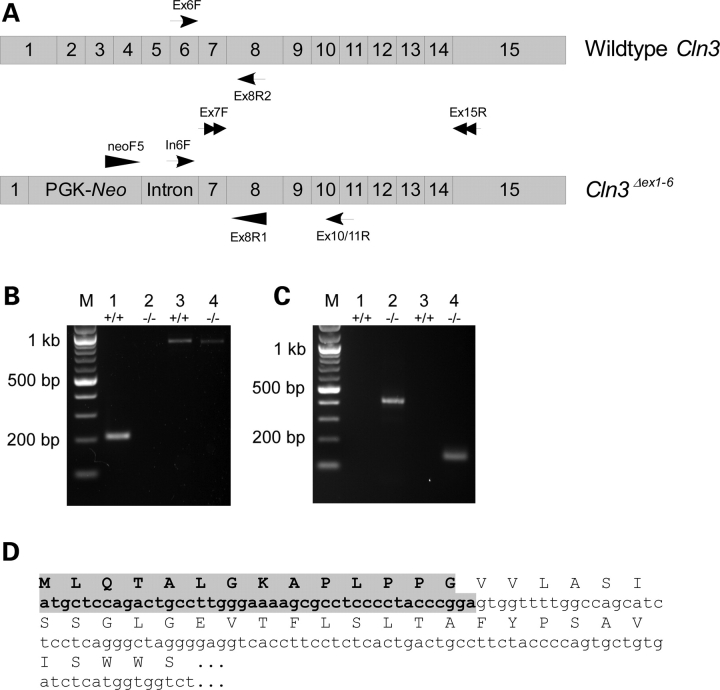
The Cln3Δex1-6 knockout mouse (7,8) is the most widely utilized model animal for JNCL research, containing a null mutation designed to ablate Cln3 function by the replacement of the majority of exon 1 (including the AUG start condon) through exon 6 of the genomic sequence with a Neo cassette in the reverse orientation. Homozygous Cln3Δex1-6 mice were previously shown to express Neo but not Cln3 exons 1 and 2 demonstrating the fidelity of the deletion (7). However, extensive transcript analysis and genomic sequencing have not been conducted in this mouse model. To address the question of whether residual Cln3 transcripts may be expressed, we performed RT–PCR using primers (Ex7F and Ex15R) specific for the non-deleted 3′ region of Cln3 (Fig. 2A). We demonstrate that transcripts containing portions of Cln3 from exon 7 through exon 15 are present in these mice (Fig. 2B), whose expression is presumably driven by the intact endogenous Cln3 promoter region upstream of the reverse Neo cassette. We next used quantitative RT–PCR to determine whether the mutant transcripts were expressed at the same abundance as in wild-type controls. Using primers directed toward the undeleted 3′ region of Cln3 mRNA sequence that is present in both wild-type and Cln3Δex1-6 mice (mCln3ex11F and mCln3ex13R), we show that the expression of the mutant transcript is approximately 2.5-fold lower in comparison with wild-type controls (P < 0.001) (Table 4).
Figure 2.
Reverse-transcriptase-polymerase chain reaction (RT–PCR) analysis of expressed transcripts in the Cln3Δex1-6 mouse. (A) Schematic diagram of predicted transcripts from the endogenous Cln3 promoter in wild type and Cln3Δex1-6 mice. Primer pairs used for RT–PCR analysis are indicated by matching arrows. (B) Primers directed towards exon 6–8 of Cln3 (Ex6F and Ex8R2) detect transcript in wild type (Lane 1) but not in Cln3Δex1-6 mouse cerebellum (Lane 2). The non-deleted region of Cln3 (exons 7–15, primers Ex7F and Ex15R)) is expressed in both wild type (Lane 3) and the Cln3Δex1-6 mouse (Lane 4). No PCR products were observed in Cln3Δex1-6 mouse using an exon 1 primer located upstream of the deleted region and an exon 7 reverse primer (data not shown). (C) Primers against intron 6 or the Neo cassette (In6F or NeoF5), in combination reverse primers (Ex1010/11R and Ex8R1, respectively) in the non-deleted region of Cln3 demonstrate that alternative splicing events in the Cln3Δex1-6 mouse generate transcripts containing intron 6 (Lane 2) or the Neo cassette (Lane 4) fused to exons 7–15. No alternate transcripts were observed in wild type mice (Lanes 1 and 3). (D) Sequence analysis indicates the presence of an in-frame ATG codon derived from the reverse Neo cassette (shaded region) located upstream of Cln3 exon 7–15.
The presence of a partial Cln3 transcript suggests that the endogenous Cln3 promoter is able to drive transcription since the PGK promoter driving the expression of the Neo gene is in the reverse orientation to Cln3 and thus would not account for the presence of this transcript. With this in mind, we next sought to determine whether the transcript existed as a spliced transcript comprising of only exons 7–15 of Cln3, or whether it is expressed as a fused transcript with elements of the reverse Neo cassette. To this end, we used forward primers located in either the Neo cassette (NeoF5) or intron 6 (In6F) of genomic Cln3 in combination with reverse primers (Ex8R1 and Ex10/11R, respectively) located in one of the expressed exons of Cln3 (Fig. 2A). By RT–PCR, we show that two partial Cln3 transcripts appear to be present, one that is preceded at the 5′ end with portions of the Neo cassette, and the other by intron 6 of the genomic sequence (Fig. 2C). We also performed RT–PCR using a forward primer located in the retained portion of exon 1 but did not observe any PCR product indicating that exon 1 is spliced out of the final transcripts (not shown). These results suggest that multiple splicing events may occur due to the replacement of the 5′ region of the Cln3 coding sequence resulting in alternate mRNA transcripts.
The presence of a hybrid Cln3 transcript may be significant if the additional 5′ sequence were to donate an in-frame AUG start codon such that a fusion truncated Cln3 protein product could be expressed. Therefore, we performed an in silico analysis of the transcript comprising intron 6 fused to exons 7–15 of Cln3 using published genomic sequence data. Our analysis did not show the presence of any additional in-frame AUG start codons upstream of exon 7 and we conclude that such a fusion transcript, if it exists, is unlikely to produce functional partial CLN3 protein products. When the same analysis was performed using the predicted sequence of the Neo cassette-Cln3 hybrid, the presence of an alternate in-frame AUG donated by the Neo cassette fragment was detected. We sought to confirm that this putative AUG codon was in the correct reading frame by sequencing the RT–PCR product obtained when using the neoF5 and Ex8R1 primers. The results indicated that an in-frame AUG codon is located 40 bp upstream of the beginning of exon 7 of Cln3. If translated, the protein product would comprise the C-terminal 284 amino acid residues of CLN3 shown in Figure 1, with the addition of 14 novel amino acids at the N-terminus. However, in silico analysis of the RNA sequence surrounding this novel AUG start site indicates the lack of a Kozak consensus sequence and it is therefore unlikely to be translated efficiently (Fig. 2D). Moreover, only severely truncated proteins are predicted to be encoded by this C-terminal 284 amino acid portion of CLN3, as discussed above.
DISCUSSION
Juvenile Neuronal Ceroid Lipofuscinosis or Batten disease, arises from mutations in the CLN3 gene. Over 40 mutations have been identified, including a common 1 kb deletion mutation arising from a Founder effect. Mutations in CLN3 are generally thought to result in a null mutation with loss of CLN3 protein function leading to the disease. However, it has recently been proposed that the 1 kb deletion, the most common mutation in JNCL patients, may give rise to the expression of a truncated CLN3 protein that retains partial function (1). The expression of truncated CLN3 proteins was modeled in the fission yeast Sz. pombe using Btn1p, the yeast homolog of CLN3 (16,20). Previous studies have demonstrated the utility of Sz. pombe as a model for JNCL, as Sz. pombe deleted for Btn1p (Btn1-Δ) demonstrate a vacuolar phenotype similar to that observed in JNCL patient cell lines, and could be rescued by the expression of human CLN3 (16). Kitzmuller et al. observed that Btn1p constructs with mutations homologous to truncated CLN3 encoded by the exons 1–6 (Btnp102fsX5/CLN31–153) and a second minor transcript lacking exons 7–9 (Btn1p102-208del/CLN3154-263del) mimicking that expressed in 1 kb deletion JNCL fibroblasts were able to rescue the vacuolar phenotype exhibited by the btn1-Δ Sz. pombe null allele model (1). In addition, it was shown that Btn1p constructs harboring a mutation equivalent to Btn1p103–396/CLN3154–438, purportedly expressed in the Cln3Δex1-6 mouse, had a similar effect. Parallel experiments in JNCL fibroblasts using the truncated protein (CLN31–153) resulted in the reduction in lysosome size and, therefore, it was concluded that the truncated CLN31–153 retained partial function (1). It is noteworthy that these results suggest that both the N- and C-terminal portions of CLN3, when expressed independently, have a similar function to the full-length protein, yet there is no significant sequence homology between the two fragments. Based on these results, Kitzmuller et al. have proposed that JNCL is a mutation-specific disorder that arises through partial loss of CLN3 function, thus establishing a genotype–phenotype correlation. It is, however, questionable whether a truncated protein, encoded by the 1 kb deletion, is actually expressed in JNCL patients or the Cln3Δex7/8 Cotman model that recapitulates this mutation. The lack of specific antibodies that detect endogenous levels of CLN3, or truncated CLN3, has hampered the efforts to answer this important question and it remains to be seen whether aberrant CLN3 proteins are present in JNCL patients.
Our current study demonstrates that expression of mutant mRNA transcripts is reduced in both 1 kb deletion JNCL lymphoblasts and in Cln3Δex7/8 Cotman cerebellum tissue. Whether such truncated transcripts would actually be translated is not clear. The deletion of exons 7 and 8 is predicted to cause a frameshift after amino acid 153, resulting in the addition of 28 novel amino acids (11 in mouse) followed by a premature stop codon. This mutation also makes the transcript an ideal candidate for the nonsense-mediated mRNA decay (NMD) pathway, an mRNA surveillance mechanism that selectively degrades mRNA sequences containing a premature termination codon [reviewed in (21)]. The presence of a nonsense codon >50–55 bp upstream of an exon–exon junction is sufficient to trigger NMD as would be the case in Cln3 transcripts lacking exons 7 and 8. Therefore, as transcripts would presumably undergo degradation before translation occurs, it is likely that any truncated protein, if present at all, would be at much reduced levels. Previous functional studies suggest it would also be retained in the endoplasmic reticulum (ER) rather than being trafficked correctly to the lysosome (1,6). Furthermore, it is conceivable that the presence of the novel 28 amino acids at the C-terminus of the truncated protein could cause misfolding and subsequent degradation in the ER (22). Thus, it is unlikely that sufficient levels of truncated protein would be present to have a significant biological function capable of influencing disease progression. Several years ago we used an in vitro approach to investigate the possible effect of the 1 kb CLN3 truncation and its novel sequence by thoroughly screening this novel polypeptide for protein interactors using yeast two-hybrid analysis. Compared with the full-length protein, the truncated CLN3 does not interact with any novel proteins, making it unlikely that it could interfere with other protein functions, if it were expressed and not degraded (Weimer and Pearce, unpublished). While we cannot rule out the possibility that alternative splicing may result in additional aberrant CLN3 proteins as has been shown in previous studies (1,9,10), such proteins are likely to be present at levels far below than that of normal CLN3, as those studies showed them to be minor transcripts in comparison with the exons 7/8-deleted mRNA.
Genetically engineered murine models of human disease are an invaluable tool for understanding the consequences of protein loss of function and the development of effective disease treatments. The field of JNCL research benefits from the availability of not less than four Cln3 mouse models of the human disease, each with differing attributes, but all designed to provide a glimpse into the mechanisms of this devastating disease. The suitability of one of these models, namely the Cln3Δex1-6 mouse, as a loss of function model for JNCL was recently called into question in a study by Kitzmuller et al. (1). Based upon studies conducted in Sz. pombe, the authors proposed that the Cln3Δex1-6 mouse expresses a mutant Cln3 transcript that encodes a partially functional CLN3 protein, yet offered no experimental evidence to support the existence of such transcripts or protein products. Furthermore, in arriving at their conclusions, the authors of this study appear to ignore several fundamental aspects governing translational regulation. First, in order for the purported mutant Cln3Δex1-6 allele (Kitzmuller Btn1p103–396/CLN3154–438 construct) to be expressed, translational initiation would have to begin at the +3 base of exon 7, yet the authors offer no explanation regarding how this may be achieved. Secondly, the lack of an AUG start codon at the 5′ end of exon 7 would preclude the translation of the mutant CLN3154–438 protein. It is important to note that the Btn1p expression constructs used in the study were fusion constructs tagged with green fluorescence protein at the N-terminus, and the authors themselves cautioned that such constructs may interfere with the expressed proteins.
Since the Cln3Δex1-6 mouse has been used extensively in JNCL research, the ramifications of any residual function could impact upon the interpretation of the results obtained with this model. It is therefore important to ascertain whether mutant protein(s) is expressed in this mouse. In our current study, we demonstrate the existence of two alternate splice variants of a partial Cln3 mRNA sequence encoding exons 7–15 with a novel leader sequence consisting of either intron 6 or an exogenous sequence derived from the PGK-neo cassette used to generate the null allele (Fig. 2). The presence of a partial Cln3 transcript could lead to the possibility of an aberrant CLN3 protein. However, this is unlikely based upon sequence analysis of the expressed transcripts. Translational initiation depends upon the presence of a suitable AUG start codon in addition to the specific Kozak consensus sequences (17–19). The scanning model for translational initiation dictates that the AUG codon is preceded by a purine (A/G) in the −3 position, and followed by a G in the +4 position (17). The presence of these two elements has been shown to greatly enhance the efficiency of translation and is regarded to be a pre-requisite for translational initiation (18,19). Our analysis indicates that of the four possible in-frame AUG codons, only the AUG (M409) closest to the 3′ end has a Kozak consensus sequence required for translational initiation, resulting in a protein of only 30 amino acids in length. Recently, it has been shown that the +4 G may not be important for translation initiation (23,24) although the −3 A/G is still required for initiation, precluding the translation of longer CLN3 proteins, as none of the upstream AUG codons (M−14, M216 and M324) fulfil this requirement. Thus, despite the presence of partial Cln3 transcripts, it is unlikely that they are translated based upon the current knowledge of translational mechanisms in eukaryotes. We therefore believe that the Cln3Δex1-6 mouse does indeed represent a true loss-of-function model, and that the milder disease phenotype often associated with mouse models likely results from species differences between man and mouse.
In summary, we contend that the mutation hypothesis presented in the recent study by Kitzmuller et al. (1) must be viewed with caution because of the assumptions that were made in arriving at their conclusions. Importantly, when considering whether a mutant protein is present in a cell, the key principles of gene transcription and translation need to be fully considered.
FUNDING
Supported in part by NIH R01 NS044310 and the Beat Batten Foundation.
ACKNOWLEDGEMENTS
Thanks to Dr Lynne Maquat for useful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kitzmuller C., Haines R.L., Codlin S., Cutler D.F., Mole S.E. A function retained by the common mutant CLN3 protein is responsible for the late onset of juvenile neuronal ceroid lipofuscinosis. Hum. Mol. Genet. 2008;17:303–312. doi: 10.1093/hmg/ddm306. [DOI] [PubMed] [Google Scholar]
- 2.Mole S.E. The genetic spectrum of human neuronal ceroid-lipofuscinoses. Brain Pathol. 2004;14:70–76. doi: 10.1111/j.1750-3639.2004.tb00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mole S.E., Williams R.E., Goebel H.H. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics. 2005;6:107–126. doi: 10.1007/s10048-005-0218-3. [DOI] [PubMed] [Google Scholar]
- 4.Siintola E., Lehesjoki A.E., Mole S.E. Molecular genetics of the NCLs—status and perspectives. Biochim. Biophys. Acta. 2006;1762:857–864. doi: 10.1016/j.bbadis.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Munroe P.B., Mitchison H.M., O'Rawe A.M., Anderson J.W., Boustany R.M., Lerner T.J., Taschner P.E., de Vos N., Breuning M.H., Gardiner R.M., et al. Spectrum of mutations in the Batten disease gene, CLN3. Am. J. Hum. Genet. 1997;61:310–316. doi: 10.1086/514846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarvela I., Lehtovirta M., Tikkanen R., Kyttala A., Jalanko A. Defective intracellular transport of CLN3 is the molecular basis of Batten disease (JNCL) Hum. Mol. Genet. 1999;8:1091–1098. doi: 10.1093/hmg/8.6.1091. [DOI] [PubMed] [Google Scholar]
- 7.Mitchison H.M., Bernard D.J., Greene N.D., Cooper J.D., Junaid M.A., Pullarkat R.K., de Vos N., Breuning M.H., Owens J.W., Mobley W.C., et al. Targeted disruption of the Cln3 gene provides a mouse model for Batten disease. The Batten Mouse Model Consortium (corrected) Neurobiol. Dis. 1999;6:321–334. doi: 10.1006/nbdi.1999.0267. [DOI] [PubMed] [Google Scholar]
- 8.Greene N.D., Bernard D.L., Taschner P.E., Lake B.D., de Vos N., Breuning M.H., Gardiner R.M., Mole S.E., Nussbaum R.L., Mitchison H.M. A murine model for juvenile NCL: gene targeting of mouse Cln3. Mol. Genet. Metab. 1999;66:309–313. doi: 10.1006/mgme.1999.2828. [DOI] [PubMed] [Google Scholar]
- 9.Katz M.L., Shibuya H., Liu P.C., Kaur S., Gao C.L., Johnson G.S. A mouse gene knockout model for juvenile ceroid-lipofuscinosis (Batten disease) J. Neurosci. Res. 1999;57:551–556. [PubMed] [Google Scholar]
- 10.Cotman S.L., Vrbanac V., Lebel L.A., Lee R.L., Johnson K.A., Donahue L.R., Teed A.M., Antonellis K., Bronson R.T., Lerner T.J., et al. Cln3(Deltaex7/8) knock-in mice with the common JNCL mutation exhibit progressive neurologic disease that begins before birth. Hum. Mol. Genet. 2002;11:2709–2721. doi: 10.1093/hmg/11.22.2709. [DOI] [PubMed] [Google Scholar]
- 11.Eliason S.L., Stein C.S., Mao Q., Tecedor L., Ding S.L., Gaines D.M., Davidson B.L. A knock-in reporter model of Batten disease. J. Neurosci. 2007;27:9826–9834. doi: 10.1523/JNEUROSCI.1710-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messer A., Manley K., Plummer J.A. An early-onset congenic strain of the motor neuron degeneration (MND) mouse. Mol. Genet. Metab. 1999;66:393–397. doi: 10.1006/mgme.1999.2817. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez-Montealegre D., Pearce D.A. Defective lysosomal arginine transport in juvenile Batten disease. Hum. Mol. Genet. 2005;14:3759–3773. doi: 10.1093/hmg/ddi406. [DOI] [PubMed] [Google Scholar]
- 14.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isolation of a novel gene underlying Batten disease, CLN3. The International Batten Disease Consortium. Cell. 1995;82:949–957. doi: 10.1016/0092-8674(95)90274-0. [DOI] [PubMed] [Google Scholar]
- 16.Gachet Y., Codlin S., Hyams J.S., Mole S.E. btn1, the Schizosaccharomyces pombe homologue of the human Batten disease gene CLN3, regulates vacuole homeostasis. J. Cell Sci. 2005;118:5525–5536. doi: 10.1242/jcs.02656. [DOI] [PubMed] [Google Scholar]
- 17.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984;308:241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 20.Pearce D.A., Ferea T., Nosel S.A., Das B., Sherman F. Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 1999;22:55–58. doi: 10.1038/8861. [DOI] [PubMed] [Google Scholar]
- 21.Isken O., Maquat L.E. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 22.Hebert D.N., Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa S., Niimura Y., Gojobori T., Tanaka H., Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–871. doi: 10.1093/nar/gkm1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia X. The +4G site in Kozak consensus is not related to the efficiency of translation initiation. PLoS ONE. 2007;2:e188. doi: 10.1371/journal.pone.0000188. [DOI] [PMC free article] [PubMed] [Google Scholar]