Abstract
The coancestry coefficient, also known as the population structure parameter, is of great interest in population genetics. It can be thought of as the intraclass correlation of pairs of alleles within populations and it can serve as a measure of genetic distance between populations. For a general class of evolutionary models it determines the distribution of allele frequencies among populations. Under more restrictive models it can be regarded as the probability of identity by descent of any pair alleles at a locus within a random mating population. In this paper we review estimation procedures that use the method of moments or are maximum likelihood under the assumption of normally distributed allele frequencies. We then consider the problem of testing hypotheses about this parameter. In addition to parametric and non-parametric bootstrap tests we present an asymptotically-distributed chi-square test. This test reduces to the contingency-table test for equal sample sizes across populations. Our new test appears to be more powerful than previous tests, especially for loci with multiple alleles. We apply our methods to HapMap SNP data to confirm that the coancestry coefficient for humans is strictly positive.
Keywords: coancestry coefficient, F-statistics, parametric bootstrap, population structure, genetic drift, HapMap data
Introduction
When populations occur as geographic isolates there is general population-genetic interest in quantifying the resulting degree of genetic differentiation. This differentiation may point to the actions of natural selection, for example, (Akey et al., 2002) or it may reflect patterns of migration. In this second case, however, McCauley and Whitlock (1999) point to the need for caution. Other situations arise where the nature of population subdivision is not known but must be accommodated: for example the probability that two people share the same forensic genetic profile depends on allele frequencies in the subpopulation to which they both belong but estimated frequencies may be available only for a larger population. The remedy is to apply an adjustment for population structure (Weir, 2007). These and other applications of measures of population differentiation have been reviewed recently by Holsinger and Weir (2009). In this article we concentrate on estimating a relevant parameter and testing hypotheses about its value.
Definitions and Notation
Our discussion is framed in terms of pairs of alleles for the same gene. Although genetic data for diploids are collected for genotypes we will assume that populations are mating at random so that a sample of n/2 genotypes can be regarded as a sample of n copies of a gene and we treat a copy as a sampling unit. We will use “allele” for the sampling unit, as in “a sample of n alleles“ and also as an alternative form of a gene, as in “a gene with m alleles.” The meaning will be clear in any context. For any two alleles in a population we work with a measure θ. This quantity is most generally defined as the correlation of alleles (Wright, 1931) although, as Holsinger and Weir (2009) emphasize, Wright was concerned with defining and using the parameter rather than with estimation or testing hypotheses. The essential nature of θ in capturing variation among populations is best revealed with the indicator variables of Cockerham (1969). If xj is an indicator variable for the jth allele in a population, defined as 1 if that allele is of type A1 and 0 otherwise, then xj has expectation p1, the allele frequency for A1. Expectation here refers to an average over all alleles in the population and over all populations within some defined framework. For convenience in this discussion our framework will be all those populations with a common ancestral population, although more complex situations may be considered (Weir and Hill, 2002). The variance of xj is p1(1−p1) and the covariance for distinct alleles j, j′ in the same population is θp1(1−p1) to define θ. Evidently θ is the correlation of xj and xj′ and was termed by Wright (1931) the correlation between gametes chosen randomly from within the same subpopulation relative to the entire population. He used the symbol FST. We will generally use the word “population” for an interbreeding group of individuals rather than “subpopulation.”
We wish to draw inferences about θ from a set of sample allele frequencies. Estimation of θ and its analogs have been discussed widely in the literature (Robertson and Hill, 1984; Weir and Cockerham, 1984; Balding, 2003). The frequentist approaches of the method of moments and maximum likelihood as well as Bayesian methods have been used. The frequentist methods are computationally less intensive whereas the Bayesian approaches have the benefit of systematic incorporation of prior information. Robertson and Hill (1984) and Weir and Cockerham (1984) discussed moment estimators. Their methods do not account for linkage disequilibrium between loci in combining the information over loci and the best way of combining information over loci depends on the magnitude of θ (Weir and Cockerham, 1984). Weir and Hill (2002) proposed a maximum likelihood estimator of θ assuming a multivariate normal distribution across populations for allele frequencies. Other authors (e.g. Holsinger, 1999; Balding and Nichols, 1997; Balding, 2003) have worked with a Dirichlet distribution. All authors invoke multinomial sampling within populations. In this paper we will discuss both moment and maximum likelihood estimators of θ and compare their performance using simulation studies.
For testing hypotheses, appeal has been made to bootstrapping (Dodds, 1986) and permutation methods (Roff and Bentzen, 1989; Raymond and Rousset, 1995). Weir and Hill (2002) found a chi-square test statistic by assuming normally-distributed allele frequencies. Li (1996) and Samanta (2006) gave alternative derivations of chi-square statistics. In this paper we propose two testing methods. The first approach is based on a parametric bootstrap method and works better for small sample sizes whereas the second approach is based on the large sample properties of allele frequencies. Both the approaches are defined for any number of allelic forms at a particular locus. We show that these new test procedures are better than existing testing methods in terms of power.
For a set of populations indexed by i we extend the definition of indicator variables to xij for the jth allele in the ith population and restrict ourselves to the case of independent populations. This means that xij is uncorrelated with xi′ j′ when i ≠ i′. A sample of ni alleles from the ith population provides the A1 sample allele frequency . For a collection of samples from r different populations, the allele frequencies can be set out in an analysis of variance format, as shown in Table 1. In that table p̄1 is the weighted average of the sample frequencies for allele A1: . For loci with multiple alleles, indexed by u, another convenient way to set out the data is as a contingency table, as in Table 2. The counts for alleles Au in a sample from the ith population are written as niu and these sum over alleles to give ni = Σu niu and then over samples to give rn̄ = Σi ni.
Table 1.
Analysis of variance format for frequencies of allele A1.
| Source | d.f. | Mean Square | Expected Mean Square | ||
|---|---|---|---|---|---|
| Among Populations | r − 1 |
|
p1(1 − p1)(l − θ) +ncθp1(1 − p1) | ||
| Within Populations |
|
|
p1(l − p1)(l − θ) |
Table 2.
Contingency table format for counts of alleles Au.
| Population | Allele 1 | ··· | Allele u | ··· | Total | |||
|---|---|---|---|---|---|---|---|---|
| 1 | n11 = n1p̃11 | ··· | n1u = n1p̃1u | ··· |
|
|||
| ··· | ··· | ··· | ··· | ··· | ··· | |||
| i | ni1 = nip̃i1 | ··· | niu = nip̃iu | ··· |
|
|||
| ··· | ··· | ··· | ··· | ··· | ··· | |||
|
| ||||||||
| Total |
|
··· |
|
··· |
|
|||
Estimation
A moment estimate of θ was given by Weir and Cockerham (1984) and, assuming normally-distributed allele frequencies, a maximum likelihood estimate was given by Weir and Hill (2002). Bayesian approaches were adopted by Balding and Nichols (1997), Holsinger (1999), and Nicholson et al. (2002). Another approach that regards θ as an overdispersion parameter, and builds on the work of Mosimann (1962) and Neerchal and Morel (2005), has been discussed by T. Tvedebrink (personal communication).
Method of Moments Estimation
The moment estimate of Weir and Cockerham (1984) follows directly from Table 1. Using allele A1 frequencies only, there are two mean squares
and the estimate can be written as θ̂1:
where . Under the assumption that θ is the same for all alleles at a locus, implying that the alleles are selectively neutral and that mutation rates are the same for all alleles, Weir and Cockerham (1984) combined estimates over alleles by summing numerators and denominators separately. If alleles are indexed by u and there are m different alleles:
If data are available from a series of loci l (l = 1, 2,…, L), there are mean squares for each allele at each locus and Weir and Cockerham (1984) combined estimates over loci to get a final moment estimate θ̂M:
The “average” sample sizes ncl are likely to be different at each locus.
It is difficult to derive expressions for the mean and variance of the moment estimate. Dodds (1986) and Weir (1996) suggested numerical resampling for obtaining the sampling distribution of θ̂M. Resampling over populations would change the structure of the data but resampling over loci exploits the assumption that (unlinked) loci provide independent replicates of the evolutionary process. Resampling was also used by Raymond & Rousset (1995). Jiang (1987) used a Taylor series expansion and approximate higher-order moments of sample allele frequencies to obtain the mean and variance of θ̂M. Li (1996) appealed to asymptotic theory to show that the mean square MSA1 has a chi-square distribution in the two-allele case,
and that the mean square MSW1 tends to a constant value of p1(1 − p1)(1 − θ). These results allowed her to derive expressions for the mean and variance of θ̂M:
| (1) |
| (2) |
The variance formula differs slightly from the variance of the intraclass correlation given by Fisher (1921), but is equal to that result for large sample sizes.
Maximum Likelihood Estimation
The maximum likelihood estimate of Weir and Hill (2002) follows from assuming the (m − 1) × 1 vector p̃i = [p̃i1,…, p̃i,m−1]′ for all but one of the alleles at a locus to be multivariate normal. If individuals, and hence genotypes, are sampled randomly from a single population their counts follow a multinomial distribution among samples from the same population. When there is random union of gametes in the population, allele counts are also multinomially distributed over samples from the population. For large samples, the multinomial distribution can be approximated by the multivariate normal distribution, and Weir and Hill (2002) assumed that the normal distribution applies also across populations. If P̃ is the vector of sample allele frequencies:
where
The vectors p̃i and p have (m − 1) components p̃iu and pu, one for each of (m − 1) of the alleles at the locus. If there is no relationship among alleles from different populations the vectors p̃i are independent and the elements of the variance matrix V are
where φi = [θ + (1 − θ)/ni]. All elements in Vii′, i ≠ i′ are zero.
From Corollary 1.7 and Theorem 3.5 of Serfling (1980), the quadratic form
has a chi-square distribution, opening the way for both estimation and hypothesis testing. Substituting the sample allele frequencies in the denominators of this expression gives the statistic T
| (3) |
For estimation, closed-form expressions are possible only if all the φi’s are the same. This will happen if all the sample sizes are equal, ni = n, φi = [θ +(1−θ)/n] or if the sample sizes are large enough that φi = θ for all i. Setting T equal to its expected value, the df, in the equal sample size situation leads to the estimate
For large sample sizes this becomes (Hill and Robertson, 1984)
| (4) |
To the extent that T has a chi-square distribution, θ̂N is an unbiased estimator of θ and it has variance
If data are available from L independent loci then the estimates are simply averaged over loci. If the lth locus has ml alleles, the sum over loci of the quadratic forms has a chi-square distribution with . In this case the estimate of θ still would be unbiased but, as expected, the variance will decrease with the number of alleles and hence with the number of loci:
| (5) |
Hypothesis Testing
Fixed Model
We will restrict attention to the hypotheses that θ is either zero or greater than zero, H0: θ = 0, H1: θ > 0, and we make the distinction (Weir, 1996) between fixed and random population models. The fixed model takes the data as being from a set of populations for which no inferences about evolutionary history are to be drawn. Instead, inferences are drawn on just the sampled set of populations. As no genetic or evolutionary model is necessary a purely statistical approach is appropriate and a contingency table test for independence of allele frequencies and populations is a very direct procedure (Raymond and Rousset, 1995; Roff and Bentzen, 1989). From Table 2, the chi-square test statistic for independence is
| (6) |
and this is assumed to have a chi-square distribution with (r−1)(m−1) df under the null hypothesis. Note that if the sample sizes are equal, ni = n̄ = n, X2 = n (r − 1) (m− 1) θ̂N, suggesting that the power of this test increases linearly with θ. Instead of appealing to the chi-square distribution, the population labels for each observed allele could be permuted and an exact null distribution of X2 obtained (Raymond and Rousset, 1995; Roff and Bentzen, 1989).
Random Model
The maximum likelihood approach in the estimation section invoked a random model in the sense that a distribution was assumed for allele frequencies over populations. There is an implicit evolutionary model that describes the relationships among populations resulting from a shared history. Under the hypothesis that θ is zero, φi = 1/ni and the test statistic becomes
| (7) |
and (Appendix of Samanta, 2006) this is distributed as . Values of T can be added over independent loci and the df are also summed. For equal sample sizes, T in Equation 7 is the same as X2 in Equation 6. Going back to Equation 3, however, would allow hypotheses for values of θ other than θ = 0 to be tested.
Li (1996) based a test on the analysis of variance framework shown in Table 1. For allele A1 she found that (r − 1) MSA1/MSW1 was asymptotically distributed as . Dodds (1986) used the moment estimate θ̂M and bootstrapped over loci to generate an empirical distribution for the estimator. This non-parametric bootstrap leads to a one-sided 100(1 − α)% confidence interval for θ. If the interval does not contain zero then the hypothesis that θ is zero is rejected at the α significance level.
Another test procedure is based on parametric bootstrap resampling, developed specifically for small sample sizes. Under the null hypothesis of zero θ the observed allele frequencies p̄u over the whole data set are maximum likelihood estimates of the parameters pu. Moreover, the allele counts in the ith population sample have a multinomial distribution with parameters ni and {pu}. A bootstrap sample consists of r sets of allele counts {niu}, each generated by multinomial sampling from a distribution with parameters ni and {p̄u}. The parameter of interest can be estimated as θ̂N. Repeated bootstrap samples provide repeated estimates and the hypothesis is rejected with level α if the estimate of θ based on the original data does not belong to the lower 100(1 − α)% of the bootstrap estimates.
Simulation Studies
Estimation
The moment estimator and the maximum likelihood estimator based on normal distribution of the allele frequencies were applied to the case of five populations that have evolved independently from a single population. We simulated data using a pure drift model. The simulation was for a single locus and 10 loci with m = 2, 3 or 4 alleles, all equally frequent initially. We assumed every population in the evolutionary processes had 500 individuals and we sampled 400 alleles for each population to estimate the coancestry coefficient θ. We consider three different current ages of all the populations, 11, 52 and 106 to give predicted θ values of 0.011, 0.051 and 0.101.
The biases and sampling errors of the estimators were calculated using 1, 000 replicates. Table 3 shows that both the moment estimator, θ̂M, and the maximum likelihood estimator based on normal distribution, θ̂N, have a small bias but a large standard error for a single locus. Information from independent loci is expected to decrease the bias and the standard errors of the estimators. From Equation 1 the bias of the moment estimator should be negative, although a full treatment of bias requires knowledge of descent measures γ, δ, and Δ for three, four and two-pairs of alleles as well as of the measure θ for two alleles (Weir and Cockerham, 1984; Jiang, 1987; Samanta, 2006). In some situations, we have found that an increase in the number of loci may cause a small increment in the bias but we believe that one or two outliers in the simulation process cause these discrepancies. On the other hand, the information from different loci always reduces the standard errors of the estimators as expected from Equation 5. The simulation studies show that the biases and standard errors of both estimators decrease as the number m of alleles per locus increases, regardless of the allelic frequencies. The values and performances of the moment and the MLE estimator are very similar.
Table 3.
Comparison of moment estimator and maximum likelihood estimator of θ.
| Single Locus |
10 Loci |
|||||||
|---|---|---|---|---|---|---|---|---|
| Alleles per locus | θ | Method | Average | Bias | SD | Average | Bias | SD |
| 2 | 0.011 | θ̂M | 0.01105 | 0.00005 | 0.00962 | 0.01087 | −0.00013 | 0.00292 |
| θ̂N | 0.01111 | 0.00011 | 0.00971 | 0.01090 | −0.00010 | 0.00294 | ||
| 0.051 | θ̂M | 0.04849 | −0.00251 | 0.03376 | 0.05064 | −0.00036 | 0.01125 | |
| θ̂N | 0.04925 | −0.00175 | 0.03473 | 0.05122 | 0.00022 | 0.01150 | ||
| 0.101 | θ̂M | 0.09590 | −0.00510 | 0.06381 | 0.09992 | −0.00108 | 0.02094 | |
| θ̂N | 0.09875 | −0.00225 | 0.06741 | 0.10205 | 0.00105 | 0.02182 | ||
|
| ||||||||
| 3 | 0.011 | θ̂M | 0.01105 | 0.00005 | 0.00655 | 0.01085 | −0.00015 | 0.00199 |
| θ̂N | 0.01107 | 0.00007 | 0.00655 | 0.01088 | −0.00012 | 0.00199 | ||
| 0.051 | θ̂M | 0.04969 | −0.00131 | 0.02476 | 0.05081 | −0.00019 | 0.00801 | |
| θ̂N | 0.05009 | −0.00091 | 0.02516 | 0.05110 | 0.00010 | 0.00807 | ||
| 0.101 | θ̂M | 0.09872 | −0.00228 | 0.04567 | 0.10090 | −0.00010 | 0.01470 | |
| θ̂N | 0.10009 | −0.00091 | 0.04660 | 0.10199 | 0.00099 | 0.01492 | ||
|
| ||||||||
| 4 | 0.011 | θ̂M | 0.01091 | −0.00009 | 0.00561 | 0.01090 | −0.00010 | 0.00172 |
| θ̂N | 0.01092 | −0.00008 | 0.00558 | 0.01090 | −0.00010 | 0.00171 | ||
| 0.051 | θ̂M | 0.05050 | −0.00050 | 0.02065 | 0.05077 | −0.00023 | 0.00656 | |
| θ̂N | 0.05074 | −0.00026 | 0.02052 | 0.05080 | −0.00020 | 0.00651 | ||
| 0.101 | θ̂M | 0.09994 | −0.00106 | 0.03864 | 0.10085 | −0.00015 | 0.01253 | |
| θ̂N | 0.10039 | −0.00061 | 0.03829 | 0.10088 | −0.00012 | 0.01228 | ||
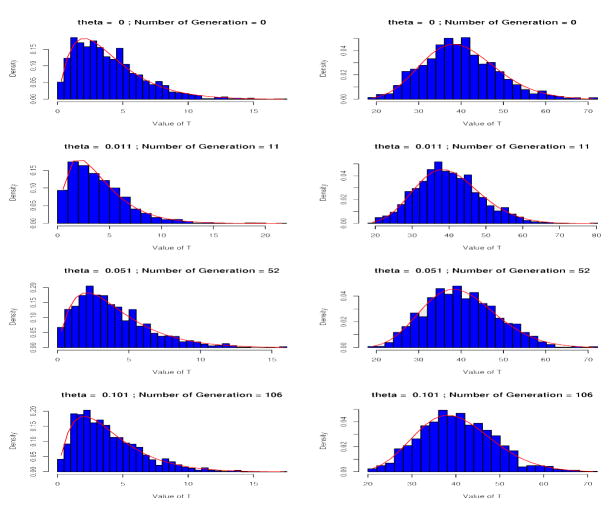
Figure 1 shows that the asymptotic distribution of the scaled T is indeed chi-square with appropriate degrees of freedom as assumed earlier. The figure shows that under different values of θ and either onr or ten loci, histograms of 1,000 simulated values of T/[1+(n − 1)θ] are very similar to the density of central chi-square distributions. The non-parametric Kolmogorov-Smirnov Test produces non-significant p-values for testing the hypotheses that the empirical distribution of T is a scaled central chi-square distribution for different values of θ.
Figure 1.
Comparison Empirical Distribution of T/[1 + (n − 1) θ] with a central Chi-square distribution with appropriate degrees of freedom. Each locus has 2 alleles, equally frequent initially. The data is generated with a pure drift model. The left column of plots is for one locus and the right column of plots is for ten loci.
Testing
The testing procedures described in this paper were applied to the same simulated data as for the study of estimation, and we present the results in Tables 4 and 5. For the two bootstrap tests we performed the tests at a 5% significance level and show in Table 4 that the empirical significance level of the parametric bootstrap is always close to 5%. In some situations for small numbers of locl (L = 5), the empirical significance level of the non-parametric bootstrap test exceeded the nominal level. The power of both bootstrap tests increases with θ, with the sample size, with the number of alleles per locus, and with the number of loci. Table 4 also shows that the parametric bootstrap method generally has the higher power and we recommend its use over the non-parametric bootstrap.
Table 4.
Empirical powers of parametric and non-parametric bootstrap tests, n and L represent the number of sampled alleles and the number of sampled loci respectively in each population.
|
L = 5 |
L = 10 |
L = 20 |
||||||
|---|---|---|---|---|---|---|---|---|
| θ | allele | n | NP Boot | P Boot | NP Boot | P Boot | NP Boot | P Boot |
| 0.00 | 10 | 0.065 | 0.057 | 0.043 | 0.054 | 0.041 | 0.050 | |
| 2 | 25 | 0.047 | 0.042 | 0.045 | 0.044 | 0.044 | 0.049 | |
| 40 | 0.044 | 0.042 | 0.043 | 0.053 | 0.041 | 0.048 | ||
| 10 | 0.080 | 0.047 | 0.057 | 0.056 | 0.049 | 0.049 | ||
| 4 | 25 | 0.082 | 0.052 | 0.056 | 0.047 | 0.055 | 0.055 | |
| 40 | 0.062 | 0.044 | 0.047 | 0.049 | 0.044 | 0.045 | ||
|
| ||||||||
| .011 | 10 | 0.114 | 0.110 | 0.111 | 0.130 | 0.151 | 0.187 | |
| 2 | 25 | 0.190 | 0.224 | 0.260 | 0.332 | 0.430 | 0.506 | |
| 40 | 0.312 | 0.365 | 0.455 | 0.553 | 0.713 | 0.797 | ||
| 10 | 0.166 | 0.139 | 0.190 | 0.200 | 0.278 | 0.303 | ||
| 4 | 25 | 0.386 | 0.389 | 0.540 | 0.595 | 0.814 | 0.842 | |
| 40 | 0.609 | 0.669 | 0.861 | 0.892 | 0.981 | 0.985 | ||
|
| ||||||||
| .051 | 10 | 0.348 | 0.409 | 0.516 | 0.611 | 0.787 | 0.838 | |
| 2 | 25 | 0.750 | 0.847 | 0.946 | 0.973 | 0.999 | 0.999 | |
| 40 | 0.911 | 0.966 | 0.994 | 0.999 | 1 | 1 | ||
| 10 | 0.696 | 0.714 | 0.903 | 0.932 | 0.997 | 0.998 | ||
| 4 | 25 | 0.991 | 0.994 | 1 | 1 | 1 | 1 | |
| 40 | 1 | 1 | 1 | 1 | 1 | 1 | ||
|
| ||||||||
| .101 | 10 | 0.658 | 0.780 | 0.893 | 0.935 | 0.993 | 0.997 | |
| 2 | 25 | 0.952 | 0.989 | 1 | 1 | 1 | 1 | |
| 40 | 0.995 | 0.999 | 1 | 1 | 1 | 1 | ||
| 10 | 0.970 | 0.981 | 1 | 1 | 1 | 1 | ||
| 4 | 25 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 40 | 1 | 1 | 1 | 1 | 1 | 1 | ||
Table 5.
Empirical powers of newly proposed chi-square test statistics with Li’s test procedure. n represents the number of sampled alleles in each population.
|
n =100 |
n =200 |
n =500 |
||||||
|---|---|---|---|---|---|---|---|---|
| θ | allele per locus | frequency | Our Test | Li’s Test | Our Test | Li’s Test | Our Test | Li’s Test |
| .000 | 2 | equal | 0.055 | 0.055 | 0.052 | 0.053 | 0.044 | 0.044 |
| 2 | 0.7 & 0.3 | 0.053 | 0.053 | 0.041 | 0.041 | 0.049 | 0.050 | |
| 2 | 0.9 & 0.1 | 0.042 | 0.042 | 0.053 | 0.053 | 0.048 | 0.048 | |
| 3 | equal | 0.050 | NA | 0.048 | NA | 0.058 | NA | |
| 4 | equal | 0.046 | NA | 0.047 | NA | 0.052 | NA | |
| 5 | equal | 0.048 | NA | 0.039 | NA | 0.056 | NA | |
|
| ||||||||
| .011 | 2 | equal | 0.313 | 0.317 | 0.566 | 0.568 | 0.825 | 0.825 |
| 2 | 0.7 & 0.3 | 0.316 | 0.321 | 0.539 | 0.543 | 0.823 | 0.823 | |
| 2 | 0.9 & 0.1 | 0.343 | 0.348 | 0.557 | 0.561 | 0.837 | 0.838 | |
| 3 | equal | 0.469 | NA | 0.774 | NA | 0.964 | NA | |
| 4 | equal | 0.566 | NA | 0.895 | NA | 0.994 | NA | |
| 5 | equal | 0.671 | NA | 0.937 | NA | 0.998 | NA | |
|
| ||||||||
| .051 | 2 | equal | 0.827 | 0.828 | 0.937 | 0.937 | 0.985 | 0.985 |
| 2 | 0.7 & 0.3 | 0.861 | 0.862 | 0.951 | 0.951 | 0.990 | 0.990 | |
| 2 | 0.9 & 0.1 | 0.861 | 0.862 | 0.951 | 0.951 | 0.990 | 0.990 | |
| 3 | equal | 0.968 | NA | 0.997 | NA | 1 | NA | |
| 4 | equal | 0.997 | NA | 1 | NA | 1 | NA | |
| 5 | equal | 1 | NA | 1 | NA | 1 | NA | |
|
| ||||||||
| .101 | 2 | equal | 0.944 | 0.944 | 0.983 | 0.983 | 0.995 | 0.995 |
| 2 | 0.7 & 0.3 | 0.951 | 0.952 | 0.985 | 0.985 | 0.996 | 0.996 | |
| 2 | 0.9 & 0.1 | 0.940 | 0.940 | 0.990 | 0.990 | 0.999 | 0.999 | |
| 3 | equal | 0.997 | NA | 1 | NA | 1 | NA | |
| 4 | equal | 1 | NA | 1 | NA | 1 | NA | |
| 5 | equal | 1 | NA | 1 | NA | 1 | NA | |
For large sample sizes we compared our new test statistic (Equation 7) with the test statistic proposed by Li (1996). The power of these two chi-square tests is shown in Table 5. For a 5% significance level, both tests have approximately 5% power when the null hypothesis is true, showing that the tests have a correct size. The power of the tests increases with the true value of θ and with the sample size. For loci with two alleles, the tests have similar power. For multiple alleles, the power of our new test increases with the number of alleles and we recommend its general use.
Application to HapMap Data
The International HapMap project (2005) generated two-allele SNP data from 270 people: Yoruba people in Ibadan, Nigeria (30 adult-and-both-parents trios), Japanese in Tokyo (45 unrelated individuals), Han Chinese in Beijing (45 unrelated individuals) and U.S. residents of northern and western European ancestry (30 trios). We applied our procedures to test if there is positive coancestry in human genome. We also used both moment and MLE estimators of θ to estimate θ in these different human populations and to quantify the heterogeneity among genome regions.
We estimated the coancestry coefficient using only those SNPs that were found to be segregating in all population samples. Due to the sampling scheme and missing data at different loci the number of alleles in the four different samples are different but the sample sizes are large enough for us to assume the same variance of allele frequencies among different populations. For maximum likelihood we used the estimator in Equation 4. Our estimates were calculated for all markers separately and also for all markers in all the 5Mb windows centered on each SNP in the autosomal genome.
There is substantial variation among estimates over the genome, even among SNPs that are very close to each other. The single-locus estimates are distributed very much like the χ2 distribution with two or three degrees of freedom. The extreme noisiness in single-locus estimates is demonstrated in Table 6, where the standard errors of the values for each chromosome are seen to be about the same size as the means. The noisiness of single-locus estimates can be reduced by combining data from several adjacent markers. The distribution of these (approximately) 1,000-locus values is close to a normal distribution (Weir et al., 2006) as expected from the chi-square distribution tending to normality as the df increase. Table 6 shows that the chromosomal standard errors of the estimates have dropped substantially for 1,000 loci. Even for the relatively large window size of 5 Mb there is substantial variation in estimates along each chromosome. Table 6 shows positive values for the coancestry coefficient, in the range of 0.1 to 0.15, and the hypothesis θ = 0 is rejected.
Table 6.
Estimates of θ based on single-locus and 1000 loci (approximately 5-Mb window) for HapMap data. (Standard errors in parentheses.)
| Single Locus |
1000 Loci |
|||
|---|---|---|---|---|
| Chromosome | θ̂M (se) | θ̂N (se) | θ̂M (se) | θ̂N (se) |
| Chromosome 1 | 0.13 (0.12) | 0.13 (0.12) | 0.14 (0.03) | 0.14 (0.03) |
| Chromosome 2 | 0.14 (0.12) | 0.14 (0.12) | 0.15 (0.02) | 0.15 (0.02) |
| Chromosome 3 | 0.13 (0.12) | 0.13 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 4 | 0.13 (0.12) | 0.13 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 5 | 0.12 (0.12) | 0.12 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 6 | 0.12 (0.11) | 0.12 (0.11) | 0.13 (0.02) | 0.13 (0.02) |
| Chromosome 7 | 0.12 (0.12) | 0.12 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 8 | 0.13 (0.12) | 0.13 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 9 | 0.12 (0.11) | 0.12 (0.11) | 0.13 (0.02) | 0.13 (0.02) |
| Chromosome 10 | 0.13 (0.12) | 0.13 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 11 | 0.12 (0.11) | 0.12 (0.11) | 0.13 (0.02) | 0.13 (0.02) |
| Chromosome 12 | 0.13 (0.12) | 0.13 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 13 | 0.12 (0.11) | 0.12 (0.11) | 0.13 (0.02) | 0.13 (0.02) |
| Chromosome 14 | 0.13 (0.11) | 0.13 (0.11) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 15 | 0.14 (0.13) | 0.14 (0.13) | 0.15 (0.03) | 0.15 (0.03) |
| Chromosome 16 | 0.13 (0.11) | 0.13 (0.11) | 0.14 (0.02) | 0.14 (0.02) |
| Chromosome 17 | 0.13 (0.13) | 0.13 (0.13) | 0.15 (0.03) | 0.15 (0.03) |
| Chromosome 18 | 0.12 (0.10) | 0.12 (0.10) | 0.12 (0.02) | 0.12 (0.02) |
| Chromosome 19 | 0.12 (0.11) | 0.12 (0.11) | 0.13 (0.01) | 0.13 (0.01) |
| Chromosome 20 | 0.12 (0.11) | 0.12 (0.11) | 0.13 (0.02) | 0.13 (0.02) |
| Chromosome 21 | 0.12 (0.11) | 0.12 (0.11) | 0.13 (0.02) | 0.13 (0.02) |
| Chromosome 22 | 0.12 (0.12) | 0.12 (0.12) | 0.14 (0.02) | 0.14 (0.02) |
Discussion
The coancestry coefficient θ is of central importance in population genetics and there is widespread interest in estimating this quantity when genetic data are available from different populations. In this article we have considered a moment estimator and a maximum likelihood estimator assuming normally distributed allele frequencies. The overall similarity of the two estimates and the advantage of having a chi-square distribution suggests general use of the maximum likelihood estimate. The moment estimator would be preferred in situations of small samples. In any event, use of simple estimators such as , where p̄ and are the sample mean and variance of allele frequencies over populations, is not recommended.
The sampling properties of both estimators were addressed with analytic expressions for the mean and variance and with simulation studies. Both approaches showed that the biases of both the estimators of θ are relatively small in magnitude, and negative in direction. The biases and variances of the estimators of θ increase as the differentiation levels increase in a total population, i.e. they increase with θ. Evolutionary variation, as measured by θ, cannot be reduced by sampling design. Increasing the number of loci sampled has a stronger effect on reducing the sampling variance of both the estimators than increasing the number of individuals sampled.
There are many factors that control the power of statistical tests. The power of the all test procedures discussed here increases with the true value of θ and with the sample size. When the true value of θ is small, then increasing the number of loci sampled has a stronger effect than increasing the number of individuals sampled. The power of the tests increases with the number of alleles per locus. Simulation studies show that our parametric bootstrap testing procedure has higher power than that of non-parametric bootstrap. Li (1996) used the central limit theorem for approximating the distribution of allele frequencies as a normal distribution and proposed a chi-square test. Here we have proposed an extension of her method that allows for an arbitrary number of alleles per locus. Our extension, captured in the statistic T, allows tests to be made about hypothesized values of θ, including θ = 0.
We have not addressed the effects of linkage or linkage disequilibrium on the two estimates. When loci are regarded as being independent it is easy to show that increasing the number of loci decreases the expected variance of the maximum likelihood estimator, and this effect was seen numerically for both estimates. The independence assumption will often be adequate. Weir et al. (2005) did allow for linkage among loci. By assuming Haldane’s mapping function they were able to predict variances of the “actual” values of θ for sets of linked loci. In this article we have written as though there is a single value of θ that applies to all genes in the genome. The stochastic nature of evolutionary forces such as mutation and the differences in actual genealogies among loci, however, means that there will be variation in actual values around the single theoretical value.
We should stress that our treatment of inference makes no assumption about the evolutionary forces that have been operating prior to the time populations were sampled to provide data. We have assumed that the mean of frequency of an allele over populations is some parameter p and that the variance of these frequencies among populations is θ p(1 − p). For maximum likelihood estimation we assumed a normal distribution of allele frequencies. In other words, maximum likelihood estimation assumes a distributional form for allele frequencies whereas the moment estimators assume the form of only the first two moments of these distributions. Other than that, our procedures hold regardless of the nature of mutation, migration, selection, population size or mating pattern. The interpretation of a particular numerical value of an estimate, on the other hand, is very much dependent on which evolutionary forces are assumed to have been acting. One of the most common uses of the estimates is to regard them as genetic distances between populations (Reynolds et al., 1983) and reconstruct the phylogeny of the populations – this application requires a drift-only model without mutation and so applies only within species.
Acknowledgments
This work was supported in part by NIH grant GM 075091. The third author recalls with gratitude the contributions of Sam Karlin to population genetics and his leadership for this journal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Balding DJ. Likelihood-based inference for genetic correlation coefficients. Theor Pop Biol. 2002;63:221–230. doi: 10.1016/s0040-5809(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Balding DJ, Nichols RA. Significant genetic correlations among Caucasians at forensic DNA loci. Heredity. 1997;78:583–589. doi: 10.1038/hdy.1997.97. [DOI] [PubMed] [Google Scholar]
- Cockerham CC. Variance of gene frequencies. Evolution. 1969;23:72–84. doi: 10.1111/j.1558-5646.1969.tb03496.x. [DOI] [PubMed] [Google Scholar]
- Dodds KG. PhD Thesis. North Carolina State University; Raleigh, NC.: 1986. Resampling Methods in Genetics and the Effect of Family Structure in Genetic Data. [Google Scholar]
- Holsinger KE. Analysis of genetic diversity in geographically structured populations: a Bayesian perspective. Hereditas. 1999;130:245–255. [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographcally structured populations: defining, estimating, and interpreting FST. Nature Reviews Genetics. 2009 doi: 10.1038/nrg2611. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;426:789–796. [Google Scholar]
- Jiang C. PhD Thesis. North Carolina State University; Raleigh, NC.: 1987. Estimation of F-statistics in subdivided populations. [Google Scholar]
- Lange K. Applications of the Dirichlet distribution to forensic match probabilities. Genetica. 1995;96:107–117. doi: 10.1007/BF01441156. [DOI] [PubMed] [Google Scholar]
- Li Y-J. PhD Thesis. North Carolina State University; Raleigh, NC.: 1996. Characterizing the Structure of Genetic Populations. [Google Scholar]
- Mosimann JE. On the compound multinomial distribution, the multivariate β-distribution and correlations among proportions. Biometrika. 1962;49:65–82. [Google Scholar]
- Nei M. Molecular Evolutionary Molecular Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Neerchal NK, Morel JG. An improved method for the computation of maximum likelihood estimates for multinomial overdispersion models. Comp Stat Data Anal. 2005;49:33–43. [Google Scholar]
- Nicholson G, Smith AV, J’onsson F, Gustafsson Ó, Stefánsson K, Donnelly P. Assessing population differentiation and isolation from single nucleotide polymorphism data. Proc Roy Stat Soc B. 2002;64:695–715. [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A, Hill WG. Deviations from Hardy-Weinberg proportions: sampling variances and use in estimation of inbreeding coefficients. Genetics. 1984;107:703–718. doi: 10.1093/genetics/107.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff DA, Bentzen P. The statistical analysis of mitochondrial polymorphisms: χ2 and the problem of small sample sizes. Molecular Biology and Evolution. 1989;6:539–545. doi: 10.1093/oxfordjournals.molbev.a040568. [DOI] [PubMed] [Google Scholar]
- Samanta S. PhD Thesis. North Carolina State University; Raleigh, NC.: 2006. A Statistical Characterization of the Genetic Structure of Populations. [Google Scholar]
- Serfling RJ. Approximation Theorems of Mathematical Statistics. Wiley; New York: 1980. [Google Scholar]
- Weir BS. Genetic Data Analysis II. Sinauer; Sunderland, MA: 1996. [Google Scholar]
- Weir BS. The rarity of DNA profiles. Annals of Applied Statistics. 2007;1:358–370. doi: 10.1214/07-AOAS128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Research. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Hill WG. Estimating F-statistics. Ann Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1931;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]