Abstract
Planar supported lipid bilayers that are stable under ambient atmospheric and ultra-high-vacuum conditions were prepared by cross-linking polymerization of bis-sorbylphosphatidylcholine (bis-SorbPC). X-ray photoelectron spectroscopy (XPS) and time-of-flight secondary ion mass spectrometry (ToF-SIMS) were employed to investigate bilayers that were cross-linked using either redox-initiated radical polymerization or ultraviolet photopolymerization. The redox method yields a more structurally intact bilayer; however, the UV method is more compatible with incorporation of transmembrane proteins. UV polymerization was therefore used to prepare cross-linked bilayers with incorporated bovine rhodopsin, a light-activated, G-protein-coupled receptor (GPCR). A previous study (Subramaniam, V.; Alves, I. D.; Salgado, G. F. J.; Lau, P. W.; Wysocki, R. J.; Salamon, Z.; Tollin, G.; Hruby, V. J.; Brown, M. F.; Saavedra, S. S. J. Am. Chem. Soc. 2005, 127, 5320–5321) showed that rhodopsin retains photoactivity after incorporation into UV-polymerized bis-SorbPC, but did not address how the protein is associated with the bilayer. In this study, we show that rhodopsin is retained in supported bilayers of poly(bis-SorbPC) under ultra-high-vacuum conditions, on the basis of the increase in the XPS nitrogen concentration and the presence of characteristic amino acid peaks in the ToF-SIMS data. Angle-resolved XPS data show that the protein is inserted into the bilayer, rather than adsorbed on the bilayer surface. This is the first study to demonstrate the use of ultra-high-vacuum techniques for structural studies of supported proteolipid bilayers.
Introduction
Artificial membrane systems such as planar supported lipid bilayers (PSLBs) have been frequently used as models of cell membranes to study interactions of lipids with proteins, cholesterol, and other biological molecules of interest.2–4 PSLBs are also being increasingly used as biocompatible coatings in biosensing and diagnostic devices.5–7 Only noncovalent interaction forces hold the lipids in these membranes together; thus, these bilayer systems are intrinsically unstable and collapse or desorb when removed from an aqueous environment and exposed to the ambient atmosphere or ultra-high-vacuum conditions.8–12 This is a major limitation for applications of PSLBs in sensors and other types of devices, as well as implantable biomaterial surfaces. Additionally, many surface-sensitive analytical techniques such as X-ray photoelectron spectroscopy (XPS) and time-of-flight secondary ion mass spectrometry (ToF-SIMS) that operate under vacuum and provide detailed chemical characterization information cannot be used to analyze PSLBs.
To address these limitations, supported bilayers composed of lipids functionalized with polymerizable dienoyl moieties have been developed. After cross-linking polymerization, these stabilized PSLBs can withstand the transition from aqueous to ambient atmospheric conditions, as well as ultrahigh vacuum.13–15 Ellipsometry, atomic force microscopy (AFM), XPS, and other surface analytical methods were used to investigate the surface coverage, structure, and defects of these bilayers.14,15 The results showed that, after drying, the poly(PSLB)s were highly uniform over surface areas of several square centimeters, with a surface composition and thickness that were consistent with values expected for a uniform bilayer composed of polymerized lipids with phosphorylcholine (PC) headgroups.
These cross-linked PSLBs were also found to be highly resistant to nonspecific adsorption of proteins, similar to fluid bilayers composed of PC lipids.15 The repulsion of nonspecific protein adsorption is essential for the detection and quantification of interacting biomolecules and possible subsequent interaction with other biomolecules (e.g., antibody/antigens, receptors, etc.).16–18 In a more recent study,1 a transmembrane protein, bovine rhodopsin (Rho),19,20 was reconstituted into polymerizable PSLBs which were subsequently cross-linked using UV photopolymerization. Using plasmon waveguide resonance (PWR) spectroscopy to measure the conformational change that accompanies photoactivation of Rho, it was shown that the activity of the protein was fully maintained in the poly(PSLB). The finding that Rho, a model for G-protein-coupled receptors, is fully functional in a poly(lipid) bilayer is potentially significant for the development of molecular devices based on stable artificial membranes reconstituted with transmembrane proteins. However, this study1 did not provide any information on how Rho is associated with the PSLB, e.g., whether it is inserted into the bilayer or merely adsorbed on its surface. This important issue can be addressed using angle-resolved XPS.
Both ToF-SIMS and XPS are surface-sensitive techniques, yielding information about the top few nanometers.21–24 While XPS is quantitative, ToF-SIMS yields information on the structure of the adsorbed organic overlayers and the underlying substrate.25–27 Here we focus on utilizing these ultra-high vacuum techniques to structurally characterize PSLBs composed of bis-sorbylphosphatidylcholine (bis-SorbPC) that are polymerized using either redox radical initiation or UV irradiation. The physical and chemical properties of poly (bis-SorbPC) bilayers produced by these two different polymerization methods have been studied extensively in lipid vesicles,28 but comparatively little is known about the corresponding PSLB architectures. Moreover, structural details of rhodopsin reconstituted into poly(PSLB)s have not been examined before. In this study XPS and ToF-SIMS are used to investigate these systems in more detail. Additionally, angle-resolved XPS is employed to investigate the location of Rho in the PSLB, an issue that has not been addressed in any prior studies of surfactant-assisted reconstitution of transmembrane proteins into PSLBs.
Materials and Methods
Bis-SorbPC (1,2-bis[10-(2′,4′-hexadienoloxy)decanoyl]-sn-glycero-3-phosphocholine) was synthesized as described previously.29 Silicon wafer substrates were purchased from Wacker-Chemie GmbH (Munich, Germany). All other reagents were purchased from standard commercial sources and used as received. Purification of retinal rod membranes and solubilization of Rho were carried out as described previously.30 Deionized water was obtained from a Barnstead Nanopure system (Barnstead/Thermolyne, Dubuque, IA) with a measured resistivity of 18 MΩ.
PSLB Formation
Detailed procedures for preparation of polymerized PSLBs of bis-SorbPC on silicon wafers have been published previously.14,15 Briefly, supported lipid bilayers were formed on cleaned substrates by fusion of small unilamellar vesicles (0.5 mg/mL solution in 10 mM phosphate buffer, pH 7.2). PSLBs were then subjected to UV photopolymerization by exposure to light for 30 min from a low-pressure mercury pen lamp with a rated intensity of 4500 µW/cm2 at 254 nm. Alternatively, PSLBs were redox polymerized for 2 h under positive argon pressure with deoxygenated solutions of potassium persulfate (0.02 M) and sodium bisulfite (0.01 M). After polymerization the PSLBs were removed from solution, rinsed with deionized water, and dried under nitrogen. For both polymerization methods, >95% of bis-SorbPC monomers react to form a polymer, on the basis of UV–vis absorbance measurements.14,15,31
Rho was reconstituted into PSLBs before polymerization using a surfactant dilution method.1 An aliquot of Rho (2 mg/mL) solublized in 30 mM octyl glucoside (OG) was injected into the aqueous buffer (10 mM phosphate, pH 7.2) above an unpolymerized PSLB in a custom-made cell. Injection caused dilution of the aliquot to final bulk protein and OG concentrations of ca. 5 µM and 3 mM, respectively. Diluting the OG below its critical micelle concentration (cmc) of 25 mM is presumed to cause spontaneous transfer of Rho from OG micelles into the PSLB.1 The solution above the PSLB was mixed gently and allowed to equilibrate for 5 h, after which the cell was rinsed with excess buffer to remove the detergent. The PSLB was then UV polymerized by exposure to the Hg pen lamp for 45 min through a band-pass filter (Hoya U-330) to remove visible light that activates Rho.30 Rho reconstitution and PSLB polymerization were performed in dim red light (Kodak safelight filter no. 1). Under these polymerization conditions: (i) >95% of bis-SorbPC monomers react to form a polymer, on the basis of UV–vis absorbance measurements;1,14,15,31 (ii) Rho retains its native UV–vis absorption spectrum including the 500 nm band indicative of a protonated retinylidene Schiff base.1
XPS Analysis
XPS measurements on polymerized PSLBs with and without reconstituted protein were carried out on a Kratos AXIS Ultra DLD instrument (Kratos, Manchester, England), using a monochromatic A1 Kα X-ray source. Angle-resolved XPS measurements were conducted at 0°, 55°, and 70° takeoff angles (TOAs). The photoelectron TOA is defined as the angle between the surface normal and the axis of the analyzer of the lens. A compositional survey and detailed scans (P 2s, N 1s, O 1s, Si 2p, and C 1s) were acquired using a pass energy of 80 eV. High-resolution spectra (C 1s) were acquired using a pass energy of 20 eV. All binding energies (BEs) were referenced to the hydrocarbon C 1s peak at 285 eV. Three spots on two or more replicates of each sample type were analyzed. The compositional data are an average of the values determined at each spot. Data analysis was performed with the Kratos Vision Processing and CASA XPS (www.casaxps.cwc.net) data reduction software.
ToF-SIMS Analysis
A model 7200 Physical Electronics instrument (PHI, Eden Prairie, MN) was used for static ToF-SIMS data acquisition. The instrument has an 8 keV Cs+ion source, a reflectron time-of-flight mass analyzer, chevron-type multichannel plates (MCPs), and a time-to-digital converter (TDC). Positive and negative secondary ion mass spectra were acquired over a mass range from m/z = 0 to m/z = 1000. The area of analysis for each spectrum was 100 µm × 100 µm, and the total ion dose used to acquire each spectrum was less than 1 × 1012 ions/cm2. The mass resolution (m/Δm) of the secondary ion peaks in the positive spectra was typically between 4000 and 6000. The ion beam was moved to a different spot on the sample for each spectrum. The mass scales of the positive secondary ion spectra were calibrated using the CH3+, C2H3+, C3H5+, and C5H10N+ peaks. The mass scales of the negative secondary ion spectra were calibrated using the CH−, OH−, and C2H− peaks. At least two replicates were analyzed for each sample type, with three positive and negative spectra acquired on each replicate. Negative ion results were not shown in this study as they did not significantly contribute any additional information to the overall results.
Principal Component Analysis (PCA)
PCA of the ToF-SIMS data was performed using the PLS Toolbox V 2.0 (Eigenvector Research, Manson, WA) for MATLAB (The Mathworks Inc., Natick, MA). Prior to this multivariate analysis, a peak set was created by assigning a centroid peak value and area of integration window for each peak detected in all spectra. The areas of these peaks were then calculated for every spectrum and normalized to the total intensity of all peaks in each individual spectrum to correct for differences in total secondary ion yield between spectra. This process yielded a matrix of n peaks and m samples, representative of all spectra collected. These data were mean centered (set to the origin) to ensure that the variance in the dataset was due to differences in sample variances rather than differences in sample means.
Results and Discussion
XPS of UV-and Redox-Polymerized Bis-SorbPC Bilayers
XPS was used to investigate UV- and redox-polymerized bis-SorbPC bilayers. Small but distinct differences in the composition of the two types of bilayers were observed. As both polymerization methods were applied to the same lipids, these differences arise from the polymerization process. Although cross-linked PSLBs are formed by both methods, previous studies on poly(bis-SorbPC) vesicles have shown that redox initiation produces a greater degree of polymerization (Xn = 50–500) than UV irradiation (Xn = 3–10).32–34 A similar trend likely occurs in bis-SorbPC PSLBs polymerized by these two methods, and significant differences in their respective structural properties have been observed.14,15 The origin of these differences may be different propagation mechanisms for the two methods. 1H NMR analysis of redox-polymerized bis-SorbPC vesicles indicates that the polymerization proceeds via a 1,4-addition reaction.35,36 The mechanism of UV photopolymerization of these lipids has not been established.
Table 1 shows the XPS-determined atomic concentrations of the PSLB-modified surfaces. The redox-polymerized bilayers exhibit slightly higher carbon concentrations compared to the UV-polymerized bilayers. Similarly, slightly higher percentages of silicon and oxygen, indicative of the underlying substrate, were found for the UV-polymerized bilayer. To remove the contribution of the silicon wafer from the atomic composition calculations, the average peak area ratio of elemental silicon (Simet) to SiO2 (Siox) was calculated for each analysis spot. This was done using the detailed Si 2p spectra to measure the relative percentages of the Simet and Siox peaks (data not shown). Then the stoichiometric 2 parts of oxygen corresponding to the Siox peak were subtracted from the total oxygen concentration. In addition to this, a second factor was incorporated into the calculation to allow for the differences in mean free path (λ) between Si (λSi) and O (λO). The final equation used for calculating the SiO2 free oxygen concentration (atom %) was
| (1) |
where λO/λSi = 3.39 nm/4.09 nm = 0.83.
Table 1.
Averages and Standard Deviations for XPS-Determined Atomic Concentrations (atom %) for UV- and Redox-Polymerized Bis-SorbPC Bilayersa
| C 1s | O 1s | N 1s | P 2p | Si 2p | |
|---|---|---|---|---|---|
| UV | |||||
| av | 55.3 | 24.4 | 1.1 | 2.3 | 16.8 |
| std dev | 2.0 | 1.3 | 0.4 | 0.4 | 0.6 |
| Redox | |||||
| av | 63.2 | 21.6 | 1.1 | 1.9 | 12.3 |
| std dev | 1.5 | 0.8 | 0.2 | 0.4 | 0.7 |
Comparison of the Si signal indicates that the UV-polymerized layer is thinner than the redox-polymerized sample.
Table 2 shows the corrected values without the underlying silicon substrate contributions. For the UV-polymerized samples, the C, O, and P XPS-determined concentrations were all consistent with the values expected from theory, but the N content was lower than predicted. The redox-polymerized samples exhibited slightly higher carbon and lower oxygen concentrations than expected. Comparison of the Si concentrations in Table 1 indicates that the UV-polymerized layer (higher Si signal) is either thinner than the redox sample or is not continuous. The former possibility is consistent with prior studies of bis-SorbPC PSLBs polymerized by these two methods. Ross and co-workers demonstrated that redox-initiated radical polymerization preserves the overall bilayer structure after drying, with very few structural defects as measured by AFM and ellipsometry.14,15 The bilayer structure of UV-polymerized PSLBs is also largely preserved after drying; however, these membranes were thinner and had a higher density of small defects, likely caused by desorption of low molecular weight oligomers upon removal from water. Nevertheless, UV-polymerized bilayers show greatly enhanced stability compared to unpolymerized, fluid lipid bilayers which are completely desorbed from the substrate upon removal from water.14,15
Table 2.
Adjusted XPS Atomic Concentrations (atom %) for UV- and Redox-Polymerized Bis-SorbPC Bilayers with the Si Wafer Contributions Removed Using Eq 1 and the PSLB Concentrations Renormalized to 100 atom %a
| C 1s | O 1s | N 1s | P 2p | |
|---|---|---|---|---|
| UV | ||||
| av | 75.2 | 21.5 | 1.1 | 2.2 |
| std dev | 2.5 | 1.7 | 0.5 | 0.5 |
| Redox | ||||
| av | 79.9 | 16.4 | 1.3 | 2.4 |
| std dev | 1.9 | 1.0 | 0.2 | 0.5 |
| Theory | ||||
| 74 | 22.2 | 1.9 | 1.9 | |
The theoretical values were determined from the stoichiometry of the bis-SorbPC molecule.
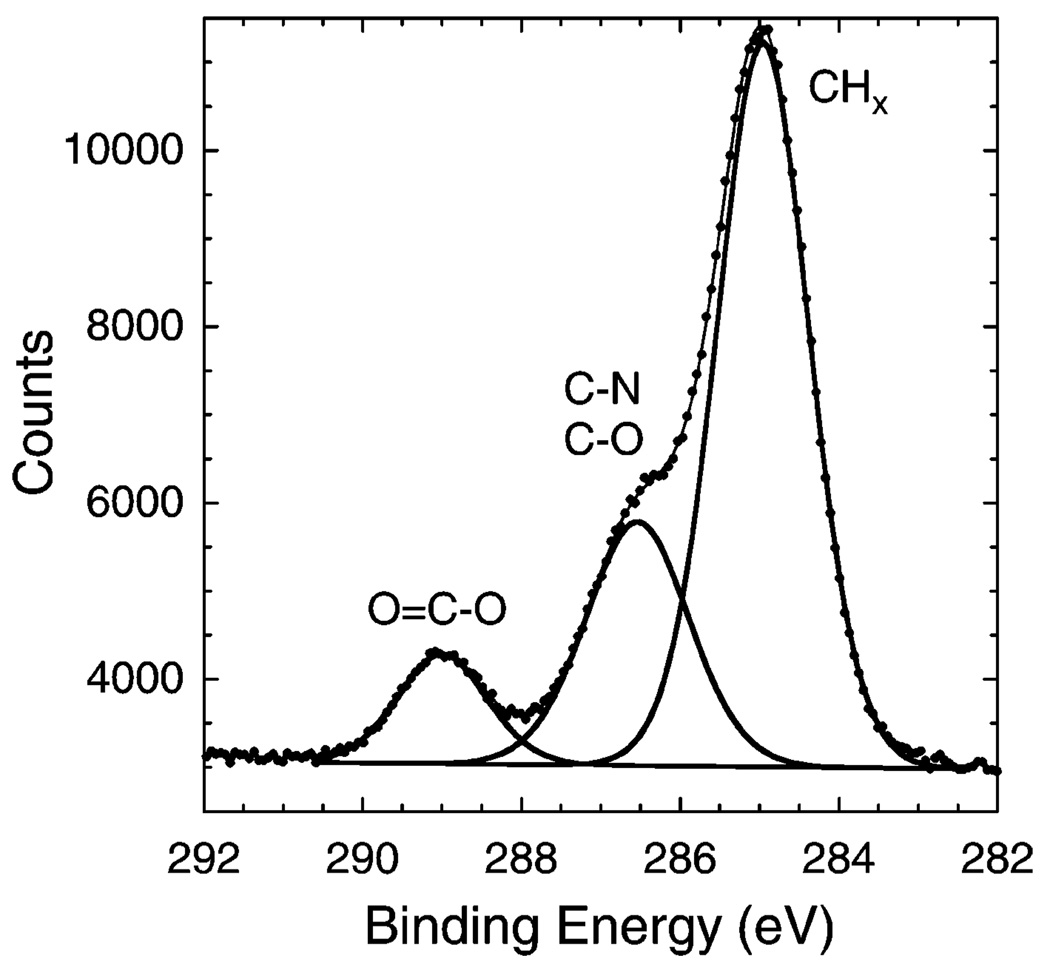
In addition to analyzing the atomic composition, peak fitting was performed on the high-resolution C 1s spectra. Figure 1 shows a representative three-peak fit to the spectra with CHx (285.0 eV), C−N and C−O (~286.5 eV), and O−C = O (289.0 eV) carbon species. The C−N (286.1 eV) and C−O (286.7 eV) contributions were too close to separately quantify and were therefore summed together. The C 1s results for both UV- and redox-polymerized PSLBs matched the theoretical values well.
Figure 1.
XPS high-resolution C 1s spectrum with the peak fit obtained from a redox-polymerized bilayer of bis-SorbPC.
ToF-SIMS and Multivariate Analysis of UV- and Redox-Polymerized Bilayers
PCA was used to examine the differences between the ToF-SIMS data of UV- and redox-polymerized PSLBs. PCA reduces the multidimensional (multipeak) aspect of each spectrum to new variables called principal components (PCs) that describe the largest variation in the sample set. The original data are thus reduced to a cross-product of two smaller matrices, termed scores and loadings. The resulting PCs are linear combinations of all original variables. Scores give the relationships between the samples for each PC, while loadings relate the original variables (ToF-SIMS peaks) to the new PCs.
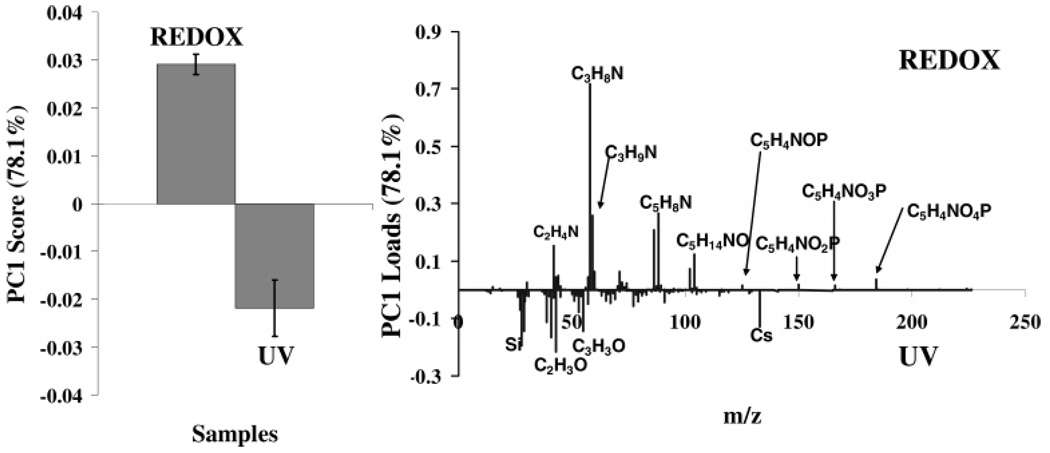
PCA indicated a distinct separation of the UV- and redox-polymerized PSLBs, with PC 1 capturing 78% of the variation in the dataset, as shown in the scores plot (Figure 2, left). Analysis of the loadings plot for PC 1 indicated that the variations between the sample types could be related to the surface composition (Figure 2, right). For both samples, the same peaks were present in the ToF-SIMS fragmentation pattern, but the relative intensities of these peaks were different. Peaks associated with the lipid head group loaded more strongly with the redox samples, while peaks associated with the lipid tails and the silicon wafer loaded more strongly with the UV samples. This result suggests that there may be a greater degree of order associated with the redox samples, resulting in a higher yield of the head group ions. Lower surface coverage or the presence of defects in the UV samples is indicated by the increased presence of tail group and substrate signals within the ToF-SIMS sampling area (~1–2 nm depth of analysis and 100 µm × 100 µm area of analysis). These results are consistent with the XPS composition results discussed above and with previous studies14,15 showing that UV polymerization produces thinner bilayers with a higher density of defects. However, we cannot rule out the possibility that the UV polymerization may cause a minor amount of lipid photodegradation. Lipid photodegradation has been shown to occur when significantly more aggressive UV illumination conditions are used.37,38
Figure 2.
PCA scores (left) and loadings (right) plot comparing positive ion ToF-SIMS data from UV- and redox-polymerized bilayers. While the same peaks are found on both types of surfaces, peaks associated with the lipid head group (C3H8N+, C5H4NO4P+, etc.) loaded more strongly with the redox-polymerized PSLBs. Peaks associated with the lipid tails (C3H8O+, etc.) and silicon (Si+, SiOH+, etc.) loaded more strongly with the UV-polymerized PSLBs.
XPS Study of Rhodopsin Incorporated into UV-Polymerized Bilayers
Rhodopsin reconstitution followed by redox polymerization of bis-SorbPC PSLBs was attempted, but the protein was found to precipitate under the harsh polymerization conditions (data not shown). This may be because the acidity of the redox solution (pH 2) causes protein denaturation or because the hydroxyl radicals generated in the process may react with protein side chains. A similar result was observed when the polymerization reaction was performed in a neutral buffer with high buffering capacity, possibly due to high salt concentrations as well as radical generation. We are currently exploring the possibility of optimizing the conditions of this redox system as well as other redox couples to permit transmembrane protein reconstitution. For the current study, only UV-polymerized bis-SorbPC PSLBs containing Rho were analyzed with XPS and ToF-SIMS.
Table 4 shows the XPS-determined compositional differences between poly(PSLBs) with and without reconstituted Rho. There is little difference in the carbon, oxygen, and silicon elemental contributions. The nitrogen signal, however, is significantly higher in the PSLBs containing Rho, indicating the presence of nitrogen-rich protein molecules. The somewhat lower phosphorus contribution is attributed to the lower surface coverage of lipids in the proteolipid film, due to some fraction of the surface being occupied by Rho.
Table 4.
XPS Compositional Averages and Standard Deviations (atom %) of UV-Polymerized PSLBs with and without Incorporated Rhoa
| C 1s | O 1s | Si 2p | N 1s | P 2p | |
|---|---|---|---|---|---|
| UV without Rho | |||||
| av | 55.3 | 24.4 | 16.8 | 1.1 | 2.3 |
| std dev | 2.0 | 1.3 | 0.6 | 0.4 | 0.4 |
| UV with Rho | |||||
| av | 55.8 | 22.7 | 15.1 | 5.1 | 1.2 |
| std dev | 3.1 | 1.0 | 2.1 | 0.8 | 0.1 |
Similar compositions are observed for both types of bilayers. However, the proteolipid bilayers have a lower phosphorus content (likely from a lower lipid surface coverage due to some surface area being occupied by Rho) and a significantly higher nitrogen content (due to the incorporated Rho).
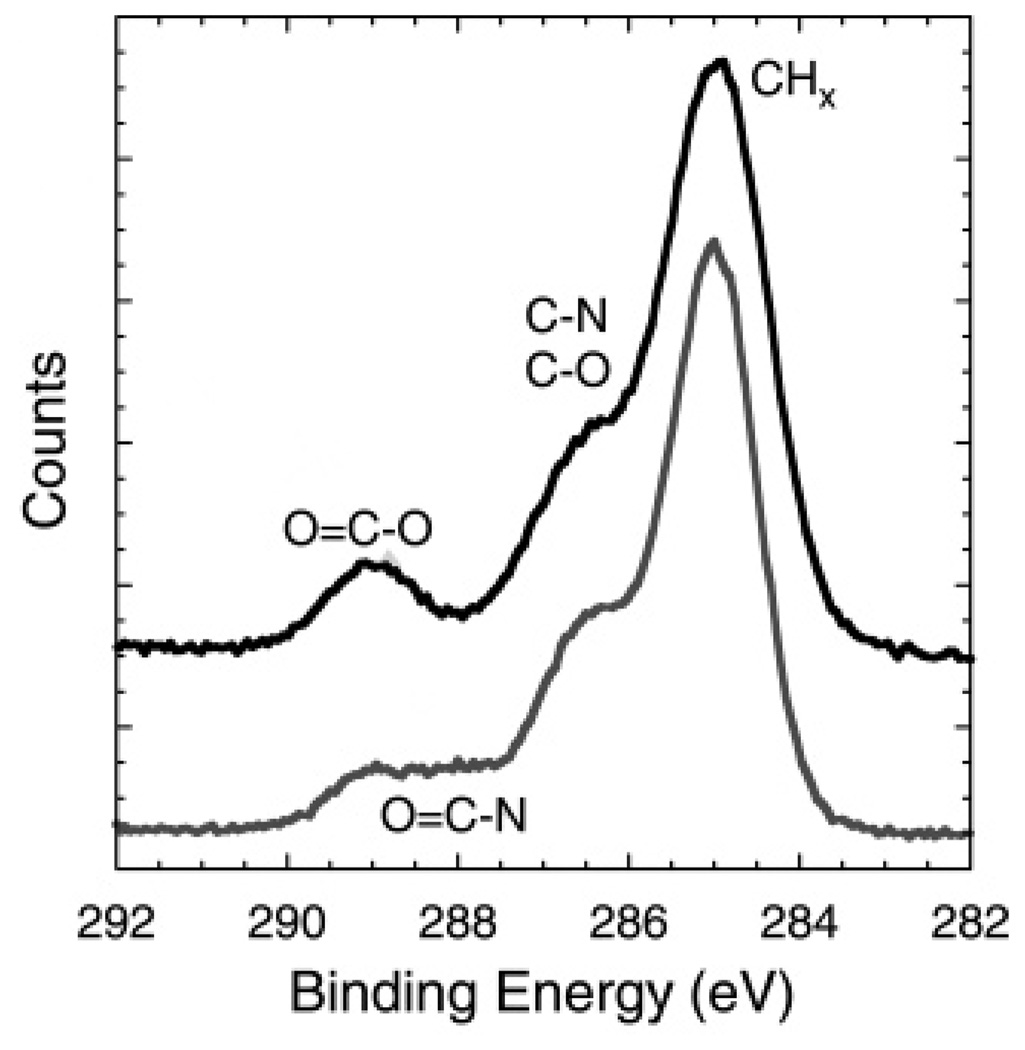
The high-resolution C 1s results from UV-polymerized PSLBs with and without incorporated Rho are shown in Figure 3 and Table 5. As can be seen by visual inspection of Figure 3 and supported by the fitting results in Table 5, Rho incorporation results in a decrease in the CHx and O−C = O species and an increase in the N−C = O species. This is expected since the N−C = O species is from the protein peptide backbone and the CHx and O−C = O species are primarily from bis-SorbPC. Thus, both the XPS composition and high-resolution spectral results are consistent with the proteolipid films having a lower lipid surface coverage after Rho incorporation.
Figure 3.
XPS high-resolution C 1s spectra comparing UV-polymerized bis-SorbPC bilayers lacking Rho (black, top) and containing Rho (gray, bottom). There is a distinct decrease of the O−C=O contribution (due to lipid tails) and an increase of the N−C=O contribution (due to Rho) in the proteolipid membrane. The top spectrum has been offset for clarity.
Table 5.
XPS High-resolution C 1s Peak Fitting Results for the UV-Polymerized Bis-SorbPC PSLBs with and without Incorporated Rho, Showing a Decrease of the C−C and O−C=0 Contributions (Mainly from the Lipid Tails and Head Groups, Respectively) and an Increase of the N−C=O Contribution (from the Amino Acids of Rho)a
| binding E (eV) | 285.0 | 286.1, 286.7 | 288.0 | 289.0 |
|---|---|---|---|---|
| type | CHx | C−N, C−O | N−C=O | O−C=O |
| UV without Rho | 61.1 | 28.4 | 0.0 | 10.7 |
| UV with Rho | 57.1 | 29.3 | 4.2 | 7.2 |
All data are shown as the relative percentage of the total C 1s peak area.
Angle-resolved XPS measurements were used to investigate the location of Rho in UV-polymerized bilayers. As described above, Rho was reconstituted into PSLBs using a surfactant dilution method, which is thought to involve spontaneous transfer of Rho from OG micelles into the PSLB.1 However, the actual mechanism is unknown, as is the nature of the association between Rho and the lipid bilayer. For example, it is clear from prior studies1 and the XPS data in Table 4 that Rho is bound to the PSLB and is photoactive, but it is not clear whether Rho is inserted into the PSLB or merely adsorbed onto its surface.
To determine the location of the protein in the bilayer, the compositions of all elements (C, O, Si, N, and P) were measured at three TOAs: 0° (perpendicular to the sample surface), 55°, and 70° (glancing angle). The silicon substrate signals were subtracted using eq 1, as described above, after determination of the percentages of silicon (Simet) and SiO2 (Siox) from the Si 2p spectra, which varied greatly depending on the TOA. The ratio of Simet to Siox was much higher at lower TOAs (data not shown). Then, twice the Siox contribution was subtracted from the overall oxygen signal, and the remaining oxygen (from the bilayer and protein), plus carbon, nitrogen, and phosphorus, was renormalized to 100%. Table 6 lists the corrected compositions of UV-polymerized PSLBs with and without Rho at the three different TOAs. The sample lacking Rho shows a slight decrease in the oxygen signal and a slight increase in carbon with increasing TOA. At a 0° TOA, the entire thickness of the bilayer is analyzed, and the experimental values for C, O, P, and N are close to the theoretical values calculated for a bis-SorbPC bilayer. The XPS compositional data at the larger glancing angles detect more of the outer leaflet of the PSLB, thereby attenuating the P, O, and N signals from the inner leaflet that is adjacent to the substrate. A similar result is found in the case of the UV-polymerized PSLB containing Rho, with the exception of overall lower C, O, and P values due to the increase of the nitrogen signal from protein incorporation. A notable finding is that the nitrogen signal remains practically constant across all TOAs. If the protein molecules (with a 4–5 times higher percentage of nitrogen compared to that of the lipid) were adsorbed on top of the PSLB, the nitrogen signal would increase significantly with increasing TOA, while the oxygen and carbon signals from the underlying lipid molecules would be attenuated. The fact that this was not observed shows that the protein is inserted into the bilayer (i.e., uniformly distributed throughout its thickness) rather than adsorbed on top of it. The results presented here and published previously1,14,15 clearly establish that these PSLBs maintain the photoactivity of incorporated Rho and their overall structure after drying.
Table 6.
Angle-Resolved XPS Results (atom %) for Bis-SorbPC Bilayers with and without Rho Determined at TOAs of 0°, 55°, and 70°, after the Subtraction of the Silicon Substrate Contributions (Silicon, SiO2)a
| sample | N 1s | C 1s | P 2p | O 1s |
|---|---|---|---|---|
| UV without Rho, 0° | 0.8 | 75.0 | 2.1 | 22.1 |
| UV without Rho 55° | 0.9 | 79.9 | 1.8 | 17.4 |
| UV without Rho 70° | 0.6 | 82.0 | 1.2 | 16.2 |
| UV with Rho 0° | 7.3 | 73.0 | 1.7 | 18.1 |
| UV with Rho 55° | 7.6 | 76.8 | 0.8 | 14.7 |
| UV with Rho 70° | 7.6 | 79.8 | 0.9 | 11.7 |
A slightly higher concentration of Rho was chosen for this experiment to enhance the effect of angle-dependent XPS analysis. The constant nitrogen concentration at all TOAs indicates that the nitrogen distribution is homogeneous across the lipid and protein layer, which shows that the nitrogen-rich protein is incorporated into, rather than adsorbed on top of, the PSLB.
ToF-SIMS Study of Rho Incorporated in UV-Polymerized Bilayers
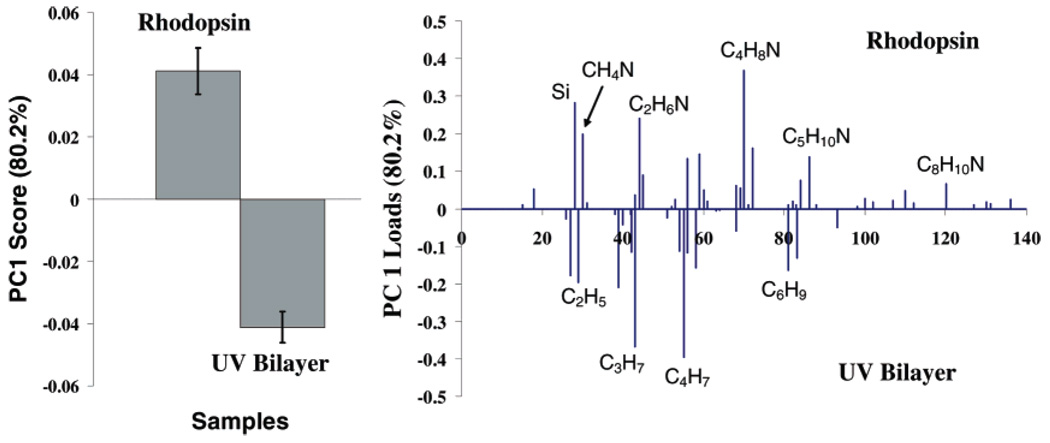
PCA was used to examine the differences in the ToF-SIMS analysis of UV-polymerized bilayers with and without incorporated Rho, using the entire positive secondary ion dataset. PC 1, which captured 80% of the overall variance in the dataset, showed that peaks occurring from the amino acids loaded more strongly with the samples containing Rho. Many of the amino acid fragments (CH4N, C2H6N, etc.) found could also originate from the lipid head groups, but are found at much higher intensities in proteins compared to lipids.39 Fragments originating from the lipid tails, on the other hand, loaded more strongly with polymerized bilayers that lacked Rho (Figure 4). These results are consistent with the XPS results discussed above.
Figure 4.
PCA scores (left) and loadings (right) plot comparing the positive ion ToF-SIMS data from UV-polymerized PSLBs with (“Rhodopsin”) and without (“UV Bilayer”) incorporated Rho. Nitrogen-containing peaks associated with amino acids loaded more strongly with the Rhodopsin sample, and peaks associated with the lipid tails (hydrocarbons) loaded more strongly with the UV Bilayer sample.
Conclusions
Two methods of preparing cross-linked PSLBs from bis-SorbPC were compared and evaluated by XPS and ToF-SIMS. Redox-initated radical polymerization produced a more structurally intact PSLB than UV photopolymerization, which is consistent with prior studies14,15 on application of these two polymerization methods to bis-SorbPC bilayers. However, the redox method was found to deactivate Rho. The UV polymerization method, which has been shown to maintain the photo-activity of incorporated Rho,1 was therefore used to prepare polymerized proteolipid membranes for further studies. The presence of protein in polymerized PSLBs was confirmed and quantified with XPS and ToF-SIMS. Angle-resolved XPS was employed to determine whether the method used to incorporate Rho results in adsorption onto or insertion into the PSLB. The results show that Rho is inserted, and previous studies1 have shown that it is also photoactive when hydrated. It is unknown whether its photoactivity is retained after the PSLB is dried; this topic will be a subject of future studies. Finally, the results of this study suggest the feasibility of reconstituting and stabilizing other types of transmembrane proteins, such as ligand-activated GPCRs and ion channels, into poly (bis-SorbPC) PSLBs to permit structural analysis using ultra-high-vacuum techniques, as demonstrated here for Rho.
Table 3.
XPS High-Resolution Carbon (C 1s) Peak Fitting Results for the UV- and Redox-Polymerized PSLBsa
| binding E (eV) | 285.0 | 286.1, 286.7 | 289.0 |
|---|---|---|---|
| type | CHx | C−N, C−O | O−C=O |
| UV | |||
| av | 61.8 | 26.7 | 11.5 |
| std dev | 1.0 | 1.2 | 0.7 |
| Redox | |||
| av | 60.5 | 29.4 | 10.2 |
| std dev | 1.4 | 1.0 | 0.7 |
| Theory | |||
| 65 | 25 | 10 | |
All data are shown as the relative percentage of the total C 1s peak area. The fitted data for both the UV- and redox-polymerized bilayers agree well with the theoretical values calculated from the stoichiometry of the bis-SorbPC molecule.
Acknowledgment
This work was supported by NIBIB Grant EB-002027 to the National ESCA and Surface Analysis Center for Biomedical Problems (NESAC/BIO), NSF Grant CHE-0518702, NIH Grant EB007047, and NIH Grant EY12049. We acknowledge Dr. Daniel Graham for insightful discussions and Pick-Wei Lau for assistance with the preliminary data.
References
- 1.Subramaniam V, Alves ID, Salgado GFJ, Lau PW, Wysocki RJ, Salamon Z, Tollin G, Hruby VJ, Brown MF, Saavedra SS. J. Am. Chem. Soc. 2005;127:5320–5321. doi: 10.1021/ja0423973. [DOI] [PubMed] [Google Scholar]
- 2.Brockman JM, Nelson BP, Corn RM. Annu. Rev. Phys. Chem. 2000;51:41–63. doi: 10.1146/annurev.physchem.51.1.41. [DOI] [PubMed] [Google Scholar]
- 3.Steinem C, Janshoff A, Ulrich WP, Sieber M, Galla HJ. Biochim. Biophys. Acta—Biomembranes. 1996;1279:169–180. doi: 10.1016/0005-2736(95)00274-x. [DOI] [PubMed] [Google Scholar]
- 4.Salamon Z, Brown MF, Tollin G. Trends Biochem. Sci. 1999;24:213–219. doi: 10.1016/s0968-0004(99)01394-8. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt C, Mayer M, Vogel H. Angew. Chem., Int. Ed. 2000;39:3137–3140. doi: 10.1002/1521-3773(20000901)39:17<3137::aid-anie3137>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 6.Song XD, Swanson BI. Anal. Chem. 1999;71:2097–2107. doi: 10.1021/ac981145f. [DOI] [PubMed] [Google Scholar]
- 7.Stelzle M, Weissmuller G, Sackmann E. J. Phys. Chem. 1993;97:2974–2981. [Google Scholar]
- 8.Sackmann E, Tanaka M. Trends Biotechnol. 2000;18:58–64. doi: 10.1016/s0167-7799(99)01412-2. [DOI] [PubMed] [Google Scholar]
- 9.Plant AL. Langmuir. 1999;15:5128–5135. [Google Scholar]
- 10.Sackmann E. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 11.Cremer PS, Boxer SG. J. Phys. Chem. B. 1999;103:2554–2559. [Google Scholar]
- 12.Holden MA, Jung SY, Yang TL, Castellana ET, Cremer PS. J. Am. Chem. Soc. 2004;126:6512–6513. doi: 10.1021/ja048504a. [DOI] [PubMed] [Google Scholar]
- 13.Ross EE, Bondurant B, Spratt T, Conboy JC, O’Brien DF, Saavedra SS. Langmuir. 2001;17:2305–2307. [Google Scholar]
- 14.Ross EE, Rozanski LJ, Spratt T, Liu SC, O’Brien DF, Saavedra SS. Langmuir. 2003;19:1752–1765. [Google Scholar]
- 15.Ross EE, Spratt T, Liu SC, Rozanski LJ, O’Brien DF, Saavedra SS. Langmuir. 2003;19:1766–1774. [Google Scholar]
- 16.Huang NP, Voros J, De Paul SM, Textor M, Spencer ND. Langmuir. 2002;18:220–230. [Google Scholar]
- 17.Larsson C, Rodahl M, Hook F. Anal. Chem. 2003;75:5080–5087. doi: 10.1021/ac034269n. [DOI] [PubMed] [Google Scholar]
- 18.Vermette P, Gengenbach T, Divisekera U, Kambouris PA, Griesser HJ, Meagher L. J. Colloid Interface Sci. 2003;259:13–26. doi: 10.1016/s0021-9797(02)00185-6. [DOI] [PubMed] [Google Scholar]
- 19.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 20.Papermas Ds, Dreyer WJ. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- 21.Cornelio-Clark PA, Gardella JA. Langmuir. 1991;7:2279–2286. [Google Scholar]
- 22.Paynter RW, Ratner BD, Horbett TA, Thomas HR. J. Colloid Interface Sci. 1984;101:233–245. [Google Scholar]
- 23.Castner DG, Ratner BD. Electron Spectroscopy for Chemical Analysis. Chichester, UK: John Wiley and Sons; 1997. [Google Scholar]
- 24.Belu AM, Graham DJ, Castner DG. Biomaterials. 2003;24:3635–3653. doi: 10.1016/s0142-9612(03)00159-5. [DOI] [PubMed] [Google Scholar]
- 25.Xia N, May CJ, McArthur SL, Castner DG. Langmuir. 2002;18:4090–4097. [Google Scholar]
- 26.Wagner MS, Castner DG. Langmuir. 2001;17:4649–4660. [Google Scholar]
- 27.Tidwell CD, Castner DG, Golledge SL, Ratner BD, Meyer K, Hagenhoff B, Benninghoven A. Surf. Interface Anal. 2001;31:724–733. [Google Scholar]
- 28.Lamparski HG, O’Brien DF. Macromolecules. 1995;28:1786–1794. [Google Scholar]
- 29.Sells TD, Obrien DF. Macromolecules. 1994;27:226–233. [Google Scholar]
- 30.Botelho AV, Gibson NJ, Thurmond RL, Wang Y, Brown MF. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 31.Conboy JC, Liu SC, Obrien DF, Saavedra SS. Biomacromolecules. 2003;4:841–849. doi: 10.1021/bm0256193. [DOI] [PubMed] [Google Scholar]
- 32.Liu SC, O’Brien DF. Macromolecules. 1999;32:5519–5524. [Google Scholar]
- 33.Lei JT, Sisson TM, Lamparski HG, O’Brien DF. Macromolecules. 1999;32:73–78. [Google Scholar]
- 34.Sisson TM, Lamparski HG, Kolchens S, Elayadi A, O’Brien DF. Macromolecules. 1996;29:8321–8329. [Google Scholar]
- 35.O’Brien DF, Armitage B, Benedicto A, Bennett DE, Lamparski HG, Lee YS, Srisiri W, Sisson TM. Acc. Chem. Res. 1998;31:861–868. [Google Scholar]
- 36.Clapp PJ, Armitage BA, Obrien DF. Macromolecules. 1997;30:32–41. [Google Scholar]
- 37.Yee CK, Amweg ML, Parikh AN. J. Am. Chem. Soc. 2004;126:13962–13972. doi: 10.1021/ja047714k. [DOI] [PubMed] [Google Scholar]
- 38.Yee CK, Amweg ML, Parikh AN. Adv. Mater. 2004;16:1184–1189. [Google Scholar]
- 39.Ostrowski SG, Szakal C, Kozole J, Roddy TP, Xu JY, Ewing AG, Winograd N. Anal. Chem. 2005;77:6190–6196. doi: 10.1021/ac0508189. [DOI] [PubMed] [Google Scholar]