Abstract
Retrospective analyses of clinical dynamic contrast-enhanced (DCE) MRI studies may be limited by failure to measure the longitudinal relaxation rate constant (R1) initially, which is necessary for quantitative analysis. In addition, errors in R1 estimation in each individual experiment can cause inconsistent results in derivations of pharmacokinetic parameters, Ktrans and ve, by kinetic modeling of the DCE-MRI time course data. A total of 18 patients with lower extremity osteosarcomas underwent multislice DCE-MRI prior to surgery. For the individual R1 measurement approach, the R1 time course was obtained using the two-point R1 determination method. For the average R10 (pre-contrast R1) approach, the R1 time course was derived using the DCE-MRI pulse sequence signal intensity equation and the average R10 value of this population. The whole tumor and histogram median Ktrans (0.57 ± 0.37 and 0.45 ± 0.32 min−1) and ve (0.59 ± 0.20 and 0.56 ± 0.17) obtained with the individual R1 measurement approach are not significantly different (paired t test) from those (Ktrans: 0.61 ± 0.46 and 0.44 ± 0.33 min−1; ve: 0.61 ± 0.19 and 0.55 ± 0.14) obtained with the average R10 approach. The results suggest that it is feasible, as well as practical, to use a limited-population-based average R10 for pharmacokinetic modeling of osteosarcoma DCE-MRI data.
Keywords: dynamic contrast-enhanced MRI, osteosarcoma, R1, pharmacokinetic modeling, Ktrans, ve
Introduction
T1-weighted dynamic contrast-enhanced (DCE) MRI has in recent years been widely utilized in studies of many different types of cancer (1), frequently for purposes of diagnosis (2–5) and assessment of the effectiveness of antiangiogenic therapies (6–10). For interpretation of the DCE-MRI signal time course, there are generally three approaches (11): a) qualitative assessment of the curve shape, such as wash-out, plateau, and persistence; b) empirical quantitation, such as maximum slope, percent signal intensity change; and c) analytical pharmacokinetic modeling. The results from the first two approaches depend on DCE-MRI pulse sequence parameters, contrast agent dose and injection rate, magnetic field strength, and vendor platform, etc (11–13). These data acquisition details vary greatly between institutions, thus limiting DCE-MRI study reproducibility and comparability between different imaging sites. The analytical approach is more sophisticated, and also the more desirable. Unlike the first two approaches, analytical modeling of DCE-MRI data extracts pharmacokinetic parameters that are in principle independent of data acquisition details mentioned above. This data analysis method, therefore, should improve study reproducibility, enable meaningful comparison of results from different groups, and be especially important for multi-center trial studies. The extracted pharmacokinetic parameters from analytical modeling of DCE-MRI time course data are usually variants of: Ktrans, a rate constant for contrast agent plasma/interstitium transfer, and ve, the interstitial space volume fraction (the putative contrast agent distribution volume).
Using a semi-quantitative approach, we reported that the histogram amplitude of initial slope of DCE-MRI signal time course correlated significantly with necrosis percentage of osteogenic and Ewing sarcoma, which is an important indicator of the effectiveness of chemotherapy (14). For pharmacokinetic modeling of DCE-MRI data, both the determination of arterial input function (AIF) and the measurement of longitudinal relaxation rate constant, R1 (= 1/T1) [either region of interest (ROI) or pixel by pixel R1], time course are critical to the accuracy of absolute quantitation of Ktrans and ve (11–13). In a previous study (11), we have shown that it is feasible and practical to use a limited-population-based average AIF for kinetic modeling of osteosarcoma DCE-MRI data of a larger population. For R1 measurement, two methods are often used: a) two-point R1 determination method (15, 16) by comparing signal intensities of the T1-weighted DCE-MRI images with those of the proton density images acquired prior to contrast injection or b) multiple flip angle method (17). In the former approach, however, proton density images are sometimes not collected due to time constraints in the clinical setting or poor planning for a clinical DCE-MRI study. This results in great difficulty in retrospective quantitative analysis of DCE-MRI data. Furthermore, possible patient motion between the acquisitions of the proton density and the DCE-MRI series may cause errors in R1 estimation or require motion correction when comparing signal intensities of the two image series. In the latter approach, any inconsistency between flip angle images such as scale factor mis-calibration or motion can lead to major systematic errors in R1 measurement (18). A recent study (18) demonstrates that fixing the pre-contrast R1, R10, makes it possible to obtain more stable measures of the vascular properties using DCE-MRI, while still yielding pharmacokinetic results consistent with those when actual R10 values were measured.
In this study, R10 values were measured using the two-point R1 determination method in a limited population of patients with osteosarcomas. We sought to assess the feasibility of calculating R1 values of the DCE-MRI time course using a limited-population-based average R10 for the purpose of pharmacokinetic modeling of osteosarcoma DCE-MRI data of a larger population.
Materials and Methods
Patients
Prior to definitive surgery, 18 patients (mean age: 16 years, range: 10–29 years) with osteosarcomas in the lower extremity underwent a routine clinical MRI protocol, in which a DCE-MRI scan was added for the purpose of evaluating the efficacy of chemotherapy in inducing tumor necrosis. The DCE-MRI study was conducted under an Institutional Review Board-approved protocol, and the written consent was obtained from each patient prior to the DCE-MRI scan.
Data Acquisition
All the MRI studies were performed with a 1.5T GE Excite system (General Electric Medical Systems, Milwaukee, WI, USA). An extremity knee coil was used for RF transmission and reception. Before the DCE-MRI study was conducted, a standard clinical MRI exam was performed through the tumor. Axial T1-weighted and fat-suppressed fast spin-echo T2-weighted images were obtained with as small a field of view (FOV) as possible. Longitudinal (coronal and/or sagittal) T1-weighted and fat-suppressed fast spin-echo T2-weighted images through the entire bone were also obtained with a small FOV. These were followed by proton density MRI and the T1-weighted DCE-MRI study in the sagittal plane, and then post-contrast axial fat-suppressed T1-weighted MRI. For DCE-MRI data acquisition, a fast multiplanar spoiled gradient echo sequence was employed with a 30o flip angle (α), 2.9 ms TE, 7.5–9.0 ms TR, 20–24 cm FOV, and 256×128 matrix size zero filled to 256×256 during image reconstruction. The entire tumor was imaged with 8–11 sagittal slices of 10–12 mm thickness and zero gap. The total DCE-MRI acquisition time was about 5–10 min with 7–10 sec temporal resolution and 30–60 time course data points. At the beginning of the sixth image set (data point) acquisition, Gd-DTPA contrast agent (Magnevist; Berlex Laboratories, Wayne, NJ, USA) at a dose of 0.1 mmol/kg was administered intravenously at a rate of 1 cc/sec or 2 cc/sec by an MR-compatible programmable power injector (Spectris; Medrad, Indianola, PA, USA). The patient often arrived at the MRI suite with the IV catheter already in place because he/she was undergoing other clinical procedures during the same visit. The injection rate was determined according to the location and the size of the IV catheter. Proton density images were acquired for the purpose of determining R1 for each DCE-MRI data point, using the same pulse sequence with a 30o flip angle, 2.0 ms TE, 350 ms TR, and DCE-MRI-matching slice number, thickness and location.
DCE-MRI Data Analysis
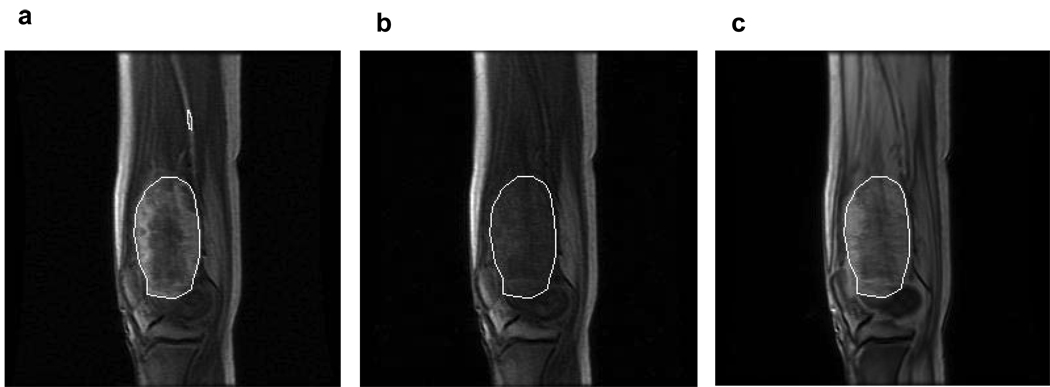
One ROI was manually drawn by an experienced radiologist (DMP) on each post-contrast DCE-MRI image slice that showed contrast enhancement in the tumor region (white ROI in Figure 1a). The ROI circumscribed the entire contrast-enhanced tumor area and was positioned in the same spatial location on the corresponding pre-contrast (baseline) DCE-MRI image slice (Figure 1b), as well as the proton density image slice (Figure 1c). If needed, motion corrections were performed by aligning the images based on anatomic features of skin, bone, and knee joint. The signal intensity from each pixel within the ROI was obtained from the proton density images and the five pre-contrast DCE-MRI images. The average of the latter was considered as the single pre-contrast intensity value. Assuming TE << T2, the signal intensity (S) of a spoiled gradient echo sequence is given by (19):
| [1] |
where S0 is a constant proportional to the proton density of the sample. By comparing the corresponding pixel S values between the pre-contrast DCE-MRI and the proton density images, pixel R10 values can be theoretically calculated using Eq. [1]. To correct for possible errors in T1 calculation likely caused by imperfect slice profile, a calibration curve of signal intensity ratio of T1-weighted image over proton density image versus T1 was constructed using a method introduced by Parker et al (15). Twelve agar gel phantoms doped with various concentrations of Gd-DTPA were imaged with the same pulse sequence and acquisition parameters as those used for DCE and proton density MRI. The T1 values for each phantom were first measured using an inversion recovery spectroscopy sequence, covering a range of 105 to 2224 msec. The twelve data points were empirically fitted with a biexponential function with offset (15) to generate the calibration curve. The pixel R10 (1/T10) values within the multi-slice tumor ROIs were obtained from the calibration curve. The average of these values gave the average R10 value for one tumor region. Measurement of tumor R10 for each of the 18 patients resulted in R10 = 0.87 ± 0.29 s−1 (mean ± SD) for this population of lower extremity osteosarcomas with a range of 0.58 to 1.62 s−1 . For pharmacokinetic modeling of the DCE-MRI data, the R1 value for each time course data point, R1(t), was converted to Gd-DTPA concentration using the following linear equation:
| [2] |
where Ct(t) is the tumor tissue Gd-DTPA concentration at time t, and r1 is the contrast agent relaxivity which was taken to be 4.1 sec−1 (mmol/L)−1 at 1.5T (20).
Figure 1.
Sagittal images from a patient with an osteosarcoma in the distal femur: (a) A post-contrast image extracted from a multi-slice dynamic contrast-enhanced (DCE) MRI acquisition, with the white ROI circumscribing the contrast-enhanced tumor. The yellow ROI was placed within the adjacent femoral artery for arterial input function (AIF) data sampling. (b) The pre-contrast image from the same DCE-MRI series with the same location as in panel a. (c) A proton density image acquired prior to DCE-MRI with the same location and slice thickness as panels a and b. The white ROIs in panels b and c were positioned in the same spatial location as in panel a.
For the individual R1 measurement approach, the R1 values for all the DCE-MRI time course data points, including both pre- and post-contrast phases, were obtained with the two-point R1 determination method (15, 16) by comparing signal intensities of the DCE-MRI images with those of the proton density images and using the T1 calibration curve. For the average R10 approach, the R1 value for each DCE-MRI time point was calculated using the following equation derived from Eq. [1], assuming for each patient R10 was uniformly equal to the average value, 0.87 s−1 , for each ROI and each pixel within the ROI:
| [3] |
where Spre is the pre-contrast S.
The biexponential AIF was constructed from data sampled in a ROI placed within a femoral artery (yellow ROI in Figure 1a) that was adjacent to the tumor (11). The Ct(t) time course (obtained through either the individual R1 measurement or the average R10 approach) and an average AIF [obtained from individual measurements in five patients, Figure 2 of (11)] based on 2 cc/sec contrast injection rate were subjected to kinetic modeling using the Toft’s model (21). We have shown that it is feasible and reasonable to use limited-population-based average AIF for quantitative analysis of lower extremity osteosarcoma DCE-MRI data obtained with either 1 or 2 cc/sec contrast injection rate (11). An in-house IDL (6.0 version; Research Systems, Boulder, CO, USA) program was used to fit the Ct(t) time course for the extraction of the Ktrans and ve parameters, as shown in the following Kety-Schmidt type of rate law equation:
| [4] |
where Cp(t’) is the plasma Gd-DTPA concentration time course, i.e. AIF. The term that includes plasma volume fraction (vp), vpCp(t), is ignored on the right hand side of this equation. Generally, ve is significantly greater than vp. Simulations have shown that, for a relatively long DCE-MRI acquisition, such as the one in this study, the signal change during the DCE time course is affected mostly by the Ktrans and ve terms (22). The vp term contributes mainly only to the early signal rising part of the time course and becomes important for curve fitting when the DCE time course is short (22). Further, it has been shown that when there is sufficient contrast agent extravasation from plasma to interstitium, such as in tumor tissue, the Ktrans and ve parameters are adequate for pharmacokinetic modeling of DCE-MRI data (22). Whole tumor Ktrans and ve values were calculated by averaging ROI Ktrans and ve values on each image slice, respectively, weighted by the number of pixels in each ROI. Histogram analyses (14) of the pixel Ktrans and ve values within the entire tumor were also performed and the median values of these parameters were calculated.
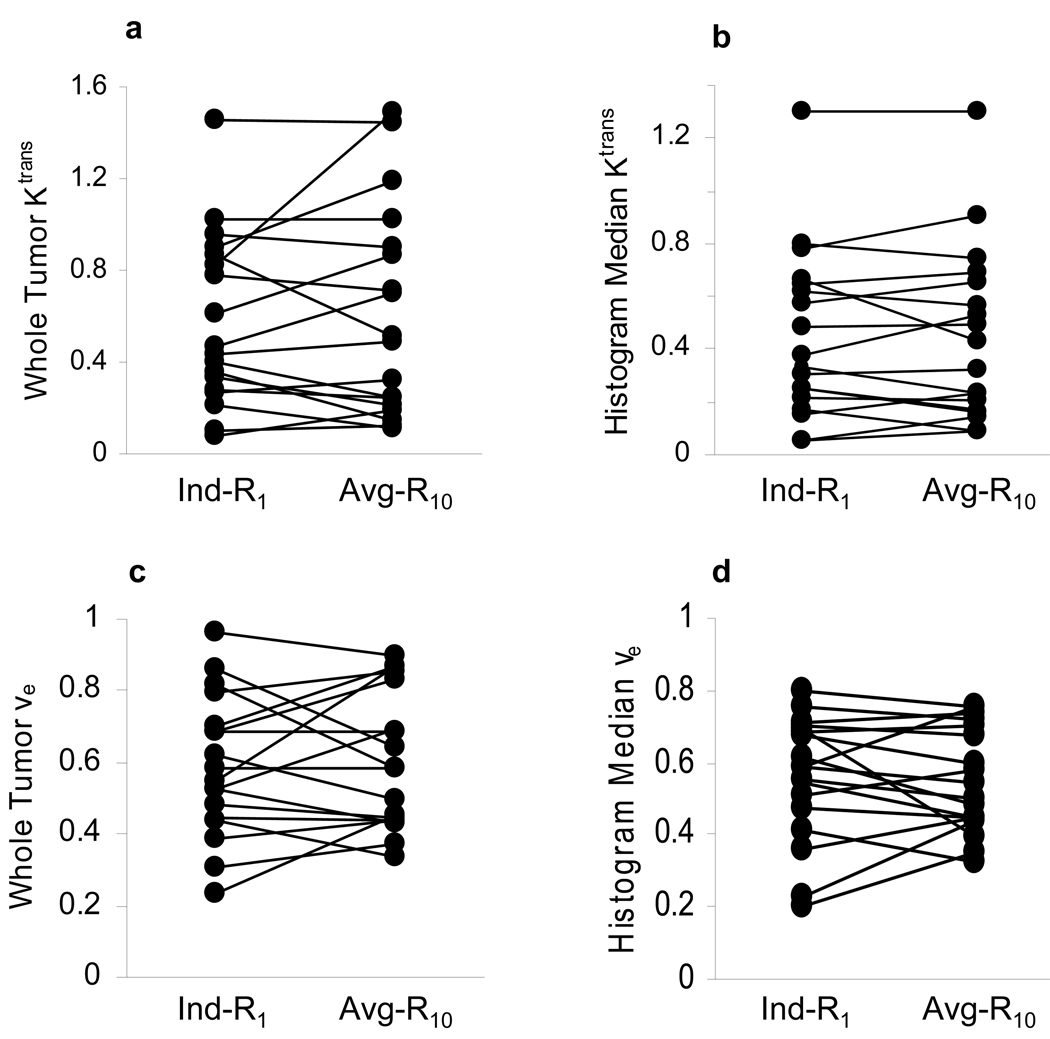
Figure 2.
Scatter plots of Ktrans (min−1) and ve parameters obtained from pharmacokinetic modeling analyses of DCE-MRI data from eighteen patients with lower extremity osteosarcomas. Kinetic analyses with the individual R1 measurement (Ind-R1) approach and the average R10 (Avg-R10) approach were performed for each study, yielding whole tumor ROI Ktrans (a) and ve (c), and median values from histogram analyses of pixel Ktrans (b) and ve (d) parameters within the tumor. The straight lines connect data points from the same patient.
Student paired t test was used to evaluate differences in pharmacokinetic parameters resulted from the individual R1 measurement approach and the average R10 approach.
Simulations
To further validate the average R10 approach, simulations were performed for kinetic modeling of the DCE-MRI signal time courses from three patients. The measured tumor R10 values for the two patients were 0.58 s−1 and 1.62 s−1, respectively, representing the lowest and highest ends of the R10 range for the study population. The R10 value for the other patient was 0.92 s−1, approximately the average R10 (0.87 s−1) of the group. The time course data from each patient was fitted with the fixed average AIF and assumed R10 values varied from 0.58 s−1 to 1.62 s−1 with 0.04 s−1 increment. Whole tumor Ktrans and ve values were derived for each assumed R10 value.
Results
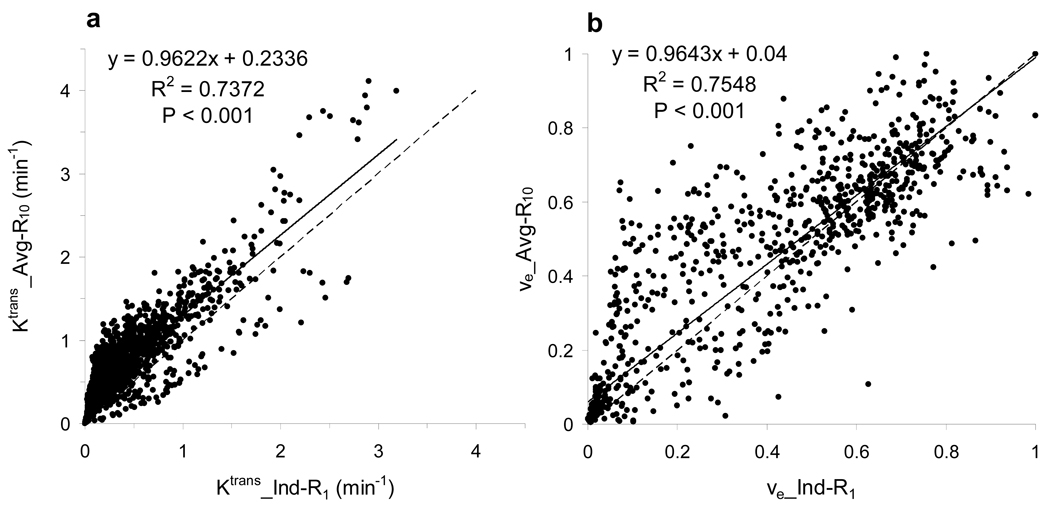
Figure 2 shows scatter plots of (a) whole tumor Ktrans, (b) histogram median Ktrans, (c) whole tumor ve, and (d) histogram median ve. The left column represents the parameter values derived from kinetic modeling of the DCE-MRI data with the R1 time courses measured individually (Ind-R1) using the two-point R1 determination method, while the right column represents those obtained with the R1 time courses calculated from Eq. [3] using the average R10 (Avg-R10). The straight lines connect the data points from the same patient. There are no statistically significant differences between Ktrans parameters derived with the Ind-R1 approach and those derived with the Avg-R10 approach (p = 0.55 for whole tumor Ktrans and 0.87 for histogram median Ktrans, paired t test). The same conclusion applied to the ve values (p = 0.64 for whole tumor ve and 0.72 for histogram median ve, paired t test). The comparisons are summarized in Table 1. No significant changes in Ktrans and ve parameters occurred when the Avg-R10 approach was used for R1 time course determination. Figure 3 displays the representative graphs from one patient study, showing pixel Ktrans (a) and ve (b) values within the whole tumor obtained with the Avg-R10 approach plotted against those obtained with the Ind-R1 approach. There were a total of 2174 pixels within the whole tumor. Both plots demonstrate significant linear correlations (p < 0.001) with the slope values close to one (0.962 for the Ktrans plot and 0.964 for the ve plot). Similar results were obtained from the other seventeen patients. The above results indicate that the use of Avg-R10 for R1 time course determination works equally well for both ROI and pixel-by-pixel data analyses.
Table 1.
Comparisons of Ktrans and ve Obtained with Ind-R1 and Avg-R10 Approaches
| R1 Determination | Ind-R1 | Avg- R10 |
|---|---|---|
| Whole Tumor Ktrans (min−1) | 0.57 ± 0.37 | 0.61 ± 0.46a |
| Histogram Median Ktrans (min−1) | 0.45 ± 0.32 | 0.44 ± 0.33b |
| Whole Tumor ve | 0.59 ± 0.20 | 0.61 ± 0.19c |
| Histogram Median ve | 56 ± 0.17 | 0.55 ± 0.14d |
Mean ± SD; Student paired t test for Ktrans and ve values obtained with the Ind-R1 and the Avg-R10 approaches:
p=0.55
p=0.87
p=0.64
p=0.72.
Figure 3.
Scatter plots of pixel (a) Ktrans and (b) ve parameters from one patient with lower extremity osteosarcoma. The Ktrans and ve parameters derived with the average R10 (Avg-R10) approach are plotted against those derived with the individual R1 measurement (Ind-R1) approach. The analyzed pixels (n = 2174) are within the contrast-enhanced tumor tissue region. The solid straight lines represent linear correlations, while the dashed ones are lines of identity.
Table 2 lists the whole tumor Ktrans and ve ranges and the standard deviations (SDs) of the study population obtained with the Ind-R1 approach, and the simulation results of the time course data from the three tumors. Compared to the experimental results, the Ktrans and ve ranges and SDs from simulations with presumed R10 value across the entire R10 range were much smaller, whether the actually measured R10 was at the lowest or highest end of the spectrum, or near the mean.
Table 2.
Experimental and Simulation Results for Whole Tumor Pharmacokinetic Analysis
| Ind-R1 | Simulation 1 | Simulation 2 | Simulation 3 | |
|---|---|---|---|---|
| Ktrans range (min−1) | 0.08–1.45 | 0.09–0.44 | 0.53–1.28 | 0.30–1.02 |
| Ktrans SD (min−1) | 0.37 | 0.11 | 0.23 | 0.22 |
| ve range ve | 0.24–0.96 | 0.27–0.62 | 0.70–0.94 | 0.46–0.79 |
| SD | .20 | 0.10 | 0.07 | 0.10 |
SD: standard deviation; Simulations 1, 2, and 3 represent simulations performed with the DCE-MRI time course data from the tumors with measured R10 values of 0.58, 0.92, and 1.62 s−1, respectively.
Discussion
Using the two-point R1 determination method (15, 16), individual R10 values were measured from 18 patients with lower extremity osteosarcomas. Post-surgical pathology analyses of this patient population revealed a wide range of tumor necrosis percentage, 10 – 100%. Consequently, the measured R10 values had a relatively broad range: 0.58 – 1.62 s−1. This is expected, as the more solid tumor tissue and the more fluid-like necrotic region can have quite different R1 values. However, the results of this study show that pharmacokinetic modeling of the DCE-MRI data yielded pharmacokinetic parameters that were not significantly different whether individual R1 value was measured or a fixed R10 (average R10) was used to calculate the R1 value for the DCE-MRI time course. The experimental results were further validated by simulations of the time course data from three tumors whose measured R10 values were at the lowest and highest ends of the R10 range, and near the mean, respectively. With smaller SDs, the Ktrans and ve ranges from the simulations were smaller than, and within those obtained from the individual R1 measurement approach. Therefore, both the experimental and simulation results indicate the feasibility of using a fixed, limited-population-based average R10 for pharmacokinetic modeling (ROI or pixel-by-pixel analysis) of lower extremity osteosarcoma DCE-MRI data from a larger population.
In pharmacokinetic modeling of the DCE-MRI data, the tumor Ct(t) time course is submitted to quantitative analysis along with the AIF. As shown in Eq. [2], Ct(t) is determined by ΔR1(t) which is [R1(t) - R10]. Therefore, the accuracy of AR1(t) determination directly affects the accuracies of derived pharmacokinetic parameters. With the average R10 approach, R1(t) is calculated from Eq. [3] using a fixed average R10 value. For an individual DCE-MRI study of one specific tumor, the difference between the actual R10 and the average R10 could be quite substantial, potentially causing significant errors in calculated R1(t). However, the errors in both R1(t) and R10 determinations may originate from the same source and therefore partially negate each other when ΔR1(t) is calculated. This possibly explains why the pharmacokinetic parameters obtained with the individual R1 measurement and the average R10 approaches are not significantly different from each other. This study was conducted in a population of 18 patients with lower extremity osteosarcomas, and with one specific set of flip angle (α) and TR for DCE-MRI acquisition. Further validation of the average R10 approach with different types of cancer and different data acquisition parameters is desirable. In a breast DCE-MRI study (23), Tofts et al. pointed out the importance of R10 measurement for kinetic modeling of the signal time-course data, showing that errors in assumed parameter values, such as R10, r1, TR, α, contrast dose, caused propagational errors in derived Ktrans and ve values. Thus, the rigorous approach of R10 measurement should always be adopted when reliable measurements can be carried out. However, in that study the TR (50 ms) and α (60°) values used in simulation and actual data acquisition are quite different from those used in this study. The types of cancer studied are also different. Our simulation and experimental results show that with the set of acquisition parameters employed for this study, the errors in estimations of Ktrans and ve caused by the use of average R10 in place of measured individual R10, if any, are not significant. It is worth noting that a breast DCE-MRI study using a fixed average R10 for pharmacokinetic modeling has been showing consistent and promising results of discriminating malignant and benign breast tumors using the Ktrans parameter (4, 24, 25).
Using multiple flip angle method for R1 measurement, Haacke et al. (18) demonstrated, with both simulated data and actual liver and muscle DCE-MRI data, that noise in the experiments led to spread of R10 estimates and unstable Ct(t) curves, which could be effectively eliminated by forcing R10 to a fixed value. These authors have also shown that fixing R10 not only yields more consistent results when evaluating DCE-MRI data, but also is useful in analyzing data collected with wrong flip angle scale factors or patient motion prior to DCE-MRI data acquisition. The results from this study are consistent with such findings even though the two-point R1 determination method was used instead of the multiple flip angle approach. In our osteosarcoma DCE-MRI protocol, the proton density images are acquired immediately before the DCE-MRI series. Any inconsistency in flip angle scaling between the two acquisitions can lead to errors in R1 estimation. Furthermore, since the tumor cannot be easily delineated on proton density image, any slight patient movement, which is more likely to happen between scans than during a scan, can cause difficulty in spatially registering the tumor ROI on the proton density and the DCE-MRI images for the purpose of measuring R1 by comparing signal intensity in the same ROI or pixel. This will result in errors in R1 measurement as well. Thus, it is quite reasonable, and sometimes necessary to use the average R10 approach for determination of the R1(t) time course.
DCE-MRI examination is often included in clinical MRI protocols to study cancer. In such clinical studies, data collection schemes are usually planned solely for the purpose of qualitative or semi-quantitative analysis of DCE-MRI data. Thus, data may not be available for actual R1 measurement. One clear advantage of the average R10 approach is to enable retrospective pharmacokinetic modeling of such data by using a fixed average R10 and Eq. [3] if a spoiled gradient echo type of sequence was used for DCE-MRI acquisition. However, it is important to identify the appropriate situation to use a fixed average R10 value for DCE-MRI data analysis. For instance, in a longitudinal DCE-MRI study to evaluate the effects of chemotherapy through monitoring the changes in tumor necrosis, the individual R1 measurement approach may be a wiser choice if can be done accurately, as R10 values can be quite different pre- and post-treatment. Use of the average R10 approach may lead to errors in estimating changes of pharmacokinetic parameters in such a study.
Acknowledgments
We thank Drs. Amita Dave and Yousef Mazaheri Tehrani for providing the data for the T1 calibration curve, Ms. Maayan Korenblit and Ms. Melissa Potuzak for managing the patient data base and the IRB protocol. This study was conducted with support from National Cancer Institute/National Institutes of Health (grant number 1 R01 CA104754).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leach MO, Brindle KM, Evelhoch JL, Griffiths JR, Horsman MR, Jackson A, Jayson GC, Judson IR, Knopp MV, Maxwell RJ, McIntyre D, Padhani AR, Price P, Rathbone R, Rustin GJ, Tofts PS, Tozer GM, Vennart W, Waterton JC, Williams SR, Workman P. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. Br J Cancer. 2005;92:1599–1610. doi: 10.1038/sj.bjc.6602550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knopp MV, Weiss E, Sinn HP, Mattern J, Junkermann H, Radeleff J, Magener A, Brix G, Delorme S, Zuna I, van Kaick G. Pathophysiologic basis of contrast enhancement in breast tumors. JMRI. 1999;10:260–266. doi: 10.1002/(sici)1522-2586(199909)10:3<260::aid-jmri6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Buckley DL, Roberts C, Parker GJM, Logue JP, Hutchinson CE. Prostate cancer: evaluation of vascular characteristics with dynamic contrast-enhanced T1-weighted MR imaging - initial experience. Radiology. 2004;233:709–715. doi: 10.1148/radiol.2333032098. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Huang W, Yankeelov TE, Tudorica A, Rooney WD, Springer CS. Shutter-speed analysis of contrast reagent bolus-tracking data: preliminary observations in benign and malignant breast disease. Magn Reson Med. 2005;53:724–729. doi: 10.1002/mrm.20405. [DOI] [PubMed] [Google Scholar]
- 5.Chang EY, Li X, Jerosch-Herold M, Priest RA, Enestvedt CK, Xu J, Springer CS, Jr, Jobe BA. The evaluation of esophageal adenocarcinoma using dynamic contrast-enhanced magnetic resonance imaging. J Gastrointest Surg. 2007;11:166–175. doi: 10.1007/s11605-007-0253-5. [DOI] [PubMed] [Google Scholar]
- 6.Galbraith SM, Rustin GJ, Lodge MA, Taylor NJ, Stirling JJ, Jameson M, Thompson P, Hough D, Gumbrell L, Padhani AR. Effects of 5,6-dimethylxanthenone-4-acetic acid on human tumor microcirculation assessed by dynamic contrast-enhanced magnetic resonance imaging. J Clin Oncol. 2002;20:3826–3840. doi: 10.1200/JCO.2002.09.144. [DOI] [PubMed] [Google Scholar]
- 7.Morgan B, Thomas AL, Drevs J, Hennig J, Buchert M, Jivan A, Horsfield MA, Mross K, Ball HA, Lee L, Mietlowski W, Fuxius S, Unger C, O’Byrne K, Henry A, Cherryman GR, Laurent D, Dugan M, Marme D, Steward WP. Dynamic contrast-enhanced magnetic resonance imaging as a biomarker for the pharmacological response of PTK787/ZK 222584, an inhibitor of the vascular endothelial growth factor receptor tyrosine kinases, in patients with advanced colorectal cancer and liver metastases: results from two phase I studies. J Clin Oncol. 2003;21:3955–3964. doi: 10.1200/JCO.2003.08.092. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Rugo HS, Wilding G, McShane TM, Evelhoch JL, Ng C, Jackson E, Kelcz F, Yeh BM, Lee FT, Jr, Charnsangavej C, Park JW, Ashton EA, Steinfeldt HM, Pithavala YK, Reich SD, Herbst RS. Dynamic contrast-enhanced magnetic resonance imaging as a pharmacodynamic measure of response after acute dosing of AG-013736, an oral angiogenesis inhibitor, in patients with advanced solid tumors: results from a phase I study. J Clin Oncol. 2005;23:5464–5473. doi: 10.1200/JCO.2005.04.143. [DOI] [PubMed] [Google Scholar]
- 9.Mross K, Drevs J, Muller M, Medinger M, Marme D, Hennig J, Morgan B, Lebwohl D, Masson E, Ho YY, Gunther C, Laurent D, Unger C. Phase I clinical and pharmacokinetic study of PTK/ZK, a multiple VEGF receptor inhibitor, in patients with liver metastases from solid tumors. Eur J Cancer. 2005;41:p1291–p1299. doi: 10.1016/j.ejca.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Thomas AL, Morgan B, Horsfield MA, Higginson A, Kay A, Lee L, Masson E, Puccio-Pick M, Laurent D, Steward WP. Phase I study of the safety, tolerability, pharmacokinetics, and pharmacodynamics of PTK787/ZK 222584 administered twice daily in patients with advanced cancer. J Clin Oncol. 2005;23:4162–4171. doi: 10.1200/JCO.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Huang W, Panicek DM, Schwartz LH, Koutcher JA. Feasibility of Using Limited-Population-Based Arterial Input Function for Pharmacokinetic Modeling of Osteosarcoma Dynamic Contrast-Enhanced MRI Data. Magn Reson Med. 2008;59:1183–1189. doi: 10.1002/mrm.21432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou R, Pickup S, Yankeelov TE, Springer CS, Glickson JD. Simultaneous measurement of arterial input function and tumor pharmacokinetics in mice by dynamic contrast enhanced imaging: effects of transcytolemmal water exchange. Magn Reson Med. 2004;52:248–257. doi: 10.1002/mrm.20143. [DOI] [PubMed] [Google Scholar]
- 13.Yankeelov TE, Rooney WD, Li X, Springer CS. Variation of the relaxographic “Shutter-Speed” for transcytolemmal water exchange affects the CR bolus-tracking curve shape. Magn Reson Med. 2003;50:1151–1169. doi: 10.1002/mrm.10624. [DOI] [PubMed] [Google Scholar]
- 14.Dyke JP, Panicek DM, Healey JH, Meyers PA, Huvos AG, Schwartz LH, Thaler HT, Tofts PS, Gorlick R, Koutcher JA, Ballon D. Osteogenic and Ewing sarcomas: estimation of necrotic fraction during induction chemotherapy with dynamic contrast-enhanced MR imaging. Radiology. 2003;228:271–278. doi: 10.1148/radiol.2281011651. [DOI] [PubMed] [Google Scholar]
- 15.Parker GJM, Suckling J, Tanner SF, Padhani AR, Revell PB, Husband JE, Leach MO. Probing tumor microvascularity by measurement, analysis and display of contrast agent uptake kinetics. J Magn Reson Imaging. 1997;7:564–574. doi: 10.1002/jmri.1880070318. [DOI] [PubMed] [Google Scholar]
- 16.Parker GJ, Tofts PS. Pharmacokinetic analysis of neoplasms using contrast-enhanced dynamic magnetic resonance imaging. Top Magn Reson Imaging. 1999;10:130–142. doi: 10.1097/00002142-199904000-00006. [DOI] [PubMed] [Google Scholar]
- 17.Li KL, Zhu XP, Waterton J, Jackson A. Improved 3D quantitative mapping of blood volume and endothelial permeability in brain tumors. J Magn Reson Imaging. 2000;12:347–357. doi: 10.1002/1522-2586(200008)12:2<347::aid-jmri19>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Haacke EM, Filleti CL, Gattu R, Ciulla C, Al-Bashir A, Suryanarayanan K, Li M, Latif Z, DelProposto Z, Sehgal V, Li T, Torquato V, Kanaparti R, Jiang J, Neelavalli J. New algorithm for quantifying vascular changes in dynamic contrast-enhanced MRI independent of absolute T1 values. Magn Reson Med. 2007;58:463–472. doi: 10.1002/mrm.21358. [DOI] [PubMed] [Google Scholar]
- 19.Haase A. Snapshot FLASH MRI. Application to T1, T2, and chemical-shift imaging. Magn Reson Med. 1990;13:77–89. doi: 10.1002/mrm.1910130109. [DOI] [PubMed] [Google Scholar]
- 20.Yankeelov TE, Rooney WD, Huang W, Dyke JP, Li X, Tudorica A, Lee J-H, Koutcher JA, Springer CS. Evidence for shutter-speed variation in CR bolus-tracking studies of human pathology. NMR Biomed. 2005;18:173–185. doi: 10.1002/nbm.938. [DOI] [PubMed] [Google Scholar]
- 21.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7:91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Rooney WD, Springer CS. A unified magnetic resonance imaging pharmacokinetic theory: intravascular and extracellular contrast reagents. Magn Reson Med. 2005;54:1351–1359. doi: 10.1002/mrm.20684. [DOI] [PubMed] [Google Scholar]
- 23.Tofts PS, Berkowitz B, Schnall MD. Quantitative analysis of dynamic Gd-DTPA enhancement in breast tumors using a permeability model. Magn Reson Med. 1995;33:564–568. doi: 10.1002/mrm.1910330416. [DOI] [PubMed] [Google Scholar]
- 24.Huang W, Li X, Morris EA, Tudorica LA, Venkatraman ES, Wang Y, Xu J, Springer CS. Shutter-Speed DCE MRI Pharmacokinetic Analyses Facilitate the Discrimination of Malignant and Benign Breast Disease. Proc Int Soc Magn Reson Med. 2007;15:141. [Google Scholar]
- 25.Li X, Huang W, Morris EA, Tudorica LA, Rooney WD, Wang Y, Xu J, Springer CS. NMR Shutter-Speed Discrimination of Malignant and Benign Breast Tumors Using ROI Data. Proc Int Soc Magn Reson Med. 2008;16:584. [Google Scholar]