Abstract
Co-amplification at chromosomes 8p11-8p12 and 11q12-11q14 occurs often in breast tumors, suggesting possible cooperation between genes in these regions in oncogenesis. We used high resolution array comparative genomic hybridization (array CGH) to map the minimal amplified regions. The 8p and 11q amplicons are complex and consist of at least four amplicon cores at each site. Candidate oncogenes mapping to these regions were identified by combining copy number and RNA and protein expression analyses. These studies also suggested that CCND1 at 11q13 induced expression of ZNF703 mapping at 8p12, which was subsequently shown to be mediated via the Rb/E2F pathway. Nine candidate oncogenes from 8p12 and four from 11q13 were further evaluated for oncogenic function. None of the genes individually promoted colony formation in soft agar or collaborated with each other functionally. On the other hand, FGFR1 and DDHD2 at 8p12 cooperated functionally with MYC, while CCND1 and ZNF703 cooperated with a dominant negative form of TP53. These observations highlight the complexity and functional consequences of the genomic rearrangements that occur in these breast cancer amplicons, including transcriptional cross-talk between genes in the 8p and 11q amplicons, as well as their cooperation with major pathways of tumorigenesis.
Introduction
Amplifications involving chromosomes 8p (RAB11FIP1, FGFR1), 11q (CCND1) and 17q (ERBB2) are among the most common high level copy number aberrations in breast tumors, occurring, for example, in one study, in 22.8%, 19.6% and 9.9% of tumors, respectively (Letessier et al., 2006). Amplification of 8p and 11q are most often observed in estrogen receptor positive tumors, while amplification of 17q (ERBB2) occurs in both estrogen receptor positive and negative tumors (Fridlyand et al., 2006; Letessier et al., 2006; Loo et al., 2004). Poor prognosis is associated with the presence of these amplicons in breast cancer. Thus, overexpressed genes within amplicons are attractive targets for therapy, as exemplified by the targeted use of herceptin to treat patients with tumors with ERBB2 amplification.
Frequently, two or more of these amplicons are present in a given tumor, suggesting that genes in the two amplified regions may collaborate in formation of the transformed phenotype (Yang, Moffa, Haddad, Streicher and Ethier, 2007). Of interest in this regard, amplification of FGFR1 at 8p12 and CCND1 at 11q13 occurs preferentially in breast cancers, with amplification of 8p12 having been reported in 30–40% of tumors with CCND1 amplification. Co-amplification of these genes is associated with significantly reduced patient survival (Cuny et al., 2000). In some cases, cytogenetic findings are consistent with physical co-amplification of the regions (Bautista and Theillet, 1998; Paterson et al., 2007), but selection for elevated copy number of a translocation-fusion gene is not thought to drive amplification (Paterson et al., 2007). Amplification and overexpression of a number of candidate oncogenes mapping at 8p12 and 11q13 are associated with the luminal breast cancer subtype (Adelaide et al., 2007). These observations raise the question as to whether collaboration between oncogenes within the two amplified regions from each chromosome in formation of the transformed phenotype provides a selective advantage for co-amplification. Here, we investigated this question by comprehensively profiling the 8p12 and 11q13 amplicons to define the recurrent regions of amplification and to identify candidate oncogenes. Subsequently, we evaluated these genes for their ability to (a) transform MCF10A cells by themselves, (b) in combination with genes from the other chromosome and (c) with genes involved in major pathways of cancer.
Results
Copy number profiling of tumors with amplicons at 8p12 and 11q13
We assembled a microarray with 64 BACs providing near tiling path coverage of the 9 Mb region of chromosome 8p11-p12 from RNF122 to the centromere and 101 BACs at 11q12-q14. The 8p12 or 11q13 amplicons identified in tumors and cell lines using our standard genome scanning arrays of 2464 BACs (Snijders et al., 2001) were then fine mapped on this 8p11q array. The analysis of six tumors with 8p amplification from our previous study (Fridlyand et al., 2006) allowed us to define four amplicon cores in the 8p11-p12 region (Figure 1A, Supplementary Table 1). Further analysis of a second set of breast tumors (Climent et al., 2007) in which 8p amplification was present in nine tumors yielded a similar result (Supplementary Table 1). The minimal amplified regions are summarized in Table 1. Amplicon cores A1 and A2 occurred most frequently, each being present in 7% of tumors (n=360) from three breast cancer array CGH datasets (Chin et al., 2006; Climent et al., 2007; Fridlyand et al., 2006). The minimal amplicon core A1 spanned 0.4 Mb as defined by tumor S0131 and cell line SUM225 (Supplementary Table 1). Amplification of the two more centromere proximal amplicon cores, A3 and A4 occurred less frequently (5% of tumors, n=360).
Figure 1.
Copy number profiles of 8p and 11q amplicons. Copy number profiles of selected tumors and cell lines are shown as determined by array CGH on the 8p11q BAC array. The centers of amplicon cores are indicated together with genes in each core region. (A) Amplicons at 8p11-p12. (B) Amplicons at 11q13-q14.
Table 1.
Minimal amplicon cores at 8p12 and 11q13
| Amplicon | Flanking clone | Genes in (or partially in) flanking clone or amplicon core |
|---|---|---|
| 8p A1 | RP11-101H15 | no genes |
| ZNF703, ERLIN2, PROSC, GPR124, BRF2, RAP11FIP1 | ||
| CTC-497A2 | GOT1L1, ADRB3, EIF4EBP1 | |
|
| ||
| 8p A2 | RP11-90P5 | ASH2L, STAR, LSM1, BAG4, (DDHD2) |
| DDHD2, PPAPDC1B, WHSC1L1, FGFR1 | ||
| RP11-647P1 | TACC1, (PLEKHA2) | |
|
| ||
| 8p A3 | RP11769N8 | ADAM18 |
| ADAM2, INDO1, INDOL1, C8orf4 | ||
| RP11-51K12 | ZMAT4 | |
|
| ||
| 8p A4 | RP11-407N24 | GOLGA7, GINS4, (AGAT6) |
| AGAT6, NKX6-3, ANK1 | ||
| RP11-109A10 | MYST3 | |
|
| ||
| 11q A1 | RP11-40B5 | TPCN2 |
| MEOV, OCIM, CCND1 | ||
| CTD-3190C8 | ORAOV1, FGF19, BF510715, FGF4, FGF3 | |
|
| ||
| 11q A2 | RP11-368I20 | no genes |
| TMEM16A, FADD, PPFIA1, CTTN | ||
| RP11-916J3 | SHANK2 | |
|
| ||
| 11q A3.1 | RP11-133F12 | UVRAG |
| WNT11, PRKRIR, C11orf30 | ||
| RP11-115O9 | LRRC32 | |
|
| ||
| 11q A3.2 | CTD-2501F13 | C11orf30 |
| LRRC32, TSKU | ||
| RP11-259H21 | PHCA | |
|
| ||
| 11q A4 | CTD-2504L8 | PHCA, B3GNT6, OMP, CAPN5 |
| MYO7A, GDPD4, PAK1, AQP11, CLNS1A, RSF1, C11orf67 | ||
| RP11-7I15 | INTS4, KCTD14, THRSP, NDUFC2 | |
Eighteen tumors with amplification at 11q13 were analyzed on the 8p11q array and the 11q13-q14 copy number profiles generally showed four regions of amplification (Table 1) extending distally from the region encompassing CCND1 (Figure 1B). An amplicon core mapping more proximally to CCND1 was also present. Variably positioned amplicons between A2 and A3 were also observed. The A3 amplicon was further subdivided into two sub amplicon cores, A3.1 and A3.2. Three tumors (J3891, S0065 and S0081, Supplementary Table 1, Supplementary Figure 1) defined a minimal amplified region bracketed by UVRAG and LRRC32 including C11orf30, whereas a minimal amplified region bracketed by C11orf30 and PHCA, including LRRC32 was present in two other tumors (J1144 and J5683, Supplementary Table 1, Supplementary Figure 1). Amplification at 11q13 varied in the number of cores which were present (Supplementary Figure 2). For example, in five tumors, J3981, J1901, S0132, S1534 and S1598 all four cores were present, whereas only A1 and A2 were amplified in J1333. In two tumors (J363 and J665), high level copy number was restricted to only A2 (Supplementary Table 1 and Supplementary Figures 2 and 3). Thus, the amplicon structure at 11q13 is highly complex; amplification of the four cores can occur independently, suggesting the presence of several driver genes for amplification.
Selection of candidate driver genes for amplification
We subjected genes in the amplicons to four tests for candidacy as driver oncogenes with the rationale that candidate oncogenes should be overexpressed when amplified, although overexpression may also occur by other means. Thus, we evaluated genes for copy number and expression by first accessioning a dataset of 90 breast tumors for which both DNA copy number and expression measurements were available (Chin et al., 2006). We then asked if expression levels of genes showed significant correlation with amplification at 8p12 or 11q13 (Supplementary Table 2). In addition, because co-amplification of 8p12 and 11q13 is frequent, we asked whether expression levels of genes mapping on 8p12 or 11q13 were responsive to copy number of the other chromosome by assessing correlation of expression of genes on the one chromosome with copy number of the other. In the second test, we carried out the same analysis on breast cancer cell lines previously profiled for transcript levels and DNA copy number (Neve et al., 2006). In the third and fourth assays, we compared transcript and protein levels in breast cancer cell lines with amplification at 8p12 alone, 11q13 alone or 8p12 and 11q13 to MCF10A (Figures 2 and 3 and Supplementary Table 3). The cell lines were profiled on the 8p11q array and included BT549 with a gain of 8p, SUM225 with amplification of 8p12 alone (Willmarth, Albertson and Ethier, 2004; Yang, Albertson and Ethier, 2004), 600MPE and MDA-MB-175 with amplification of 11q13 (Garcia et al., 2005; Snijders et al., 2001), SUM52, SUM44 and MDA-MB-134 with amplification of 8p12 and 11q13 (Garcia et al., 2005; Yang, Albertson and Ethier, 2004) (Supplementary Table 1).
Figure 2.
Expression of selected genes in the 8p12 amplicon. Transcript levels for each gene were determined by quantitative RT-PCR and normalized to GUSB. The PCR reaction conditions and primers and probes for each gene are given in Supplementary Table 4. The data are displayed as expression levels relative to MCF10A (histograms) or as expression relative to GUSB if no expression was detected in MCF10A (FKSG2, PLEKHA2, TM2D2, ADAM18, ADAM2, ZMAT4). No expression of HTRA4 was detected. Protein expression levels were determined if suitable antibodies were available (Supplementary Table 5). Western blots are shown below the RT-PCR histograms. Equal loading of lanes was confirmed by expression of β-actin (data not shown). Genes are displayed according to inclusion in 8p amplicons A1-A4 and the copy number of the locus in each cell line is shown below the name of the cell line (white = average copy number, light gray = gain, and dark gray = amplification).
Figure 3.
Expression of selected genes in the 11q13 amplicon. Transcript levels for each gene were determined by quantitative RT-PCR and normalized to GUSB. The PCR reaction conditions and primers and probes for each gene are given in Supplementary Table 4. The data are displayed as expression levels relative to MCF10A (histograms) or as expression relative to GUSB if no expression was detected in MCF10A (FGF4 and LRRC32). No expression of MRGD or FGF3 was detected in any cell line. Protein expression levels were determined if suitable antibodies were available (Supplementary Table 5). Western blots are shown below the RT-PCR histograms. Equal loading of lanes was confirmed by expression of β-actin (data not shown). Genes are displayed according to inclusion in 11q13 amplicons A1–A4 and the copy number of the locus in each cell line is shown below the name of the cell line (white = average copy number, light gray = gain, and dark gray = amplification).
The cell line, MCF10A is immortal, but not transformed and does not form colonies in soft agar. We considered genes to be overexpressed at the transcript level if levels were at least five times that in MCF10A. This cutoff was selected because cell lines typically showed amplification levels greater than four-fold (e.g. SUM225 and MDA-MB-134 are amplified eight-fold on 8p relative to the median copy number for the cell line, Figure 1A). Similarly, we expected protein expression levels of candidate driver genes to be higher in breast cancer cell lines compared to MCF10A and also to be expressed at low levels in MCF10A. The outcome of these analyses is summarized in Figure 4. In addition, three genes mapping at 8p12 (ADAM9, ADAM2 and ANK1) showed significant correlation of expression with copy number at 11q13, but no association of chromosome 11q gene expression with copy number at 8p was found (Supplementary Table 2).
Figure 4.
Expression of candidate oncogenes in 8p12 and 11q13 amplicons. Genes in the amplicons are shown in genome order. The results of the expression analyses described in the text are shown from left right, correlation of copy number with expression in the breast tumor dataset, correlation of copy number with expression in the cell line dataset, transcript and protein levels in cell lines. + = significant positive correlation, transcript overexpression or protein overexpression, 0 = no correlation or overexpression, and N = not done.
In the 8p amplicon, 10 genes met the criteria to be classified as candidate oncogenes for all assays for which they were tested, although the number of tests to which they were subjected varied from all four to only one. Thus, ZNF703, ERLIN2 (C8orf2), PROSC, RAB11FIP1, ASH2L, LSM1, BAG4, DDHD2, PPAPDC1B and WHSC1L1 were considered good candidates. We consider FGFR1 also to be a reasonably strong candidate, because it met the criteria for three of the four tests. Eight of these genes mapped within the minimal amplicon cores A1 and A2, providing further support for their candidacy as driver genes for these amplicons. Additional possible candidates include TAAC1, TM2D2 (BLP1) and ADAM18, because they variably showed some increase in expression in cell lines by quantitative RT-PCR and/or expression levels were correlated with copy number in breast tumors or cell lines. We ruled out TM2D2 (BLP1), because it maps outside the minimal amplicon cores, but TAAC1 and ADAM18 map at the distal edge of the A2 and A3 amplicon cores, respectively, and so remain possible drivers for amplification. We ruled out two additional genes, GPR124 and BRF2, because of high protein levels in MCF10A. A similar analysis of the 11q13 amplicon revealed that four genes, which were subjected to all four tests met the criteria for three of them. They included CCND1, FADD, PPFIA1 and CTTN.
Transcriptional crosstalk between chromosome 8p12 and 11q13 amplicons
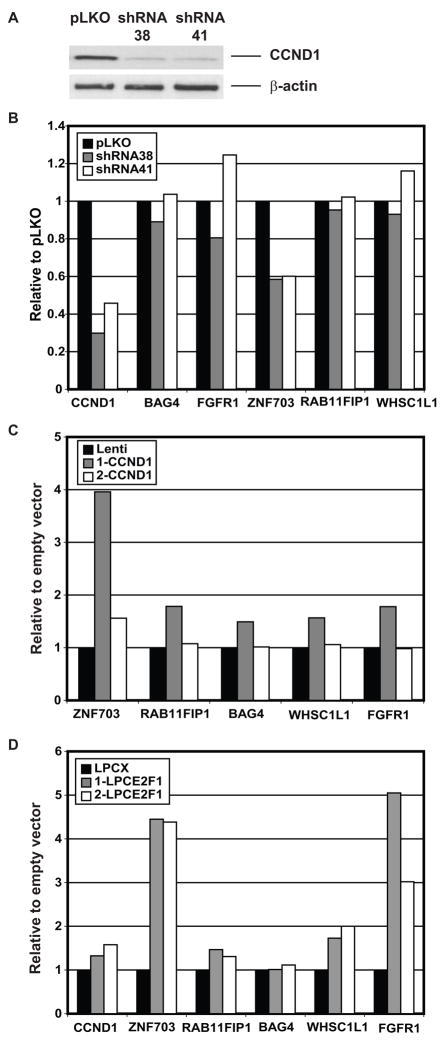
Amplification of 8p12 and 11q13 frequently occur together suggesting possible interactions between the genes in these two amplicons. Indeed, it has been reported previously that FGFR1 (at 8p12) is up-regulated by increased expression of CCND1 (at 11q13) in fibroblasts, and occurs via CCND1 mediated activation of the pRB/E2F pathway (Tashiro et al., 2003). In our evaluation of expression of genes in the 8p amplicons in cell lines, we observed increased expression of ZNF703 and RAB11FIP1 at the RNA level and BAG4, RAB11FIP1 and WHSC1L1 at the protein level in cell lines with amplification at chromosome 11q12-11q14, even when these genes, which map to 8p11-8p12 were not amplified (Figure 2). In order to determine if any of these genes could be regulated by CCND1, we assessed expression of the genes when CCND1 was knocked down in SUM44 cells, which harbor both the 8p12 and 11q13 amplicons and also following forced overexpression of CCND1 in MCF10A cells. When two different shRNAs were used to knock down the transcript and protein expression levels of CCND1 by 60% and 70%, respectively in SUM44 cells, only expression of ZNF703 decreased by 40%, while there was no change in expression of BAG4, FGFR1, RAB11FIP1, and WHSC1L1 (Figures 5A and B). This reduction in ZNF703 expression was observed at 72 and 96 hours after infection with shRNA. Nevertheless, it was not maintained after one week in cells selected for stable expression of the shRNA, even though CCND1 levels remained low compared to control infected cells (results not shown). By contrast, stable infection of MCF10A cells with a CCND1 lentivirus resulted in a maximal four-fold increase in expression of ZNF703 and 1.5-fold expression of BAG4, FGFR1, RAB11FIP1, and WHSC1L (Figure 5C). Since the reported up-regulation of FGFR1 by CCND1 occurs via activation of the pRB/E2F pathway (Tashiro et al., 2003), we investigated whether the up-regulation of ZNF703 is similarly mediated by E2F1. Overexpression of E2F1 in MCF10A consistently resulted in up-regulation of all genes tested, but much greater up-regulation of ZNF703 and FGFR1. These observations suggest that expression of at least two genes in the 8p12 amplicon, FGFR1 and ZNF703, are regulated by E2F1 and CCND1. Nevertheless, examination of a breast cancer expression array dataset comprised of all breast cancer subtypes (Chin et al., 2006) revealed no significant correlation of FGFR1 expression with either E2F1 or CCND1 expression levels (Spearman correlation < 0.25). On the other hand, when considering only luminal tumors, all four FGFR1 probesets showed some association with one or both E2F1 probesets (Spearman correlation > 0.3), and the correlation was considered significant for two of the FGFR1 probesets (Spearman correlation ≥ 0.4, FDR < 0.05), suggesting some association.
Figure 5.
Interaction between genes at 8p12 and 11q13.
A. Knockdown of CCND1 by two shRNAs. Western blot showing decreased expression of CCND1 protein in SUM44 cells stably infected with either of two different shRNA lentiviruses 38 and 41 compared to a control shRNA pLKO (upper panel) and levels of β-actin on the same membrane (lower panel).
B. Transcript levels of genes normalized to GUSB in SUM44 cells expressing CCND1 shRNAs compared to SUM44 cells infected with empty vector (pLKO).
C. Transcript levels of genes normalized to GUSB in MCF10A cells overexpressing CCND1 compared to control MCF10A cells infected with an empty vector (Lenti). The results of two separate infections of MCF10A cells (1-CCND1 and 2-CCND1) are shown. Note that expression of CCND1 in 1-CCND1 was 2.7 times the level achieved in 2- CCND1. In both experiments, expression of ZNF703 was reproducibly induced to 5% of the level of expression of CCND1.
D. Transcript levels of genes normalized to GUSB in SUM44 cells infected with E2F1 expressing retrovirus compared to control cells infected with empty vector (LPCX). The results of two separate infections of SUM44 cells (1-LPCE2F1 and 2-LPCE2F1) are shown.
Transforming activity of overexpressed genes in 8p12 and 11q13 amplicons
To evaluate possible oncogenic functions of genes in the amplicons, they were overexpressed in MCF10A and assessed for the capability to promote growth in soft agar. We selected six overexpressed genes from the A1 and A2 cores of the 8p12 amplicon (ZNF703, ERLIN2, RAB11FIP1, DDHD2, WHSC1L1 and FGFR1), three additional overexpressed genes at the boundary of A2 (ASH2L, LSM1 and BAG4), and four genes from the 11q13 amplicon (CCND1, FADD, PPFIA1 and CTTN) for study. The genes were expressed by themselves in MCF10A, and we also asked whether they collaborate with MYC or TP53, genes deregulated in breast cancer. Amplification and/or overexpression of MYC occurs in 30–50% of breast cancers and has been associated with poor prognosis and resistance to anti-estrogen therapy (Agrawal, Yang, Murphy and Agrawal, 2006). TP53 is altered in ~18% of breast cancers and can be further inactivated by deregulation of interacting genes such as MDM2 (i.e. alterations in the CDKN2A – MDM2 – TP53 pathway have been reported in 25% of breast cancers) (Ho et al., 2001). Expression of the genes individually did not promote colony formation in soft agar; however greater numbers of colonies were observed in cells co-expressing MYC and FGFR1 or DDHD2 (Figure 6A, Supplementary Figure 3). Expression of GSE56, a dominant negative mutant of TP53 together with CCND1 or ZNF703 also resulted in more than two-fold increase in numbers of colonies, while expression with CTTN enhanced colony formation, but less than two-fold compared to MCF10A cells expressing GSE56 alone (Figure 6B, Supplementary Figure 4). Because 8p12 and 11q13 are often co-amplified in breast tumors, we also assessed whether selected genes from these two amplicons cooperated with each other. Genes from 8p (ZNF703, BAG4, WHSC1L1 or FGFR1) were assayed in combination with (CCND1, FADD, CTTN or PPFIA1); however, no increase in the number of colonies was observed in any of the combinations (data not shown).
Figure 6.
Soft agar assays to evaluate the oncogenic function of genes on 8p12 and 11q13.
A. Expression of genes individually and in combination with MYC. Expression of MYC alone in MCF10A gave a few to ~40 colonies per well of a six-well plate. To control for the variability in the number of colonies in MYC expressing cells, all assays with were carried out with MYC alone control. The number of colonies from MYC expressing cells varied little within any one assay. Assays were carried out in triplicate with two different cell lines expressing MYC and each gene generated from two independent infections. Shown are the numbers of colonies per six-well plate (mean and standard deviation) for MCF10A cells stably infected with vector control (empty vector LentiV6 or LPCX) or vectors expressing individual genes, and MCF10A cells stably co-infected with control pBabeH (empty vector) plus appropriate corresponding empty vector for a candidate gene or co-expressing pBabeH-MYC and the selected genes.
B. Expression of genes in combination with GSE56. Expression of GSE56 alone in MCF10A cells produced colonies in soft agar that differed greatly in size, which by western-blotting of clonal cell lines derived from the GSE56-infected MCF10A appeared to result from variable expression levels of GSE56. Therefore, candidate genes were expressed in two different clonal cell lines, one expressing a high level of GSE56 in LXSN (56SN-1) and the other a moderate level (56SN-2). Shown are the numbers of colonies per six-well plate (mean and standard deviation) for MCF10A cells stably infected with vector control (56SN-1 or 56SN-2 and either empty vectors LPCX or LentiV6, V6) or vectors expressing individual genes together with either of the two GSE56-expressing MCF10A clones (56SN-1 or 56SN-2). Cells expressing CCND1 or ZNF703 alone do not form colonies in soft agar (Supplementary Figure 2).
Discussion
The 8p12 amplicon has been the subject of a number of studies using high resolution mapping by tiling path BAC array CGH and FISH. Three studies (Garcia et al., 2005; Prentice et al., 2005) concluded that there was a single minimal amplicon encompassing ZNF703 (FLJ14299), ERLIN2 (SPFH2, C8orf2), BRF2 and RAB11FIP1 and corresponding to the A1 core defined here. On the other hand, Gelsi-Boyer and colleagues (Gelsi-Boyer et al., 2005) defined four overlapping amplicon cores at 8p11-p12, with their most telomeric amplicon cores, A1 and A2 corresponding to the A1 and A2 cores defined here. There is less agreement on the definition of the minimal amplicon cores A3 and A4 in the two studies, as we considered them to map more distally (Table 1). Amplification of cores largely similar to A1 and A2 have also been observed in familial breast cancer (Melchor et al., 2007). A partial explanation for the failure of Garcia et al. (Garcia et al., 2005) and Pole et al. (Garcia et al., 2005) to find multiple amplicons can in part be due to the use of an array in these two studies that spanned the region from WRN to ZMAT4 and so excluded the A4 core. On the other hand, Prentice et al. (Prentice et al., 2005) studied only five samples on a whole genome tiling path array and may have observed only the A1 amplicon core due to the small sample size. By contrast, Haverty and colleagues (Haverty et al., 2008) recently reported that their data on ~50 samples did not support the existence of multiple cores, as only the more frequently amplified A1 amplicon core was identified using the GISTIC algorithm (Beroukhim et al., 2007). Several lines of evidence argue against there being only a single 8p11-p12 amplicon core of importance in breast cancer, including (a) the consistency with which multiple 8p amplicon cores have been observed [this report and (Gelsi-Boyer et al., 2005; Melchor et al., 2007)], (b) the fact that the cores can be amplified independently [Figure 1A and Supplementary Table 1 and (Garcia et al., 2005; Gelsi-Boyer et al., 2005; Prentice et al., 2005; Ray et al., 2004)], (c) the association of particular amplicon cores with different breast tumor types (Reis-Filho et al., 2006), and (d) prognostic significance associated with individual cores (Gelsi-Boyer et al., 2005). Thus, it appears that aggregation of copy number data using the GISTIC procedure is insensitive to the complexity of amplified regions such as those present in breast tumors.
Amplification of 11q13 is also frequently complex in breast and other tumor types. Four to five amplicon cores have been identified in breast cancers by Southern blotting, FISH and chromosome CGH (Bekri et al., 1997; Janssen et al., 2002; Karlseder et al., 1994; Rodriguez et al., 2004), and here by high resolution array CGH. The amplicon cores may be amplified independently and their presence has been associated with different clinical features (Bekri et al., 1997; Cuny et al., 2000; Janssen et al., 2002; Karlseder et al., 1994; Rodriguez et al., 2004; Rots et al., 1999). These observations suggest that multiple driver oncogenes map to 11q13 and further that they may define different breast tumor subtypes.
Generally, studies of amplicons have identified a number of likely candidate oncogenes based on correlation of overexpression with amplification. Subsequently, a variety of assays can be used to provide evidence for oncogenic function; however the conclusions appear to be highly dependent on the assay used. For example, LSM1 and BAG4, but not PPAPDC1B were reported to be transforming by one group (Yang, Streicher, Ray, Abrams and Ethier, 2006), while another group came to the opposite conclusion and attributed the difference to the assays used by the two groups (Bernard- Pierrot et al., 2008). In our studies, none of the tested genes, including LSM1 and BAG4 enhanced colony formation in soft agar when expressed in MCF10A. In addition, when assayed in combination with other genes from 8p12 or 11q13, no evidence of cooperation was found that might have provided selective advantage for the frequent co-amplification of 8p and 11q. Similarly, we found no genes for which the expression of a gene on 8p or 11q was correlated with amplification of the other chromosome. We did find, however, that transcriptional cross-talk occurred between the two regions, as expression of ZNF703 on chromosome 8p12 was induced by expression of CCND1 on chromosome 11q13 via the Rb/E2F pathway. On the other hand, functional cooperation of genes on 8p with MYC (FGFR1 and DDHD2) and loss of TP53 (ZNF703) supports their candidacy as driver oncogenes for the 8p12 amplicons.
Little is known about ZNF703 and DDHD2, aside from the presence of conserved domains including a zinc finger domain in ZNF703, and in DDHD2, a sterile alpha motif found in signaling and nuclear proteins, as well as a DDHD domain that may be involved in metal binding. In a comparison of basal and luminal breast cancer expression profiles, ZNF703 emerged as the candidate oncogene most significantly associated with the luminal subtype (Adelaide et al., 2007). Further investigation will be required to understand the possible oncogenic functions of these genes. Similarly, although expression of FGFR1 in mammary epithelial cells induces a number of transformed properties (Xian, Schwertfeger, Vargo-Gogola and Rosen, 2005), a mechanistic understanding of the cooperation between FGFR1 and MYC in promoting a transformed phenotype is currently lacking. Cooperation between CCND1 and TP53 loss in promoting growth in soft agar also requires further investigation. Although not observed in breast cancer (Fridlyand et al., 2006), a positive correlation of TP53 mutation with CCND1 amplification is observed in oral squamous cell carcinoma (Mineta, Borg, Dictor, Wahlberg and Wennerberg, 1997; Snijders et al., 2005). Possibly, the cooperation between CCND1 and TP53 dysfunction results from suppression of growth control mechanisms similar to the p53-dependent cell cycle arrest pathway reported to function independently of RB1 in cells with CCND1 overexpression (Kan, Patton, Stark and Jackson, 2007).
The heterogeneity of breast cancer is highlighted by the discrimination of tumor subtypes by genome-wide copy number and expression profiling. Genomic copy number changes are expected to reflect cancer pathways active in the different tumor subtypes. Identification of the driver oncogenes and the functional consequences of their overexpression, together with the knowledge of their gene-gene interactions will enable tailoring of therapeutics for each tumor subtype. Our studies of the 8p12 and 11q13 amplicons have contributed to such an effort by further refining the mapping of the minimal amplicons and identifying ZNF703, DDHD2 and FGFR1, genes amplified and overexpressed at 8p12 and CCND1 at 11q13 as genes that functionally cooperate with major pathways of oncogenesis.
Materials and Methods
Cell lines and tumors
Cell lines obtained from the American Type Culture Collection (ATCC) included MCF10A, BT474, BT549, 600MPE, MDA-MB-175, and MDA-MB-134. MCF10A cells were grown in ATCC growth medium with 2% rather than 5% horse serum, since growth and colony formation in soft agar was inhibited in the higher serum concentration (Supplementary Figure 5). Moreover, propagation of cells in 5% serum promoted selection in later passages for cells with improved growth and higher frequency of spontaneous colony formation in soft agar. MDA-MB-134 cells were grown in 10% instead of 20% fetal bovine serum as we found that decreasing the serum did not affect their growth. SUM52, SUM225 and SUM44 cell lines were the kind gift of Stephen P. Ethier (Ethier, 1996; Ethier, Mahacek, Gullick, Frank and Weber, 1993; Forozan, Karhu, Kononen, Kallioniemi and Kallioniemi, 1997).
Breast tumors with amplification at 8p12 and/or 11q13 were accessioned from two sets of breast tumors previously profiled by array CGH (Climent et al., 2007; Fridlyand et al., 2006). Data from a third set of 90 tumors (Chin et al., 2006), profiled for both DNA copy number by array CGH and expression using the Affymetrix HTA system, were used for the analysis of gene expression with respect to DNA copy number.
Array CGH
The 8p11q array, providing near tiling path coverage of 8p and 11q13, was assembled with 64 BACs spanning the 8p11-p12 region, 101 BACs at 11q12-q14 and a set of 192 BACs distributed across the genome (Snijders et al., 2003). Clones covering the 8p and 11q regions were selected by reference to the genome assembly using the UCSC genome browser (July 2003 assembly). The DNA spotting solutions for the arrays were prepared using ligation mediated PCR as described previously (Snijders et al., 2001) after isolation of BAC DNA using the PhasePrep™ BAC DNA Kit (Sigma-Aldrich). Array CGH, imaging and data analysis were carried out as described previously (Snijders et al., 2001). The array CGH data for the 8p11q array were deposited in the NCBI Gene Expression Omnibus database, accession number GSE12761.
Statistical analysis of gene expression and amplification
All analyses were performed using the freely available R language. Copy number and expression data were obtained for breast tumors (Chin et al., 2006) and cell lines (Neve et al., 2006). To study the main effects of chromosome 8 and 11 amplification (as previously defined (Fridlyand et al., 2006) when multiple adjacent clones were included in the region or as log2ratio = 0.75 for single clones) on gene expression, an additive linear model was fit to each clone in the regions of amplification on chromosomes 8 and 11 with the two indicator variables as independent variables and gene expression as response. Clone-wise test of the main effect was done using moderated t-statistics (Smyth, 2004), based on empirical Bayes method of shrinkage of standard errors towards a common value, using smoothed data and adjusting for multiple testing by controlling for false discovery rate (FDR) (Hochberg, 1995). An FDR adjusted p-value less than 0.05 was used to declare a clone significant. Further, because of the higher variability in gene expression in the samples with amplifications, a robust linear model applying iterated re-weighted least squares (IWLS) method (Smyth, 2004) was applied.
To study the correlation between copy number and gene expression for the 8p12 and 11q13 regions, the gene expression probesets were mapped to the nearest BAC clone from the 8p12 and 11q13 regions. Correlation was computed for each probeset and a Spearman correlation coefficient of at least 0.4 and FDR adjusted p-value cut-off of 0.05 were used to identify clones having positive correlation between copy number and gene expression. In order to avoid spurious correlations in the absence of real copy number changes, correlations were computed for the gene transcripts whose absolute assigned copy number exceeded 0.25 in at least five samples
Expression of candidate oncogenes in MCF10A
The source of cDNA clones is given in Supplementary Table 6. Clones in pOTB7 were recombined into Gateway entry vector, pDONR221, generating the genes in pENTR221 vector (pDONR221 is renamed pENTR221 when the genes are recombined into it and the ccdB gene is recombined out). The pENTR clones were then recombined with Gateway competent pLenti-V6-dest lentiviral vector with blasticidine resistance to generate genes in the pLenti-V6-dest blasticidine vector. The LPCX vector has been converted into a Gateway entry vector by insertion of sequences for recombination and was also used to recombine with the pENTR clones to generate genes in LPCX. All recombination experiments and Gateway conversion were carried out as described in the Gateway Technology protocol (Invitrogen, Inc.). Expression of all genes was checked with western blotting when antibodies were available and/or by real-time PCR.
CCND1 shRNA
Five shRNAs against CCND1 in pLKO.1 lentiviral puro vector (Sigma-Aldrich Co. cat# TRCN0000040038, TRCN0000040039, TRCN0000040040, TRCN0000040041, TRCN0000040042) were evaluated for knockdown of CCND1 protein levels by western blotting. The two shRNAs (TRCN0000040038 and TRCN0000040041) that gave the greatest knockdown were used for further assays.
Soft Agar Assay
We observed that late passages of the MCF10A cell line developed the ability to grow in soft agar. Therefore, MCF10A cells lacking soft agar growth capability were identified by dilution-cloning. The clonal MCF10A cells were infected with retroviral or lentiviral vectors alone or with the vectors expressing the cDNA of the gene of interest. The infected cells were exposed to the appropriate antibiotics for the selectable markers present in the vectors until uninfected cells were completely dead. For doubly infected cells, the infections and selections were carried out sequentially. Following selection with the appropriate antibiotics for the selectable marker in the first construct, antibiotics were removed from the media, and the cells were infected with the second construct containing a different selectable marker and selected for infection accordingly. At least two independent infections for each construct were carried out and assayed in soft agar as described by David Bowtell (http://www.bio.com/protocolstools/protocol) with the following modifications: The bottom and top agar layers were 1.4% and 0.8% Bacto Agar (Difco), respectively. For each cell line, 2 × 104 cells were plated in each of three wells of a six well plate; however the number of cells was reduced to 1 × 104 cells per well if the cells were previously transduced with GSE56. HRAS transduced MCF10A was plated with every assay as a positive control for growth in soft agar (Wang, Soule and Miller, 1997). Culture medium on top of the agar was changed weekly to prevent the agar from drying out. Colonies, which were usually visible between two to three weeks after incubation, were fixed and stained with 0.005% crystal violet in 50% methanol/50% PBS. Photomicrographs of each well were obtained using a dissecting microscope and the number of colonies was counted using ImageJ (http://rsbweb.nih.gov/ij/).
Supplementary Material
Acknowledgments
We thank members of the UCSF Helen Diller Family Comprehensive Cancer Center Genome Analysis, Informatics, and Microarray Shared Resources for performing the TaqMan assays and printing the custom 8p11q array. This work was supported by NIH grants CA90421 and CA101359. Serena S. Kwek was the recipient of a DOD BCRP fellowship, Grant no. BC021074, DAMD17-03-1-0483.
References
- Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–75. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Yang J, Murphy RF, Agrawal DK. Regulation of the p14ARF-Mdm2-p53 pathway: an overview in breast cancer. Exp Mol Pathol. 2006;81:115–22. doi: 10.1016/j.yexmp.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Bautista S, Theillet C. CCND1 and FGFR1 coamplification results in the colocalization of 11q13 and 8p12 sequences in breast tumor nuclei. Genes Chromosomes Cancer. 1998;22:268–77. [PubMed] [Google Scholar]
- Bekri S, Adelaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, et al. Detailed map of a region commonly amplified at 11q13-->q14 in human breast carcinoma. Cytogenet Cell Genet. 1997;79:125–31. doi: 10.1159/000134699. [DOI] [PubMed] [Google Scholar]
- Bernard-Pierrot I, Gruel N, Stransky N, Vincent-Salomon A, Reyal F, Raynal V, et al. Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res. 2008;68:7165–75. doi: 10.1158/0008-5472.CAN-08-1360. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–41. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Climent J, Dimitrow P, Fridlyand J, Palacios J, Siebert R, Albertson DG, et al. Deletion of chromosome 11q predicts response to anthracycline-based chemotherapy in early breast cancer. Cancer Res. 2007;67:818–26. doi: 10.1158/0008-5472.CAN-06-3307. [DOI] [PubMed] [Google Scholar]
- Cuny M, Kramar A, Courjal F, Johannsdottir V, Iacopetta B, Fontaine H, et al. Relating genotype and phenotype in breast cancer: an analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000;60:1077–83. [PubMed] [Google Scholar]
- Ethier SP. Human breast cancer cell lines as models of growth regulation and disease progression. J Mammary Gland Biol Neoplasia. 1996;1:111–21. doi: 10.1007/BF02096306. [DOI] [PubMed] [Google Scholar]
- Ethier SP, Mahacek ML, Gullick WJ, Frank TS, Weber BL. Differential isolation of normal luminal mammary epithelial cells and breast cancer cells from primary and metastatic sites using selective media. Cancer Res. 1993;53:627–35. [PubMed] [Google Scholar]
- Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–9. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- Fridlyand J, Snijders AM, Ylstra B, Li H, Olshen A, Segraves R, et al. Breast tumor copy number aberration phenotypes and genomic instability. BMC Cancer. 2006;6:96. doi: 10.1186/1471-2407-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, et al. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–45. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–67. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- Haverty PM, Fridlyand J, Li L, Getz G, Beroukhim R, Lohr S, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–42. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- Ho GH, Calvano JE, Bisogna M, Abouezzi Z, Borgen PI, Cordon-Cardo C, et al. Genetic alterations of the p14ARF -hdm2-p53 regulatory pathway in breast carcinoma. Breast Cancer Res Treat. 2001;65:225–32. doi: 10.1023/a:1010686518990. [DOI] [PubMed] [Google Scholar]
- Hochberg YBY. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- Janssen JW, Cuny M, Orsetti B, Rodriguez C, Valles H, Bartram CR, et al. MYEOV: a candidate gene for DNA amplification events occurring centromeric to CCND1 in breast cancer. Int J Cancer. 2002;102:608–14. doi: 10.1002/ijc.10765. [DOI] [PubMed] [Google Scholar]
- Kan CE, Patton JT, Stark GR, Jackson MW. p53-mediated growth suppression in response to Nutlin-3 in cyclin D1 transformed cells occurs independently of p21. Cancer Res. 2007;67:9862–8. doi: 10.1158/0008-5472.CAN-07-0259. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Zeillinger R, Schneeberger C, Czerwenka K, Speiser P, Kubista E, et al. Patterns of DNA amplification at band q13 of chromosome 11 in human breast cancer. Genes Chromosomes Cancer. 1994;9:42–8. doi: 10.1002/gcc.2870090108. [DOI] [PubMed] [Google Scholar]
- Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V, et al. Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer. 2006;6:245. doi: 10.1186/1471-2407-6-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, et al. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–9. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- Melchor L, Garcia MJ, Honrado E, Pole JC, Alvarez S, Edwards PA, et al. Genomic analysis of the 8p11-12 amplicon in familial breast cancer. Int J Cancer. 2007;120:714–7. doi: 10.1002/ijc.22354. [DOI] [PubMed] [Google Scholar]
- Mineta H, Borg A, Dictor M, Wahlberg P, Wennerberg J. Correlation between p53 mutation and cyclin D1 amplification in head and neck squamous cell carcinoma. Oral Oncol. 1997;33:42–6. doi: 10.1016/s0964-1955(96)00039-5. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AL, Pole JC, Blood KA, Garcia MJ, Cooke SL, Teschendorff AE, et al. Co-amplification of 8p12 and 11q13 in breast cancers is not the result of a single genomic event. Genes Chromosomes Cancer. 2007;46:427–39. doi: 10.1002/gcc.20424. [DOI] [PubMed] [Google Scholar]
- Prentice LM, Shadeo A, Lestou VS, Miller MA, deLeeuw RJ, Makretsov N, et al. NRG1 gene rearrangements in clinical breast cancer: identification of an adjacent novel amplicon associated with poor prognosis. Oncogene. 2005;24:7281–9. doi: 10.1038/sj.onc.1208892. [DOI] [PubMed] [Google Scholar]
- Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, Macoska JA, et al. Genomic and expression analysis of the 8p11-12 amplicon in human breast cancer cell lines. Cancer Res. 2004;64:40–7. doi: 10.1158/0008-5472.can-03-1022. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Simpson PT, Turner NC, Lambros MB, Jones C, Mackay A, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–62. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- Rodriguez C, Hughes-Davies L, Valles H, Orsetti B, Cuny M, Ursule L, et al. Amplification of the BRCA2 pathway gene EMSY in sporadic breast cancer is related to negative outcome. Clin Cancer Res. 2004;10:5785–91. doi: 10.1158/1078-0432.CCR-03-0410. [DOI] [PubMed] [Google Scholar]
- Rots MG, Pieters R, Peters GJ, Noordhuis P, van Zantwijk CH, Kaspers GJ, et al. Role of folylpolyglutamate synthetase and folylpolyglutamate hydrolase in methotrexate accumulation and polyglutamylation in childhood leukemia. Blood. 1999;93:1677–83. [PubMed] [Google Scholar]
- Smyth GK. Statistical Applications in Genetics and Molecular Biology. 2004;3(1) doi: 10.2202/1544-6115.1027. Article 3. Available at: http://www.bepress.com/sagmb/vol3/iss1/art3. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Fridlyand J, Mans DA, Segraves R, Jain AN, Pinkel D, et al. Shaping of tumor and drug-resistant genomes by instability and selection. Oncogene. 2003;22:4370–9. doi: 10.1038/sj.onc.1206482. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–4. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, et al. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene. 2005;24:4232–42. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- Tashiro E, Maruki H, Minato Y, Doki Y, Weinstein IB, Imoto M. Overexpression of cyclin D1 contributes to malignancy by up-regulation of fibroblast growth factor receptor 1 via the pRB/E2F pathway. Cancer Res. 2003;63:424–31. [PubMed] [Google Scholar]
- Wang B, Soule HD, Miller FR. Transforming and oncogenic potential of activated c-Ha-ras in three immortalized human breast epithelial cell lines. Anticancer Res. 1997;17:4387–94. [PubMed] [Google Scholar]
- Willmarth NE, Albertson DG, Ethier SP. Chromosomal instability and lack of cyclin E regulation in hCdc4 mutant human breast cancer cells. Breast Cancer Res. 2004;6:R531–9. doi: 10.1186/bcr900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian W, Schwertfeger KL, Vargo-Gogola T, Rosen JM. Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J Cell Biol. 2005;171:663–73. doi: 10.1083/jcb.200505098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZQ, Albertson D, Ethier SP. Genomic organization of the 8p11-p12 amplicon in three breast cancer cell lines. Cancer Genet Cytogenet. 2004;155:57–62. doi: 10.1016/j.cancergencyto.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Yang ZQ, Moffa AB, Haddad R, Streicher KL, Ethier SP. Transforming properties of TC-1 in human breast cancer: interaction with FGFR2 and beta-catenin signaling pathways. Int J Cancer. 2007;121:1265–73. doi: 10.1002/ijc.22831. [DOI] [PubMed] [Google Scholar]
- Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res. 2006;66:11632–43. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.