Abstract
Genetic studies in patients with severe early-onset obesity have provided insights into the molecular and physiological pathways that regulate body weight in humans. We report a 19-year-old male with hyperphagia and severe obesity, mild learning difficulties and hypogonadism, in whom diagnostic tests for Prader–Willi syndrome (PWS) had been negative. We carried out detailed clinical and metabolic phenotyping of this patient and investigated the genetic basis of this obesity syndrome using Agilent 185 k array comparative genomic hybridization (aCGH) and Affymetrix 6.0 genotyping arrays. The identified deletion was validated using multiplex ligation-dependent probe amplification and long-range PCR, followed by breakpoint sequencing which enabled precise localization of the deletion. We identified a ∼187 kb microdeletion at chromosome 15q11–13 that encompasses non-coding small nucleolar RNAs (including HBII-85 snoRNAs) which were not expressed in peripheral lymphocytes from the patient. Characterization of the clinical phenotype revealed increased ad libitum food intake, normal basal metabolic rate when adjusted for fat-free mass, partial hypogonadotropic hypogonadism and growth failure. We have identified a novel deletion on chromosome 15q11–13 in an individual with hyperphagia, obesity, hypogonadism and other features associated with PWS, which is normally caused by deficiency of several paternally expressed imprinted transcripts within chromosome 15q11–13, a region that includes multiple protein-coding genes as well as several non-coding snoRNAs. These findings provide direct evidence for the role of a particular family of non-coding RNAs, the HBII-85 snoRNA cluster, in human energy homeostasis, growth and reproduction.
INTRODUCTION
It is well-established that severe obesity runs in families, although the vast majority of cases do not segregate with a clear Mendelian pattern of inheritance (1). There are about 30 Mendelian disorders with obesity as a clinical feature, often associated with developmental delay, dysmorphic features and organ-specific developmental abnormalities (i.e. pleiotropic syndromes) (2). Additionally, mutations in genes encoding molecules in the hypothalamic leptin–melanocortin pathway result in severe obesity in childhood without developmental pleiotropic features but often associated with neuroendocrine abnormalities (3). Significant advances in the identification and characterization of the genes implicated in these syndromes have been made in the last few years, revealing the complex molecular and physiological pathways involved in the regulation of eating behaviour and body weight in humans.
We identified a patient with severe, early-onset obesity, hypogonadism and mild learning difficulties, but who was negative for diagnostic tests for Prader–Willi syndrome (PWS). The patient had a normal male karyotype: Smith–Magenis syndrome was excluded by FISH (17p11), and a methylation study using SNRPN for PWS was normal. In addition, we excluded coding mutations in the genes encoding leptin, leptin receptor, pro-opiomelanocortin, melanocortin 4 receptor, brain derived neurotrophic factor and its receptor (NTRK2). In order to investigate the underlying genetic abnormality in this patient, we investigated genomic integrity using two different array platforms and identified a microdeletion in the imprinted region on chromosome 15 where larger deletions have been associated with PWS. The microdeletion encompasses the HBII-85 cluster of small nucleolar RNAs (snoRNAs), supporting a role for these non-coding RNAs in human appetite and body weight.
RESULTS
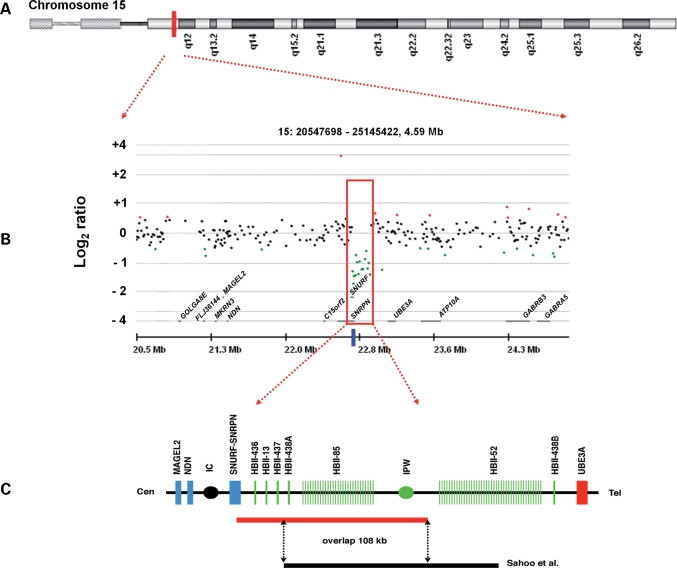
We studied a 19.5-year-old male with hyperphagia and severe obesity, mild learning difficulties and hypogonadism. Previous investigations included a normal karyotype, exclusion of Smith–Magenis syndrome by FISH (17p11) and a methylation study using SNRPN for PWS, which was normal. An initial array comparative genomic hybridization (array CGH) investigation revealed a small deletion in the chr15q11–13 region (Fig. 1). Using the ADM algorithm in the CGH Analytics 3.4 programme, a 15 probe deletion was detected with a mean log2 ratio of −1.143 across the deleted probes, suggesting a one copy loss relative to the reference sample. The genomic positions of the first and last aberrant probes were 22 761 009 and 22 940 338, thus the minimum possible size of the deletion was 179 329 bp. The maximum size, calculated from the locations of the up- and downstream undeleted probes, was 201 087 bp. The deletion was confirmed at a higher resolution on a second platform, the Affymetrix 6.0 SNP array, which enabled more precise localization of the deletion boundaries. On this platform, the deletion included 123 probes; the genomic position of the first deleted probe was 22 757 491 and of the last was 22 943 170, thus the minimum possible size of the deletion was 185 679 bp and the maximum size was 196 803 bp.
Figure 1.
Mapping of a chromosome 15 deletion in a 19-year-old male with suspected PWS. (A) Ideogram of chromosome 15, showing the approximate chromosomal position of the microdeletion as highlighted by the red block. (B) Detection of the PWS-region deletion by array CGH on the Agilent 185 k microarray, as viewed in CGH Analytics 3.4. The red box highlights the 15 probes that appear to be deleted, with an average log2 ratio of −1.143, compared with an expected ratio of 0 for a normal 2n copy number state. The blue bar below the red box shows the position of the MLPA probe (probe 1318-L07970 from the SALSA MLPA P245-A1 Microdeletion Syndromes probe mix) that was found to be deleted when compared with control probes in the patient only. (C) The red bar represents the novel deletion we have detected (chr15: 22 757 219–22 944 158), which is shown to overlap the deletion detected by Sahoo et al. (13) by ∼108 kb.
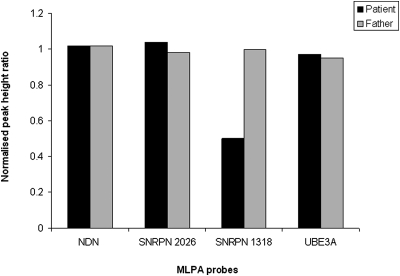
Multiplex ligation-dependent probe amplification (MLPA) was subsequently used to determine whether this deletion was inherited from the patient's father, who is of normal intelligence and body weight. MLPA confirmed the presence of the 15q11–13 deletion in the patient, but not in the patient's father: one of the probes (Fig. 1B) (SNRPN probe 1318, with a probe ligation target site at chr15:22 764 291) was deleted in only the patient when compared with control probes (Fig. 2).
Figure 2.
MLPA of patient and patient's father using the SALSA MLPA P245-A1 Microdeletion Syndromes probemix. Peak height data for each probe has been normalized against five control probes (2n). The probemix contains a total of 49 different MLPA probes, but only four are shown here: NDN probe 6282 (ligation site at chr15:21482517), SNRPN probe 2026 (chr15:22652723), SNRPN probe 1318 (chr15:22764291) and UBE3A probe 4620 (chr15:23167784). SNRPN probe 1318 is shown to be deleted in the patient but not in the father, whereas the other probes are of equal copy number in both individuals.
Long-range PCR also confirmed that the deletion was present in the patient but not his father and allowed determination, by breakpoint sequencing, of the precise size and location of the deletion. The deletion starts at position 22 757 219 and ends at 22 944 158, giving a deletion size of 186 940 bp. This overlaps several exons of the SNURF-SNRPN transcript (exons 2–10 of SNRPN and exons 2–4 of SNURF) but is downstream of the imprinting control centre, which is located in the region of the promoter and first exon of the SNURF-SNRPN gene locus (4,5). Repeat element analysis revealed that the upstream breakpoint was within the poly-A tail of a LINE1 element, and the downstream breakpoint within the poly-A tail of an Alu element (Fig. 3).
Figure 3.
Positions of the up- and downstream breakpoints in the patient's DNA relative to the human reference genome sequence (hg17_DNA). In the upstream breakpoint, the red box shows the position of a LINE1 element, and in the downstream breakpoint the blue box represents the position of an Alu element, according to RepeatMasker (30). In both cases, the deletion breakpoint occurs within the poly-A tail of the repeat element.
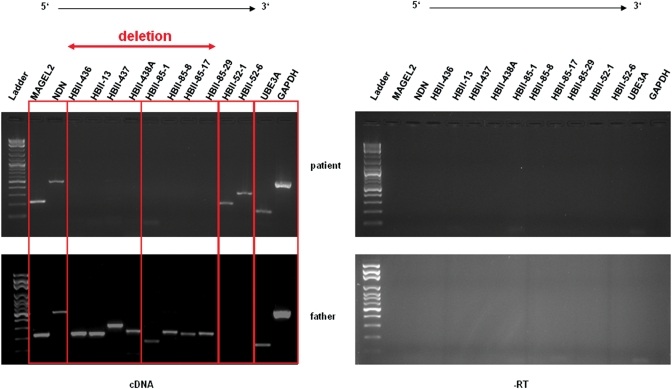
To determine the expression of transcripts within, upstream and downstream of the deleted region, we carried out RT–PCR with the RNA extracted from lymphoblasts of the affected individual and his father who did not carry the deletion. The deletion results in loss of expression of several members of the HBII-85 snoRNA cluster in the patient, but these snoRNAs are expressed in the father. However, the snoRNAs in the HBII-52 cluster, UBE3A and the genes MAGEL and NECDIN, all which lie outside the deleted region, are expressed in both the patient and his normal weight father (Fig. 4), therefore the deletion only seems to have affected expression of transcripts within the deleted region.
Figure 4.
Expression analysis using RT–PCR of lymphocyte-derived RNA obtained from the proband and his father who does not carry the deletion. The 186 940 bp deletion results in loss of expression of several members of the HBII-85 snoRNA cluster in the patient, but these snoRNAs are expressed in the father. However, UBE3A, MAGEL2 and NECDIN, which lie outside the deleted region, are expressed in both individuals. GAPDH was used as an internal control. The experiments were conducted with and without reverse transcriptase.
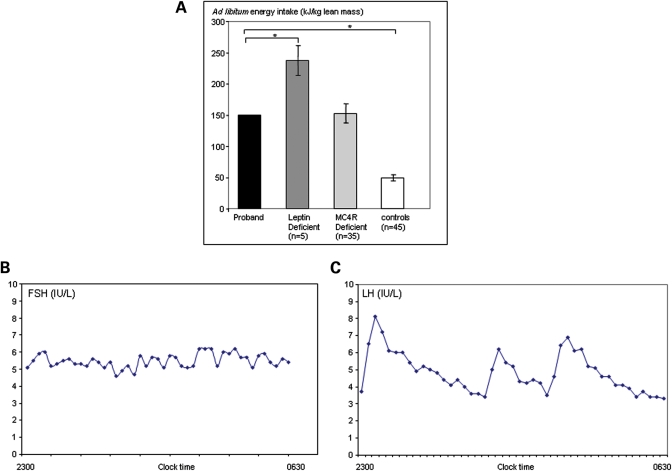
The proband was reported to be hyperphagic under free-living conditions and the hyperphagia was confirmed using an 18MJ ad libitum test meal administered after an overnight fast. The subject ate 148 kJ/kg lean body mass, three times that of control subjects (Fig. 5A). Basal metabolic rate measured by indirect calorimetery was comparable with that predicted by age and gender-specific equations (12.2 MJ/day) with a respiratory quotient in the normal range (0.84). The patient had normal blood glucose (4.8 mmol/L) but had mild fasting hyperinsulinaemia (115 pmol/L; 0–60 pmol/L; HOMA IR 4.1). Previous oral glucose tolerance tests showed impaired glucose tolerance with hyperinsulinaemia responding to metformin although currently insulin sensitivity is maintained (Table 1). Full serum lipid profile, serum leptin and thyroid function tests were in the normal range (data not shown) with a normal response to TRH stimulation (Table 2).
Figure 5.
Obesity and endocrine phenotype. (A) Food intake at an 18 MJ ad libitum test meal expressed per kg lean body mass, for the proband with the deletion affecting the HBII-85 snoRNA cluster compared with subjects with mutations in other genes (LEP and MC4R) responsible for obesity syndromes and controls. Food intake is expressed per kg lean body mass measured by dual energy X-ray absorptiometry to allow comparison between subjects of different ages (*=P < 0.05). (B and C) Gonadotropin hormone pulsatile secretion. Samples were obtained every 10 min overnight during sleep.
Table 1.
Oral glucose tolerance test (metformin had been stopped for 2 months before testing)
| Glucose |
Insulin |
||||
|---|---|---|---|---|---|
| 0 min | 4.8 mmol/l | 0 min | 115 pmol/l | ||
| 30 min | 7.6 mmol/l | 30 min | 672 pmol/l | ||
| 60 min | 7.3 mmol/l | 60 min | 937 pmol/l | ||
| 90 min | 5.4 mmol/l | 90 min | 658 pmol/l | ||
| 120 min | 6.3 mmol/l | 120 min | 318 pmol/l | ||
Table 2.
TRH/LHRH stimulation test
| FSH |
LH |
TSH |
|||
|---|---|---|---|---|---|
| 0 min | 5.8 IU/l | 0 min | 3.6 IU/l | 0 min | 0.88 IU/l |
| 30 min | 9.1 IU/l | 30 min | 17.1 IU/l | 30 min | 13.3 IU/l |
| 60 min | 9.0 IU/l | 60 min | 14 IU/l | 60 min | 8.7 IU/l |
Pubertal development was delayed with evidence of hypogonadotropic hypogonadism which required induction of puberty with testosterone. Current assessment of the hypothalamo-pituitary gonadal axis (undertaken after 2 months off testosterone replacement) shows partial hypogonadotropic hypogonadism with evidence of sub-optimal pulsatile FSH and LH secretion confirming the need for long-term therapy (Table 2 and Fig. 5B and C). There was a normal cortisol profile with a maintained circadian variation, and dynamic GH testing in childhood confirmed GH deficiency (data not shown).
DISCUSSION
We have identified a unique microdeletion of ∼187 kb on chromosome 15q11–13, which encompasses the HBII-85 snoRNA cluster that was not expressed in a patient with severe obesity, hyperphagia, hypogonadism and impaired linear growth. These features overlap those seen in PWS, which is characterized by diminished fetal activity, hypotonia, mental retardation, short stature, hypogonadotropic hypogonadism, hyperphagia and obesity (6). PWS is caused by deficiency of several paternally expressed imprinted transcripts within chromosome 15q11–13 (7,8), which includes SNURF-SNRPN and multiple small nucleolar RNAs (snoRNAs). The majority of cases of PWS (70–75%) are due to deletions on the paternally inherited chromosome 15 (9), with the remainder caused mainly by maternal uniparental disomy for region 15q11–13 (10). We have shown that the deletion is not present in the patient's father and, although we could not test DNA from the patient's mother (who was not overweight), the deletion is likely to be de novo.
Balanced translocations that cause PWS but do not disrupt the SNURF-SNRPN promoter and leave coding regions intact suggest that disruption of SNURF-SNRPN is less important (11,12), whereas a recently reported microdeletion in a child with PWS affecting the entire HBII-85 cluster, 50% of the HBII-52 cluster and one of the two copies of HBII-438 provides strong evidence for the role of snoRNAs in the aetiology of this complex phenotype (13). The only genes in common between the deletion reported here and that identified recently by Sahoo et al. (13) are the HBII-85 snoRNA cluster and the HBII-438A snoRNA. Furthermore, studies of paternally inherited chromosome 15 deletions in healthy individuals (14) and of PWS individuals with balanced translocations (11) suggest that the snoRNAs upstream and downstream of this region do not play a major role in PWS.
Sequences from HBII-85 snoRNAs are highly conserved between the human and mouse genomes, while no orthologous version of the HBII-438A snoRNA has been located in mice (15). Although it is well known that previous mouse models of the PWS deletion have failed to consistently recapitulate the human disease phenotype, mice lacking all copies of the HBII-85 snoRNA orthologues (MBII-85) have shown some phenotypes associated with human PWS, including postnatal growth retardation, delayed sexual maturation, motor learning deficit and hyperphagia (15).
Further investigation is required to determine how the loss of these non-coding sequences could lead to the range of phenotypes associated with PWS. The majority of C/D box snoRNAs are universally expressed and function in the post-transcriptional modification of ribosomal RNAs and small nuclear RNAs in the nucleolus (reviewed in 16). Both mouse (MBII-85) and human (HBII-85) snoRNAs, however, have been found to be expressed predominantly in the brain (17). Most snoRNAs contain 10–21-nt complementary regions to rRNA that can direct the processes of pseudouridylation or ribose methylation, and it is possible that the HBII-85 snoRNA cluster may have as yet unidentified targets in the brain that affect appetite, neuroendocrine and other brain functions that lead to PWS-associated phenotypes. Recently, Ender et al. (18) showed that snoRNAs can bind to the Argonaute proteins which are part of the microRNA silencing complex, suggesting additional mechanisms may contribute to effects on gene expression.
In addition to refining the critical region for PWS, we have also examined potential mechanisms of formation for this novel microdeletion through sequence analysis at the deletion breakpoints. Both the up- and downstream breakpoints of this deletion have been found to lie within the poly-A tail of a repeat element, in LINE1 and Alu elements, respectively (Fig. 3). Both Alu and LINE1 elements have been implicated in the formation of genomic deletions underlying a range of diseases, such as α-thalassaemia (19) and AML (20), as well as in common deletion copy number variants in healthy subjects (21). The absence of extended sequence similarity between the up- and downstream breakpoints of this deletion indicates that it is likely to have arisen through non-homologous end-joining following an initial double-stranded DNA breakage, possibly originating within one of the repeat element poly-A tails (21): there is no evidence of retrotransposition having formed the deletion, as comparisons with the chimpanzee (Pan troglodytes) genome sequence in Ensembl (22) showed these repeat elements to be ancestral (i.e. they are present at the same locus in both species).
In conclusion, our array CGH investigation of a patient with obesity, impaired growth and hypogonadism has led to the identification of a microdeletion in the chromosome 15q11–13 region that reduces the minimum critical region for PWS to two C/D box snoRNAs, HBII-85 cluster and HBII-438A. Given the combination of learning difficulties, hypogonadism, hyperphagia and severe obesity, it would seem likely that the clinical phenotype in this patient has resulted from a reduction in these non-coding RNAs. It is possible that snoRNAs at other loci in the genome, particularly those that are expressed predominantly in the brain, may underlie other genomic disorders for which a causative gene has yet to be determined.
MATERIALS AND METHODS
Subjects
The subject was recruited for an array comparative genomic hybridization (array CGH) study of patients with obesity and suspected genomic disorders. Molecular studies were carried out at the North West Thames Regional Genetics Service, based at Northwick Park Hospital after approval by the Harrow Research Ethics Committee. The clinical metabolic studies were performed after approval by the local–regional Ethics Committee of Cambridge. For this part of the work, the patient and his father were invited to the Wellcome Trust Clinical Research Facility at Addenbrooke's Hospital, Cambridge, UK. All clinical studies were conducted in accordance with the principles of the Declaration of Helsinki and performed as previously described (23).
Clinical description
The patient, who is the second child of non-consanguineous Indian parents, was first referred to the genetic service at the age of 4 years. He was born by lower segment caesarean section for fetal distress at term, following a normal pregnancy, with a birth weight of 2.8 kg. He fed slowly and was noted to be floppy in the neonatal period. Subsequently all his developmental milestones were delayed and he walked at approximately 2 years of age. At about the same time, his appetite improved and he started to eat ‘almost continuously’. When he was first seen in the genetic clinic at 4 years of age he was obese with upslanting palpebral fissures, myopathic facies with a tented mouth (Fig. 6) and small hands and feet. He subsequently attended a mainstream primary school with extra support, but later transferred to a school for children with moderate learning difficulties. There was no significant family history. His mother had died following the birth of her third child. Chromosome analysis, Fragile X gene testing and methylation studies for PWS, using probe PW71, were normal at that time. He was referred back for re-evaluation at the age of 11 years following an admission to ITU with a chest infection and multi-organ failure requiring ventilation and inotropic support. At that time his weight was 87 kg. His phenotype was felt to be consistent with PWS, but further PWS methylation studies using Kb17 (SNRPN) were normal. UPD15 studies were uninformative (as a sample from his mother was not available) and FISH 17p11.2 (for Smith–Magenis syndrome) was normal.
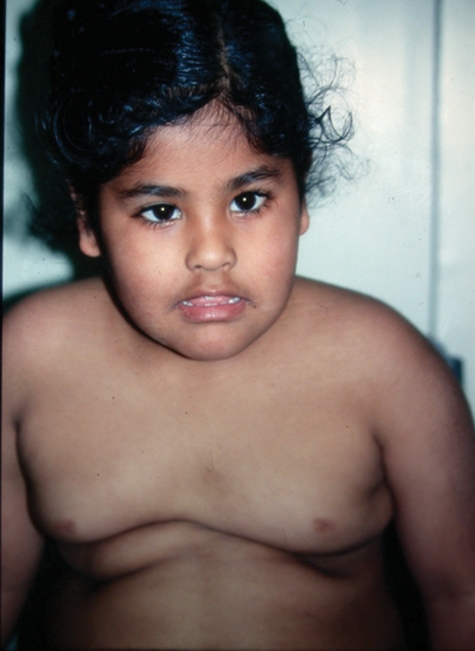
Figure 6.
Patient aged 4 years showing obesity, upslanting palpebral fissures and tented mouth.
On review aged 13 years, his weight was 101 kg with a height of 155 cm, and BMI 42 kg/m2. His phenotypes remained suggestive of PWS with skin picking and behavioural problems including lacking a sense of danger, stubbornness and temper tantrums. He had marked gynaecomastia, acanthosis nigricans on his neck, small hands and feet, and a small penis. Endocrine assessment confirmed hypogonadotropic hypogonadism which required treatment. At age 19.5 years, when clinical metabolic studies were carried out, the proband weighed 109 kg with a height of 167.5 cm (height SDS −1.41) and BMI 39 kg/m2.
Microarray comparative genomic hybridization
A prototype genome-wide scanning array, consisting of 60mer in situ synthesized oligonucleotides, was designed and manufactured by Agilent Technologies (Santa Clara, CA) (reviewed in 24). This array contained 185 000 probes, nominally spaced across the genome (average spacing of 16 kb), but biased towards genes. Array hybridization was performed according to the manufacturer's recommended protocols. In brief, restriction enzyme digestion of ∼500 ng of the patient's genomic DNA was carried out using the enzymes AluI and RsaI, followed by fluorescent labelling using the Agilent DNA Labelling kit. The patient DNA was labelled with Cyanine 3-dUTP and the reference sample, a pool of male Promega samples, with Cyanine 5-dUTP. Labelled DNA was then denatured, and Cot-1 DNA plus Agilent blocking reagent used in a pre-annealing step to block repetitive genomic regions. Hybridization was carried out in an Agilent oven at 65°C for 40 h at 20 rpm, followed by standard wash procedures.
The microarray was then scanned in an Agilent scanner at 5 µm resolution, and the array data extracted using the default CGH settings of Agilent Feature Extraction Software. For analysis of the array CGH data, Agilent CGH Analytics 3.4 software was used, along with the ADM (aberration detection module) algorithm for detecting regions of statistically significant copy number change (25). This algorithm determines genomic regions with differences in copy number between the sample and reference based on log2 ratios of the fluorescence signals from probes in the interval. The patient's DNA was also analysed with an Affymetrix Genome-wide Human SNP Array 6.0. that comprises more than 906 600 SNP probes and more than 946 000 copy number probes. The median inter-marker distance is 700 bp. The hybridization and scanning was performed by Aros (Denmark). The analysis of the array data was done using the Affymetrix Genotyping Console.
Multiplex ligation-dependent probe amplification
MLPA was used for the initial validation of the deletion detected by array CGH in the patient. We investigated both the patient and the patient's father (DNA from mother was not available) using the SALSA MLPA P245-A1 Microdeletion Syndromes probemix, which included one probe in the region shown to be deleted by array CGH (probe 1318-L07970). All MLPA reagents were obtained from MRC-Holland, and reactions were performed as described by Schouten et al. (26). MLPA products were separated using an AB 3130 Genetic Analyser (Applied Biosystems) and outputs were analysed using Gene Mapper software (Applied Biosystems) and Coffalyser (MRC Holland).
Long-range PCR validation and sequencing
Eight initial PCR primer pairs were designed in order to localize the deletion breakpoints, using Primer 3 software (27). An additional nine primer pairs were designed to further refine the position of the deletion breakpoints. Primer sequences are available on request. Long-range PCR was carried out on DNA from both the patient and the patient's father, using the Qiagen LongRange PCR Kit (see manufacturers protocol for details). All PCR products were examined by agarose gel electrophoresis. PCR products were purified using the GenElute™ PCR Clean-Up Kit (Sigma-Aldrich) before forward and reverse sequencing. DNA sequencing was carried out using an ABI 3730×l DNA Analyzer (Applied Biosystems), with the purified PCR products sequenced in both forward and reverse directions using the corresponding primers. The reference genome sequence was then retrieved using the UCSC Genome Browser (28) (March 2006 build), and a multiple alignment between the patient's deleted sequence and the corresponding reference genome sequence was carried out using ClustalW (29). Up- and downstream deletion breakpoints could, therefore, be determined, and RepeatMasker (30) was subsequently used to investigate the presence of repeat sequence elements at these breakpoints.
RNA extraction from whole blood
In order to test the expression of genes in the deleted region, total cellular RNA was extracted from whole blood and transcribed into cDNA, which was then amplified in a PCR with primer pairs specific to certain genes or snoRNAs situated in and outside the deletion region. For purification of total cellular RNA from human whole blood, the QIAamp RNA Blood Mini kit (Qiagen) was used following the manufacturer's instructions. Before synthesis of cDNA, the RNA was treated twice with DNaseI to ensure complete removal of genomic DNA. For each cDNA amplification, a negative control without reverse transcriptase was processed in parallel. To test for expression of snoRNAs and genes in the PWS region, the cDNA from patient and father was used in a PCR reaction with primer pairs specific to MAGEL2, NDN, UBE3A or one member of the snoRNA cluster. As a control for the quality of the cDNA, a primer pair amplifying the housekeeping gene GAPDH was used.
Metabolic studies
Anthropometry and whole-body dual-energy X-ray absorptiometry (DEXA) (DPX software; Lunar Corp., Madison, WI, USA) to determine body composition were performed as previously described (23). Resting metabolic rate was measured by indirect calorimetry after a 12-h fast overnight using an open-circuit, ventilated, canopy measurement system (Europa Gas Exchange Monitor; Nutren Technology Ltd, Manchester, UK). After adjustment for body composition, the resting metabolic rate was compared with that predicted according to age- and gender-specific equations. Semi-quantitative assessment of eating behaviour was undertaken using a 4300-kcal (18 MJ) meal at breakfast after an overnight fast, the contents were covertly weighed before the meal and after the patient had finished eating and total energy intake and nutrient composition were calculated. Energy intake was expressed per kilogram of lean body weight as a simple means of comparing intake among subjects of different ages and body sizes as reported previously (23). Blood samples obtained in the fasting state were analysed with the use of standard assays. Overnight pulsatility studies were performed by taking blood samples every 10 min through an indwelling cannula while the patient was asleep, as previously described (23).
FUNDING
This work was supported by the Hammersmith Hospital Charity Trustees; the NIHR Biomedical Research Centre Scheme; the Wellcome Trust, MRC; and the Cambridge NIHR Biomedical Research Centre.
ACKNOWLEDGEMENTS
We are grateful to the patient and his father for their participation in this project. We thank Mamta Purbhoosing and Jamie Studd for technical assistance in the array CGH work, and Agilent Technologies, Inc. for their collaboration on this project.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Barsh G.S., Farooqi I.S., O'Rahilly S. Genetics of body-weight regulation. Nature. 2000;404:644–651. doi: 10.1038/35007519. [DOI] [PubMed] [Google Scholar]
- 2.Blakemore A.I., Froguel P. Is obesity our genetic legacy? J. Clin. Endocrinol. Metab. 2008;93:S51–S56. doi: 10.1210/jc.2008-1676. [DOI] [PubMed] [Google Scholar]
- 3.Farooqi I.S., O'Rahilly S. Monogenic obesity in humans. Annu. Rev. Med. 2005;56:443–458. doi: 10.1146/annurev.med.56.062904.144924. [DOI] [PubMed] [Google Scholar]
- 4.Sutcliffe J.S., Nakao M., Christian S., Orstavik K.H., Tommerup N., Ledbetter D.H., Beaudet A.L. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nature Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 5.Buiting K., Saitoh S., Gross S., Dittrich B., Schwartz S., Nicholls R.D., Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nature Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- 6.Goldstone A.P. Prader-Willi syndrome: advances in genetics, pathophysiology and treatment. Trends Endocrinol. Metabol. 2004;15:12–20. doi: 10.1016/j.tem.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Butler M.G., Meaney F.J., Palmer C.G. Clinical and cytogenetic survey of 39 individuals with Prader-Labhart-Willi syndrome. Am. J. Med. Genet. 1986;23:793–809. doi: 10.1002/ajmg.1320230307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ledbetter D.H., Riccardi V.M., Airhart S.D., Strobel R.J., Keenan B.S., Crawford J.D. Deletions of chromosome 15 as a cause of the Prader-Willi syndrome. N. Engl. J. Med. 1981;304:325–329. doi: 10.1056/NEJM198102053040604. [DOI] [PubMed] [Google Scholar]
- 9.Robinson W.P., Bottani A., Xie Y.G., Balakrishman J., Binkert F., Machler M., Prader A., Schinzel A. Molecular, cytogenetic, and clinical investigations of Prader-Willi syndrome patients. Am. J. Hum. Genet. 1991;49:1219–1234. [PMC free article] [PubMed] [Google Scholar]
- 10.Mascari M.J., Gottlieb W., Rogan P.K., Butler M.G., Waller D.A., Armour J.A., Jeffreys A.J., Ladda R.L., Nicholls R.D. The frequency of uniparental disomy in Prader-Willi syndrome. Implications for molecular diagnosis. N. Engl. J. Med. 1992;326:1599–1607. doi: 10.1056/NEJM199206113262404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher R.C., Pils B., Albalwi M., Francke U. Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader-Willi syndrome. Am. J. Hum. Genet. 2002;71:669–678. doi: 10.1086/342408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schule B., Albalwi M., Northrop E., Francis D.I., Rowell M., Slater H.R., Gardner R.J., Francke U. Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader-Willi syndrome. BMC Med. Genet. 2005;6:18. doi: 10.1186/1471-2350-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahoo T., del Gaudio D., German J.R., Shinawi M., Peters S.U., Person R.E., Garnica A., Cheung S.W., Beaudet A.L. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nature Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Runte M., Varon R., Horn D., Horsthemke B., Buiting K. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Hum. Genet. 2005;116:228–230. doi: 10.1007/s00439-004-1219-2. [DOI] [PubMed] [Google Scholar]
- 15.Ding F., Li H.H., Zhang S., Solomon N.M., Camper S.A., Cohen P., Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith C.M., Steitz J.A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 17.Cavaille J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Harteveld K.L., Losekoot M., Fodde R., Giordano P.C., Bernini L.F. The involvement of Alu repeats in recombination events at the alpha-globin gene cluster: characterization of two alphazero-thalassaemia deletion breakpoints. Hum. Genet. 1997;99:528–534. doi: 10.1007/s004390050401. [DOI] [PubMed] [Google Scholar]
- 20.Schichman S.A., Caligiuri M.A., Strout M.P., Carter S.L., Gu Y., Canaani E., Bloomfield C.D., Croce C.M. ALL-1 tandem duplication in acute myeloid leukemia with a normal karyotype involves homologous recombination between Alu elements. Cancer Res. 1994;54:4277–4280. [PubMed] [Google Scholar]
- 21.de Smith A.J., Walters R.G., Coin L.J., Steinfeld I., Yakhini Z., Sladek R., Froguel P., Blakemore A.I. Small deletion variants have stable breakpoints commonly associated with alu elements. PLoS ONE. 2008;3:e3104. doi: 10.1371/journal.pone.0003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farooqi I.S., Keogh J.M., Yeo G.S., Lank E.J., Cheetham T., O'Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N. Engl. J. Med. 2003;348:1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 24.de Smith A.J., Tsalenko A., Sampas N., Scheffer A., Yamada N.A., Tsang P., Ben-Dor A., Yakhini Z., Ellis R.J., Bruhn L., et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum. Mol. Gen. 2007;16:2783–2794. doi: 10.1093/hmg/ddm208. [DOI] [PubMed] [Google Scholar]
- 25.Lipson D., Aumann Y., Ben-Dor A., Linial N., Yakhini Z. Efficient calculation of interval scores for DNA copy number data analysis. J. Comput. Biol. 2006;13:215–228. doi: 10.1089/cmb.2006.13.215. [DOI] [PubMed] [Google Scholar]
- 26.Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. (Clifton, NJ) 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 28.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smit A.F.A., Hubley R., Green P. RepeatMasker Open-3.0. :1996–2004. http://www.repeatmasker.org . [Google Scholar]