Abstract
α-Synuclein (SNCA) gene has been implicated in the development of rare forms of familial Parkinson disease (PD). Recently, it was shown that an increase in SNCA copy numbers leads to elevated levels of wild-type SNCA-mRNA and protein and is sufficient to cause early-onset, familial PD. A critical question concerning the molecular pathogenesis of PD is what contributory role, if any, is played by the SNCA gene in sporadic PD. The expansion of SNCA-Rep1, an upstream, polymorphic microsatellite of the SNCA gene, is associated with elevated risk for sporadic PD. However, whether SNCA-Rep1 is the causal variant and the underlying mechanism with which its effect is mediated by remained elusive. We report here the effects of three distinct SNCA-Rep1 variants in the brains of 72 mice transgenic for the entire human SNCA locus. Human SNCA-mRNA and protein levels were increased 1.7- and 1.25-fold, respectively, in homozygotes for the expanded, PD risk-conferring allele compared with homozygotes for the shorter, protective allele. When adjusting for the total SNCA-protein concentration (endogenous mouse and transgenic human) expressed in each brain, the expanded risk allele contributed 2.6-fold more to the SNCA steady-state than the shorter allele. Furthermore, targeted deletion of Rep1 resulted in the lowest human SNCA-mRNA and protein concentrations in murine brain. In contrast, the Rep1 effect was not observed in blood lysates from the same mice. These results demonstrate that Rep1 regulates human SNCA expression by enhancing its transcription in the adult nervous system and suggest that homozygosity for the expanded Rep1 allele may mimic locus multiplication, thereby elevating PD risk.
INTRODUCTION
α-Synuclein (SNCA) (GenBank: AF163864; Ensembl: ENSG00000145335; OMIM, Online Mendelian Inheritance in Man: MIM 163890) gene mutations (1–3) and copy number variations (4–8) have been implicated in the development of rare forms of familial Parkinson disease (PD, MIM 168600). Moreover, a recent genome-wide association study provided further support for the role of SNCA in familial PD susceptibility (9). Recently published evidence showed that an increase in SNCA copy numbers leads to elevated levels of wild-type (WT) SNCA protein in vivo and is sufficient to cause early-onset, familial PD. Whereas the triplication of the WT SNCA locus leads to 2-fold overexpression of SNCA-mRNA and protein, duplication results in a 1.5-fold elevation. Duplication results in lower penetrance than is seen with triplication, a slightly later onset of the disease and a ‘milder’ phenotype that is essentially indistinguishable from sporadic PD (5–8,10,11). Furthermore, PD, dementia with Lewy bodies (DLB, MIM 127750) and multiple system atrophy (MSA) are seen as prototypes of human ‘synucleinopathies’, which are characterized by intracellular SNCA inclusions. These aggregates have been identified in neuronal Lewy bodies and neurites, two pathological hallmark lesions of both PD and DLB (12), and in glial inclusions, a characteristic of MSA (13). Moreover, elevated SNCA-mRNA levels have been reported in midbrain tissue homogenates (14) and in dopaminergic neurons from sporadic PD cases (15). These observations emphasize the critical importance of SNCA dosage and expression levels in the pathogenesis of PD and related synucleinopathies.
Several association studies have demonstrated that genetic variability across the SNCA locus, including its promoter, is associated with altered susceptibility to sporadic PD (PDGene database, http://www.pdgene.org/) (16–19). To date, the most studied SNCA genetic variation is Rep1 (GenBank D4S3481), a complex polymorphic microsatellite repeat site located ∼10 kb upstream of the transcription start site (20,21). The overwhelming majority of these association studies, including a large meta-analysis, have shown that certain alleles of SNCA-Rep1 confer increased risk to develop late-onset, ‘idiopathic’ PD (22–25). Previously, we attempted to determine whether this genetic association correlated with any biological function. Using a reporter assay in a transiently transfected neuronal cell culture model, we demonstrated that the SNCA-Rep1 had a reproducible effect on regulating transcriptional activity; there, certain allele lengths showed a nearly 3-fold change in expression of the reporter construct (26,27). Given that multiplications of SNCA have been implicated in heritable PD, where elevated dosage increases penetrance and lowers age-of-onset, we hypothesized that a subtle increase in SNCA expression over decades confers an elevated risk for late-onset, sporadic PD. We posited that the strong genetic association of SNCA-Rep1 allele expansion with PD risk could be the consequence of a cis-acting mechanism on the transcriptional regulation of SNCA. Moreover, it was recently suggested that the SNCA-Rep1 genotype may also affect SNCA protein levels in human blood samples (28).
Here, we used a transgenic mouse model to examine three distinct Rep1 variants within the human SNCA locus to establish the relevance of the Rep1 element in the regulation of SNCA expression in vivo. We engineered the two most frequent alleles that have been reported in both Caucasian and Asian populations (22,23), i.e. the expanded, 261 bp-long Rep1 allele (∼68% in control whites) and its shorter 259 bp variant (∼27%), into a bacterial P1 artificial chromosome (PAC) containing the entire human SNCA gene. We also engineered an artificial allele consisting of a deletion of the Rep1 repeat within the same PAC. We then compared SNCA-mRNA and protein levels in the brain and blood specimens of transgenic mice carrying the 261 and 259 bp and the Rep1-deletion alleles in order to compare the effect of the different alleles on the expression of the SNCA gene in vivo.
RESULTS
Characterization of transgenic mouse lines to examine three distinct SNCA-Rep1 genotypes
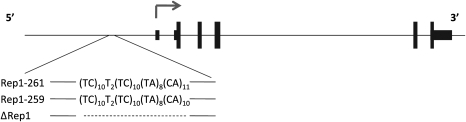
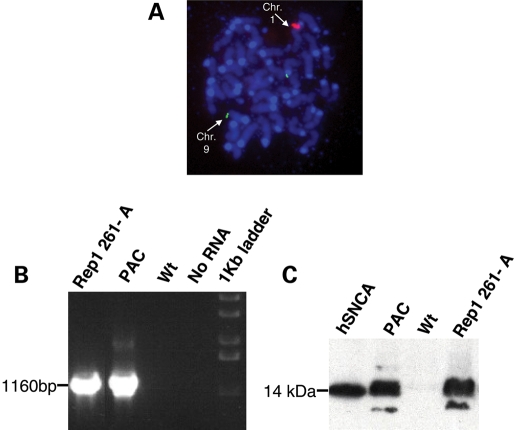
We generated transgenic mice lines for three distinct SNCA-Rep1 genotypes carrying the 146 kb-large PAC 27M07. The PAC contains the entire human SNCA gene and 34 kb of its upstream region, including the SNCA-Rep1 site located within ∼10 kb of the transcription start site (Fig. 1). Each genotype examined contained a different allele at the SNCA-Rep1 site. These included the two naturally occurring variants, Rep1-261 bp and Rep1-259 bp, which carried the following sequence combinations, (TC)10(T)2(TC)10(TA)8(CA)11 and (TC)10(T)2(TC)10(TA)8(CA)10, respectively, and one artificial allele in which the SNCA-Rep1 repeat has been deleted altogether, Δ-Rep1. Importantly, these PAC clones harbored ∼34 kb of non-coding DNA located upstream of exon 1, thereby reducing possible integration site effects on transcription. Nevertheless, to further minimize any confounding effect of genome integration site by our PAC variants on subsequent transgene expression, we decided to generate two independent lines for each of the three genotypes. Transcription and translation of human SNCA in the mouse brain was confirmed at the F1 generation, as shown, for example, in the Rep1-261/261-carrying line ‘A’ in Figure 2B and C. Fluorescent in situ hybridization (FISH) was used to determine the chromosomal integration site of each line (Supplementary Material, Table S1) and to obtain a first estimate for the range of genomic copy numbers (see, for example, 261/261 line ‘A’ in Fig. 2A). The precise copy number in each line was then determined more accurately. To this end, we employed quantitative real-time PCR using genomic DNA from animals in each line by targeting two different regions of the human PAC (29). We calculated that the relative transgene copy numbers in the different lines shown here ranged from 1 to 7 transgene copies per haploid genome (Supplementary Material, Table S1).
Figure 1.
A schematic presentation of the PAC (27M07) used to generate the transgenic mice lines. The organization of the human SNCA locus that is included in the PAC at the top panel. The three distinct SNCA-Rep1 genotypes are indicated below.
Figure 2.
Serial analyses to confirm the mouse lines in the study. Presented is an example analysis performed for transgenic line ‘A’, carrying the human SNCA-Rep1 261 bp allele. (A) FISH analysis of metaphase chromosomes obtained from the mouse's spleen was used to determine the insertion site and to estimate the range of the copy number. Red-labeled human-PAC 27M07; green-labeled mouse BAC corresponding to mouse chromosome 9, used as a reference for copy number estimation; blue-DAPI. (B) RT–PCR to the 27M07 PAC was used to confirm the RNA expression of the human SNCA in the brain of an F1 mouse. (C) Western blot using the mouse monoclonal LB509 anti-human-SNCA (Zymed), to confirm the protein expression of the human SNCA in the brain of an F1 mouse. For (B) and (C), each lane represents analysis performed using a mouse-brain sample harvested from the following mouse lines: Rep1 261-A, the 261/261 line ‘A’ transgenic; PAC, control PAC containing the original Rep1 allele transgenic (55); Wt, wild type FVB; hSNCA, recombinant human SNCA protein.
Next we analyzed human SNCA expression levels in the brain and blood of F2 homozygous transgenic mice and in the heterozygous Δ-Rep1 line ‘B’. A total of 72 F2 animals from six mouse lines, representing two F2 lines for each of the three distinct alleles (e.g. Rep1-261 bp, Rep1-259 bp and Δ-Rep1 deletion), were sacrificed at the age of 8 weeks; these mice were used for extended quantitative analyses at the mRNA and protein level to determine their variation in human SNCA expression, as controlled by each PAC transgene.
Effects of Rep1 variants on human SNCA-mRNA levels in mouse brain
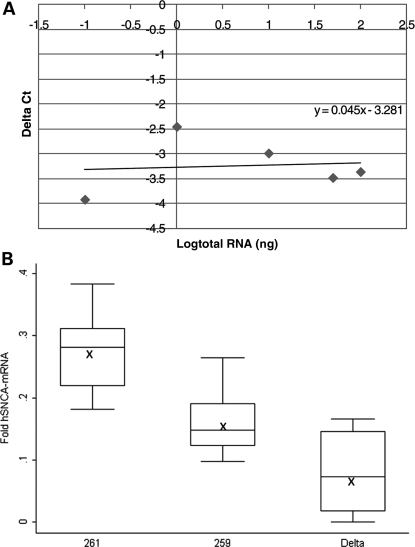
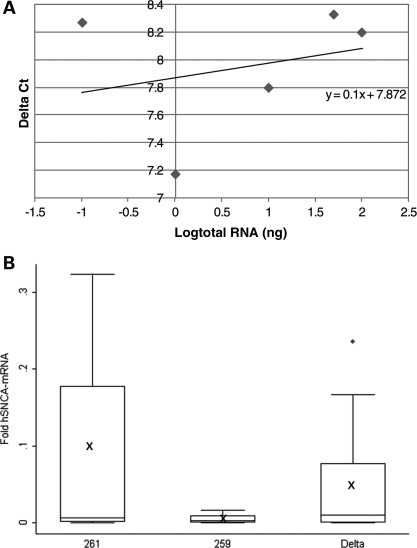
To study the effect of SNCA-Rep1 allele homozygosity on human SNCA-mRNA levels in transgenic mouse brain (n = 72), we employed a real-time PCR assay specific for the human message (no amplification was detected with the control WT mouse). All levels were measured relative to the mouse synaptophysin (Syp) endogenous reference gene by 2−ΔCt (Fig. 3A). The fold expression levels of each mouse line (Supplementary Material, Fig. S1) were then corrected according to copy number values, and the average fold expression for each of the three transgenes was determined. Our calculations, which are expressed as fold expression per diploid-copy of the transgene, showed that 261/261 transgenic mice consistently generated the highest level of SNCA-mRNA in the brain (Fig. 3B; Supplementary Material, Fig. S2). Rep1 261/261 mice demonstrated an average 0.27 ± 0.04-fold expression level relative to that of endogenous mouse Syp compared with an average 0.16 ± 0.03-fold expression level of SNCA-mRNA in the 259/259 transgenic lines and an average 0.08 ± 0.06-fold expression level in the Δ-Rep1 transgenic mice (Fig. 3B). From these results, we calculated that the mice, which carried the PD risk-associated allele Rep1-261 bp, harbored significantly elevated levels of human SNCA-mRNA, amounting to a nearly 70% increase over those mice that carried the ‘protective’ genotype, allele Rep1-259 bp (P < 0.0001). No significant correlation was seen between SNCA-mRNA levels in the brain and gender (P = 0.44).
Figure 3.
Effect of the SNCA-Rep1 promoter genotypes on human SNCA-mRNA expression levels in transgenic mouse brains. (A) Validation curve of the Δ real-time assay for relative quantization of human SNCA-mRNA relative to mouse Syp-mRNA in the brain. Relative efficiency plots of human-SNCA and mouse-Syp were formed by plotting the log input amount (ng of total RNA) versus the ΔCt = [Ct(SNCA)−Ct(Syp)]. The slope is 0.045, which indicated the validation of the ΔCt calculation in the range of 0.1–100 ng RNA. (B) Fold levels of human SNCA-mRNA were assayed by real-time RT–PCR and calculated relative to mouse Syp-mRNA reference control using the 2−ΔCt method. No detectable products of the human SNCA-mRNA reaction were observed when WT mouse brain-cDNA was used as template. The risk genotype 261/261 correlates with significant higher SNCA-mRNA levels than the protective genotype 259/259 (P < 0.0001). The deleted SNCA-Rep1 genotype correlates with significant lower SNCA-mRNA levels than genotypes 261/261 and 259/259 (P< 0.0001). In total, 72 mice were analyzed; for each genotype (261/261, 259/259 and Δ/Δ), the box plot represents the analysis performed using two transgenic lines, 12 animals from each line (6 males and 6 females), comprising 24 total brain samples per genotype, each of which was analyzed twice independently. The average values are presented by ‘X’. The box plot shows the median (horizontal line inside the box) and the 25th and 75th percentiles (horizontal borders of the box). The range between the 25th and 75th percentiles is the interquartile range. The whiskers show the minimal and maximal values inside the main data body.
In addition, we aimed to study the overall effect of the presence or absence of the Rep1 microsatellite site on the regulation of SNCA-mRNA expression. SNCA-mRNA levels in the brains of transgenic mice carrying the SNCA-Rep1 deletion were significantly lower when compared with mice carrying the naturally occurring alleles 261/261 and 259/259 (P < 0.0001; Fig. 3B; Supplementary Material, Fig. S2). The latter result suggested that in the brain tissue, Rep1 might act as an enhancer element to upregulate SNCA-mRNA levels.
Effects of Rep1 variants on human SNCA protein concentrations in mouse brain
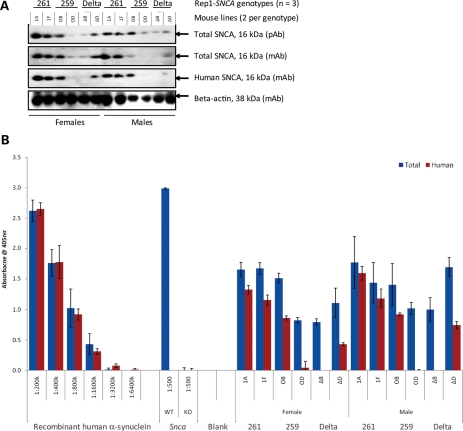
In parallel to these mRNA measurements, we also analyzed the human SNCA protein content in brain homogenates of 12 age- and gender-matched mice from each of the six lines representing the three genotypes (total n = 72). We focused on the pool of ‘soluble’ SNCA protein that are found in the neuronal cytosol and membrane-associated compartments, which encompass the overwhelming majority of available SNCA proteins in young murine brain (in contrast to the low amount that is present in ‘insoluble’ compartments at that age) (30, 31). Following the observation of variable SNCA signals in routine western blotting experiments (Fig. 4A), which represents a low throughput and non-linear method of signal detection, we employed an extensively validated, sandwich-type enzyme-linked immunoadsorbent assay (ELISA) protocol for the quantification of SNCA in each mouse brain (Fig. 4B) (31–33). Using ELISA pair [hSA3/211-B], which combines a sensitive, affinity-purified rabbit antibody [hSA3] with a human SNCA-specific monoclonal antibody (mAb) (211), we first calculated the absolute concentration (ng/µl) of human SNCA in each mouse homogenate (Supplementary Material, Fig. S3A); these values were then corrected for the gene copy number in each line to determine the protein concentration per one diploid-copy of the transgene. Consistent with our mRNA results, we recorded a 1.25-fold elevation in human SNCA levels in 261/261 transgenic mouse brain (0.71 ± 0.30 ng/µl homogenate) over the protective 259/259 allele carrying mice (0.57 ± 0.56 ng/µl) (P = 0.0004; Fig. 5A). As expected from the mRNA data (see above), the lowest concentration of human SNCA was recorded in the Δ-Rep-carrying mice (0.17 ± 0.27 ng/µl) (Fig. 5A).
Figure 4.
Analyzing SNCA proteins in F2 transgenic mouse brains. Representative western blotting (A) and ELISA (B) results from 24 age- and gender-matched F2 transgenic mouse brains are shown for the three distinct genotypes in six mouse lines. Specimens were assayed with a combination of polyclonal (pAb) and monoclonal (mAb) anti-SNCA antibodies. Equal volumes of whole-brain extracts were either directly loaded onto reducing SDS–PAGE (A) or first adjusted for their total protein content, then diluted at 1: 5000 and subsequently loaded onto ELISA plates (B). Sandwich ELISA (hSA-3/211-B; red bars to the right of each pair) and ELISA (hSA-3/Syn1-B; blue bars on the left of each pair) were carried out on the same 384-well plate in triplicates. Note the near identical monitoring of the recombinant human SNCA protein standards by both assays (left); the absence of any absorbance signal in snca-null (KO) mouse brain by either assay and the absence of signal with the human SNCA-specific ELISA in WT (non-transgenic) mouse brain (center); the fact that the total SNCA signal measuring murine and human protein (blue bars, left) is always stronger than the human-specific signal alone (red bars, right).
Figure 5.
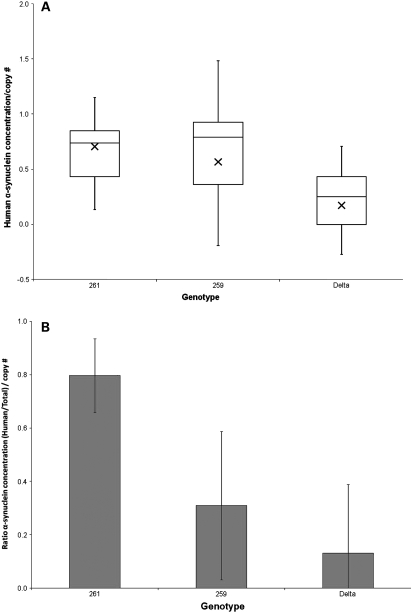
SNCA-Rep1 expansion increases SNCA protein concentration in mammalian brain. (A) Quantification of human SNCA protein in transgenic mouse brain homogenates using sandwich ELISA [hSA-3/211-B] and a 384-well plate format (triplicates; 50 µl/well); a total of 72 mice were analyzed. Concentration values (in ng/µl) were interpolated from the standard curve (Supplementary Material, Fig. S3A) and then corrected for the calculated gene copy number in each mouse line. Values were grouped according to the three genotypes examined, i.e. Rep1-261 bp; Rep1-259 bp; Δ-Rep1 (ratio: female:male, 50:50 in all lines). (B) Relative ratio of the human SNCA concentration (as shown in A) to the total SNCA protein concentration. The latter was measured using sandwich ELISA [hSA3/Syn1-B] on 384-well plates quantifying both mouse Snca and human SNCA (Supplementary Material, Fig. S3B; after correction for gene copy number: Supplementary Material, Fig. S3C). Bar graph shown in (B) represents values obtained in (A) divided by those shown in Supplementary Material, Fig. S3C.
To control for any variability in the expression of endogenous, murine snca, which could have affected transgene penetrance, we next calculated the relative abundance of human versus total (i.e. human and mouse) SNCA in the same brain homogenates. To this end, we employed our sandwich ELISA protocol [hSA3/Syn1-B], which reproducibly quantifies total SNCA from both rodent and primate sources and does not cross-react with β- or with γ-synuclein proteins (33) (Fig. 4B). Mouse lines carrying the expanded Rep1-261 bp allele showed the highest total SNCA concentration (measuring human and mouse variants, i.e. 6.17 ± 2.53 ng/µl homogenate), followed by lines that carry the protective Rep1-259 bp genotype (5.63 ± 4.74 ng/µl), followed by those carrying the Δ-Rep1 allele (4.74 ± 2.02 ng/µl) (Supplementary Material, Fig. S3B). In accordance with the above, we recorded the highest ratio of ‘human-to-total SNCA concentrations’ following copy number corrections in brains from 261/261 mice (average ratio 0.80), followed by 259/259 animals (average ratio 0.31) and Δ-Rep1 mice (average ratio 0.13) (P < 0.0001; Fig. 5B; Supplementary Material, Fig. S3C). The findings of relative and absolute SNCA concentration differences between these transgenic mouse lines remained the same when the ELISA operator was blinded to their respective genotypes (not shown). As in our mRNA studies before, we did not observe a gender effect on neural SNCA protein concentrations. We concluded from these data that the Rep1 region acts as a cis-regulatory enhancer of SNCA transcription, thereby effectively modulating the steady state of human SNCA in the mammalian brain.
Effects of Rep1 variants on human SNCA-mRNA levels in mouse blood
The effect of Rep1 on human SNCA-mRNA levels was also studied in whole-blood samples from these transgenic mice (n = 72), as obtained prior to decapitation. The quantification of SNCA proteins in peripheral blood is of interest to the PD field as a potential biomarker (32), as well as from the perspective of tissue-specific gene expression (34). To this end, we carried out a similar study in mouse blood samples to the one described above for the brain; in contrast to the brain analysis, ubiquitously expressed Gapdh was used as the endogenous reference probe to measure human SNCA-mRNA levels in murine samples (Fig. 6A). We found that mice carrying the PD risk-associated allele Rep1-261 bp showed, on average, higher human SNCA-mRNA levels in blood specimens obtained from the orbital sinus compared with mice transgenic for the protective genotype with allele Rep1-259 bp (0.1 versus 0.005-fold expression on average relative to the endogenous Gapdh mRNA levels, respectively) (Fig. 6B). However, the variability within each genotype was quite large, and the median values showed no significant differences (Fig. 6B). No correlation was observed between individual blood SNCA-mRNA levels and gender (P = 0.221).
Figure 6.
Effect of the SNCA-Rep1 promoter genotypes on human SNCA-mRNA expression levels in transgenic mouse whole blood. (A) Validation curve of the Δ real-time assay for relative quantization of human SNCA-mRNA relative to mouse Gapdh-mRNA in blood. Relative efficiency plots of human-SNCA and mouse-Gapdh were formed by plotting the log input amount (ng of total RNA) versus the ΔCt = [Ct(SNCA)−Ct(Gapdh)]. The slope is 0.1, which indicated the validation of the ΔCt calculation in the range of 0.1–100 ng RNA. (B) Fold levels of human SNCA-mRNA were assayed by real-time RT–PCR and calculated relative to mouse Gapdh-mRNA reference control using the 2−ΔCt method. No detectable products of the human SNCA-mRNA reaction were observed when WT mouse blood-cDNA was used as template. The risk genotype, 261/261, correlates with higher SNCA-mRNA levels than the protective genotype, 259/259. The deleted SNCA-Rep1 genotype shows no significant correlation with SNCA-mRNA levels in comparison with genotypes 261/261 and 259/259 (P = 0.524). In total, 72 mice were analyzed; for each genotype (261/261, 259/259 and Δ/Δ), the box plot represents the analysis performed using two transgenic lines, 12 animals from each line (6 males and 6 females); overall, 24 whole-blood samples (per genotype) were each repeated twice, independently. The average values are presented by ‘X’. The box plot shows the median (horizontal line inside the box) and the 25th and 75th percentiles (horizontal borders of the box). The range between the 25th and 75th percentiles is the interquartile range. The whiskers show the minimal and maximal values inside the main data body.
We also assessed the contributory role (if any) of the Rep1 region to SNCA-mRNA levels in these blood specimens. In contrast to the results obtained in brain homogenates, we did not observe an apparent Rep1 effect on SNCA-mRNA expression levels in these hematological specimens; there was no significant difference in SNCA-mRNA levels in blood from mice carrying the deleted Rep1 allele (0.046-fold on average relative to the endogenous Gapdh mRNA) when compared with the average value of mice carrying either allele (261 or 259) at the SNCA-Rep1 site (0.05-fold on average; P = 0.524). Here again, the median values did not differ among the three genotypes (Fig. 6B). These findings suggested that the Rep1 element did not appear to regulate the expression level of SNCA gene in the hematological precursor cells that generated these blood specimens (34,35), thereby suggesting a neuro-specific effect for the Rep1 element (27).
In order to determine whether SNCA-mRNA levels in the blood are individually concordant with mRNA levels recorded in the brain, we carried out several correlation studies in these mice; however, the comparison of the relative SNCA-mRNA levels between the brain and blood samples from each individual animal showed no correlation.
Effects of Rep1 variants on human SNCA protein concentrations in mouse blood
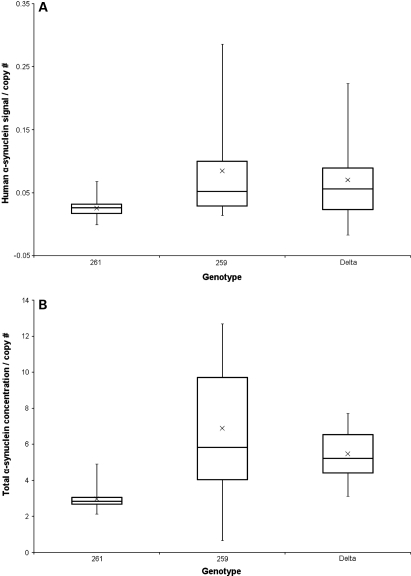
We next analyzed the effect of Rep1 on human SNCA protein levels in sister aliquots of the same blood specimens from transgenic mice carrying these three different alleles. To this end, we examined blood lysates from age- and gender-matched mice by the same two ELISA protocols employed above (Supplementary Material, Fig. S4A) and corrected the SNCA signal for the gene copy number in each animal. The parallel analysis of blood lysates (n = 72) on the same ELISA plate by an operator blinded to the genotype revealed no statistically significant difference in the levels of human SNCA (P = 0.477) among the six transgenic lines that carried the three distinct SNCA-Rep1 genotypes (Fig. 7A). In accordance with the human-specific values, the total SNCA concentration (of human and mouse SNCA) in these blood specimens did not reveal a significant difference in the Rep1-261 bp-carrying animals (Rep1-261 bp; total SNCA 20.68 ± 2.96 ng/µl) when compared with the other two genotypes (Rep1-259 bp 19.59 ± 3.14 ng/µl; Δ-Rep1 18.60 ± 2.33 ng/µl) (Supplementary Material, Fig. S4B). After copy number correction, the total SNCA concentration in the mouse blood showed a trend for the reduction in the PD risk-associated Rep1-261 bp animals, but the variability, in particular within the Rep1-259 bp genotype, was large (Fig. 7B). The calculated ratios of ‘human-to-total-SNCA levels following gene copy number correction’ were 0.85 in 261/261 mice, 1.23 in 259/259 animals and 1.28 in Δ-Rep1 mice. However, it is important to state that the overall human SNCA protein concentrations in these mouse blood specimens were markedly lower than in comparable human specimens (34,35), which may explain the failure to detect any consistent and meaningful variations. These results suggested that the human SNCA protein levels in these blood specimens did not parallel the expression levels found in the brains from the same mice.
Figure 7.
SNCA-Rep1 expansion does not lead to detectable SNCA protein concentration differences in mouse blood. (A) Quantification of human SNCA protein in transgenic mouse blood homogenates using sandwich ELISA [hSA-3/211-B] and 384-well plate format (triplicates; 50 µl/well); a total of 72 mice were analyzed. Signals were specific for human SNCA protein because whole blood from WT and snca-knockout mice did not generate ELISA signals above the background (Supplementary Material, Fig. S4A). Values were then corrected for the calculated gene copy number in each mouse line and grouped according to the three genotypes examined, i.e. Rep1-261 bp; Rep1-259 bp; Δ-Rep1 (ratio: female:male, 50:50 in all lines). (B) Quantification of total SNCA protein concentration levels in transgenic mouse blood was carried out with sandwich ELISA [hSA3/Syn1-B] and 384-well plates. This ELISA measures both mouse snca and human SNCA proteins (Supplementary Material, Fig. S4B); values (in ng/µl) were then corrected for gene copy number.
DISCUSSION
This study describes a novel approach in the field of neurodegenerative diseases for examining the role of genetic variations positioned in non-coding, putative regulatory regions of the human genome by aiming to identify causality for regulatory variants. Gene expression studies in humans, especially when they involve the analysis of post-mortem brain, often introduce several confounding variables, e.g. differences in genetic background, age, environmental exposures (including pharmacotherapy), length of illness and importantly, variability in post-mortem interval and tissue handling. All of these factors can affect mRNA and protein stability, and thus their measureable abundance (36,37). In order to overcome the combination of these confounding factors involved in studying mRNA and protein levels in human post-mortem tissue, we took the novel approach of studying human gene expression in a transgenic mouse model. Transgenic mouse models have been used, almost exclusively, to study the effect of mutations in coding gene sequences. Here, we generated a transgenic model, which carries the entire human genomic region of the SNCA gene, to study the effect of a variation located ∼10 kb upstream of the transcription start site within the putative promoter/enhancer region and to dissect its contribution to human SNCA expression levels in vivo.
A critical question concerning the molecular pathogenesis of PD is what contributory role, if any, is played by the SNCA gene in sporadic PD. Several studies conferred an association of common SNCA polymorphisms in the 5′ and 3′ regions of the gene locus with PD susceptibility (16–19) (38); however, the causal variant/s and hence the underlying mechanism/s remained elusive. We hypothesized that altered regulation of SNCA gene expression levels is important in the development of sporadic PD in a significant subset of patients; these include subjects who do not express a mutant SNCA protein and who do not carry an elevated SNCA gene dosage in their genome. One important mechanism that modulates gene expression levels is transcriptional efficiency. Markers across the 5′ region of SNCA that were associated with increased odds ratio to develop PD are in almost complete linkage disequilibrium with each other, which makes pinpointing the biologically relevant variant within an associated 5′ region challenging. In this study, we provide evidence for a contributory role of the SNCA-Rep1 variant in the transcriptional regulation of the SNCA gene and in their effects on SNCA-mRNA and protein levels in mouse brain. We found a positive correlation between SNCA-Rep1 allele lengths at the SNCA enhancer/promoter region and human SNCA-mRNA as well as protein levels in the brains of these transgenic mice. Although we are cognizant of the potential limitations by using whole-brain homogenates rather than region-specific specimens (that are of relevance to PD pathogenesis), we purposefully sought to minimize any introduction of inaccuracies stemming from variability in tissue dissection and to accommodate a larger number of animals in our quantitative assays. Our results show that allele Rep1-261 bp, which confers increased risk to develop late-onset, sporadic PD, was significantly correlated with higher SNCA levels relative to the protective allele Rep1-259 bp (P < 0.0001). Intriguingly, our collective data suggest that the carrier status of Rep1-261 bp homozygosity within an otherwise WT complement of two SNCA gene copies results in a 1.7-fold elevation of SNCA-mRNA and a 1.25-fold elevation in the protein level, suggesting that variability in translational efficiency had only a limited effect. Therefore, Rep1-261 bp homozygosity may be functionally equivalent to the situation of subjects with SNCA locus duplication who carry three SNCA copies, which results in 1.5-fold elevation. We therefore speculate that Rep1-261 bp homozygosity confers a critical gain-of-function mechanism in PD penetrance and in the expressivity of synucleinopathies (6).
Although the biological effect of the Rep1 expansion was observed in the most relevant organ to PD expression, i.e. the brain, its contribution appeared specific because its enhancer role was not recorded in the corresponding blood specimens. Of note, Fuchs et al. (28) recently reported a positive genotype-to-phenotype effect for Rep1 repeat length in human blood samples, where homozygotes of the protective, shorter allele had lower concentrations of SNCA protein in peripheral blood mononuclear cells; intriguingly, and in agreement with our finding, these authors observed no effect at the mRNA level in peripheral blood. At the current time, we cannot exclude a modulatory effect by the Rep1 genotype on SNCA protein levels in peripheral blood (as suggested by Fuchs et al.), because the human protein concentrations in our transgenic mouse blood were unexpectedly low, which could have put any significant variation below the limit of detection. The discrepancy between the Fuchs et al. and our study may also be explained by a variety of other factors; chief among them are the differences in donor species and sample sizes, the methods of blood collection and processing and, last but not least, the differences between the ELISA protocols employed.
The quantification of peripheral SNCA-mRNA levels together with the characterization of the Rep1 genotype has also been explored in the context of alcohol use-related disorders (39); the most recent study suggests an independent (but not a pairwise) association of Rep1 genotype and alcohol abuse with PD risk (40). Clearly, larger cross-sectional and longitudinal studies of carefully collected cohorts and validated, standardized protocols are now needed to further investigate these neuropsychatric conditions. This aspect is particularly relevant for PD biomarker efforts that seek to correlate variations in the transcriptome and/or proteome of venous blood with the clinical phenotype of living donors (11,41). Overall, the observed neuro-specific effect of the Rep1 element in vivo using a mouse model is consistent with our previous cellular results. Using a luciferase reporter system, we had observed that the construct harboring the expanded allele at the SNCA-Rep1 site generated the highest luciferase activity specifically in dopaminergic SH-SY5Y neuroblastoma cells (27).
Our results lend support for the overall biological significance of SNCA-Rep1, thereby suggesting that many of the observed associations between the SNCA 5′ region and PD susceptibility could possibly be explained by variations in SNCA-Rep1 genotype. We predict that the association of the 261 bp-long risk allele with sporadic PD is conferred by direct upregulation of SNCA transcription, and that the protective allele 259 bp acts through the lowering of its expression rate. Here, we chose to focus on the two most common Rep1 alleles; we acknowledge the role that other variants may play, such as the rare PD-associated 263 bp-long Rep1 variant (25). Additional Rep1 allelic variants will be analyzed using newly created transgenic mice in future studies. Likewise, it will be of interest to determine whether our PAC mice will develop any neuropathological and behavioral features of human synucleinopathies toward the end of their lifespan that are currently not seen in those transgenic models that are based solely on an SNCA cDNA construct design (42,43). At the age of 8 weeks, we have observed no readily recognizable alteration in the behavior of the mice used in this study.
In our previous work, we identified a transcription factor, poly(ADP-ribose) transferase/polymerase-1 (PARP-1, MIM 173870), which specifically bound to Rep1 and modulated SNCA transcription (44). It will be interesting to explore whether the variability in SNCA regulation (as mediated by Rep1 alleles) rests on altered binding efficiency of PARP-1 to Rep1 sequences that vary in length. Alternatively, our observed Rep1 expansion effect could also be explained by interaction/s with cis-acting elements during chromatin unfolding, which precedes SNCA gene transcription, for example, with the recently discovered GATA motif within intron 1. This intronic GATA site is selectively occupied by GATA-2 (MIM 137295) protein in neurons and by GATA-1 (MIM 305371) in erythroid cells (34).
The transgenic mouse approach we have pursued here could serve as a valuable model system for a variety of future studies in the PD field, including those that examine gene–environment interactions, such as the protective effects of smoking (45,46); our approach may also serve proof of concept studies, such as those that target the reduction of human SNCA levels in vivo either through the specific modulation of mRNA translation (47) or through the acceleration of SNCA degradation by candidate enzymes (33,48). The unequivocal evidence that puts increased steady states of SNCA at the center of PD development (10,11,47,49,50) emphasizes the importance of studies like ours in the pursuit of three important (and related) research goals: to better dissect the pathogenesis of synucleinopathies; to develop earlier markers of risk and disease state; and to find validated drug targets near the root of Parkinson's to initiate cause-directed therapy in the future. Further exploration of the mechanism by which Rep1 allele expansion enhances human SNCA gene transcription in at-risk neurons may serve all three goals.
MATERIALS AND METHODS
Recombineering PAC
A PAC containing the 146 kb PAC 27M07 (RPCI human male PAC library, BAC/PAC Resource Center, Children's Hospital, Oakland, CA, USA) was used as the substrate for recombineering by the method of Lee et al. (51). Human genomic DNA spanning the SNCA-Rep1 locus and containing either the 261 bp allele ((TC)10(T)2(TC)10(TA)8(CA)11, heretofore referred to as Rep1-261 bp) or the 259 bp allele ((TC)10(T)2(TC)10(TA)8(CA)10, heretofore referred to as Rep1-259 bp) was isolated by PCR of anonymous human DNA of the appropriate SNCA-Rep1 genotype (Centre d'Etude du Polymorphisme Humaine). Alleles were defined according to the length of the PCR product in concordance to the conventional nomenclature (20,25). The PCR fragments were sequenced and then cloned into a recombineering vector for site-directed recombination in Escherichia coli into the SNCA-Rep1 locus in PAC 27M07. A third PAC was recombineered in which the SNCA-Rep1 repeat had been precisely deleted, heretofore referred to as Δ-Rep1 (27). All three PACs were identical except at the SNCA-Rep1 locus.
Generation of transgenic lines
The three different constructs were microinjected into the pronuclei of fertilized FVB/N mouse ova by standard techniques. F1s heterozygous for the intact human PAC transgenes were crossed to produce homozygotes for each allelic variant transgene. Mice homozygous for the human transgene harboring 261 and 259 bp SNCA-Rep1 alleles and the deleted allele are referred to as 261/261, 259/259, Δ/Δ mice, respectively. Of note, one of the Δ-Rep1 lines, ‘B’, was non-viable probably due to insertion site effect, thus for this line, the analysis was performed using heterozygous mice, and the transgene copy number was corrected accordingly (see below). Two lines were generated from each genotype (i.e. a total of six lines). Six male and six female F2s homozygous from each transgenic line, 72 mice in total, were sacrificed at the age of 8 weeks for sample collection. Blood was collected from the orbital sinus, and subsequently, the whole brain was harvested. It is important to note that all mouse lines shared an identical genetic background, which included the WT murine snca locus, and were reared in the same environment. In addition, all of the mice underwent identical blood collection and brain-harvesting procedures.
DNA analysis of transgenic mice
We used DNA from mouse tails to confirm the founders. The SNCA-Rep1 genotypes were verified by direct sequence analysis of the polymorphic repeat and its flanking region. The intactness of the PAC transgenes in each founder animal and its transmission to the F1 generation were assayed by a routine series of 10 PCR assays along the transgene sequence. Primer sequences and PCR conditions for genotyping are available upon request. FISH was used to estimate transgene copy number in the F1 generation. Briefly, the human-PAC targeting construct and a mouse BAC covering comparable size sequence from mouse chromosome 9 were used as probes. Interphase preparations from leukocytes and metaphase preparations from spleen cells were generated by standard air-drying technique, and FISH was performed with labeled DNA prepared by nick translation using spectrum orange-dUTP (red) or spectrum green-dUTP (green) (Vysis, Downers Grove, IL, USA) essentially as described previously (52,53). On each slide, 100 ng of labeled probe was applied. Ten microliters of a hybridization mixture containing the labeled DNA in 50% formamide, 2× SSC and 10% dextran sulfate were denatured at 75°C for 10 min and then incubated at 37°C for 30 min for preannealing. Slides were denatured and hybridized for at least 18 h and counterstained with DAPI–Antifade before viewing. The relative number of the transgene-DNA copies was estimated by comparison of the signal intensity from the transgenic versus the endogenous probes.
Routine genotyping of all the newborn (F1 and F2 generations mice) was performed by PCR with one of the primer sets used in the founder analysis. To differentiate between F2 homozygous and heterozygous animals, we performed FISH analysis as described above.
Determination of the relative transgene copy number
To assay for transgene dosage, genomic DNA was extracted from F1 mice liver by standard Qiagen protocol and quantitative real-time PCR targeting two different regions along the human PAC, i.e. human-SNCA exons 2 and 6, was performed on the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The β-actin gene was amplified as an endogenous reference for the quantification of the human-SNCA transgene using ABI MGB probe and primer set assay-by-demand ID Mm00607939_s1 (Applied Biosystems). The assay-by-design primers and probes sequences for SNCA exon 2 and exon 6 were described previously (29). TaqMan MGB probes for human SNCA exon 2, exon 6 and mouse β-actin were labeled with 6-FAM. PCR was carried out with TaqMan Universal PCR Master Mix using 10 or 50 ng genomic DNA, 900 nm primers and 250 nm probes in a total reaction volume of 20 µl. The reactions for the test SNCA loci and the reference β-actin locus were prepared and run in parallel. PCR cycling conditions were as follows: 95°C for 10 min, 95°C for 15 s and 60°C for 1 min (40 cycles). Each genomic DNA sample was assayed in two different gDNA amounts (10 and 50 ng), running each amount in duplicate and the entire assay was repeated twice. The cycle in the log phase of PCR amplification at which a significant fluorescence threshold was reached (Ct) was used to quantify each exon. For each transgenic line, the dosage of PAC transgene relative to β-actin and normalized to control DNA was determined for each exon using the 2−ΔΔCt method. A value was considered as a single copy if it was within the range of 0.8–1.2. Greater copy numbers were calculated as ratios relative to a single copy value. A standard curve was generated using gDNA 0.1, 1, 10, 25, 50, 100 and 150 ng, and a regression curve was calculated for each assay (exon 2 and exon 6). In addition, the slope of the relative efficiency plot for SNCA exons 2 and 6 and the reference control β-actin was determined to validate the assays (Supplementary Material, Fig. S5). The transgene copy number was calculated relative to the endogenous mouse β-actin (Act) gene, and all copy number variations were expressed relative to having one copy number per haploid genome (ratio of 0.8–1.2 for homozygous transgene relative to diploid mouse β-actin gene). The copy number of the only heterozygous Δ-Rep1 transgenic line ‘B’ was adjusted accordingly.
RNA extraction and cDNA synthesis
Total RNA was extracted from half whole brain and whole blood (200 µl) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) followed by purification with RNeasy Kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol. RNA concentration was determined spectrophotometrically at 260 nm, and the 260/280 nm ratio determined the quality of the purification. In addition, the quality of the sample and the lack of significant degradation products were confirmed on an Agilent Bioanalyzer. Next, cDNA was synthesized using MultiScribe RT enzyme (Applied Biosystems) under the following conditions: 10 min at 25°C and 120 min at 37°C.
RT–PCR and real-time PCR
The expression of SNCA-RNA in the brain tissues of F1 generation mice was determined for each line by RT–PCR using the forward primer 5′-CATGGATGTATTCATGAAAGG-3′ and reverse primer 5′-GAGTGTAGGGTTAATGTTCC-3′.
Real-time PCR was used to quantify human SNCA-mRNA levels in F2 generation transgenic mice as described previously (14). Briefly, duplicates of each sample were assayed by relative quantitative real-time PCR using the ABI 7900 for the analysis of the level of SNCA message when compared in brain tissues to mRNA-encoding mouse synaptophysin (Syp), a presynaptic protein that has similar expression pattern to SNCA, and in blood to the mouse ubiquitous gene, glyceraldehyde-3-phosphate dehydrogenase (Gapdh), as internal references (14,44). Each cDNA (10 ng) was amplified in duplicate using TaqMan Universal PCR master mix reagent (Applied Biosystems) and the following conditions: 2 min at 50°C, 10 min at 95°C, 40 cycles: 15 s at 95°C, and 1 min at 60°C. The target SNCA cDNA was amplified using ABI MGB probe and primer set assay ID Hs00240906_m1, normalized to a Syp RNA control (ABI MGB probe and primer set assay ID Mm00436850_m1) for the brain samples and to a Gapdh RNA control (ABI MGB probe and primer set assay ID Mm99999915_g1) for the blood samples (Applied Biosystems). As a negative control for the specificity of the human-SNCA ‘assay-on-demand’ amplification of human message, we used whole-brain and blood RNA samples from WT FVB mouse. Data were analyzed with a threshold set in the linear range of amplification. The cycle number at which any particular sample crossed that threshold (Ct) was then used to determine the fold difference.
Fold difference was calculated as 2−ΔCt; ΔCt = [Ct(SNCA)−Ct(Syp or Gapdh)]. Of note is that three internal controls were compared: mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh Mm99999915_g1), mouse alpha-synuclein (Snca Mm00447331_m1), mouse enolase 2 (Eno2 Mm00469062_m1) and synaptophysin (Syp Mm00436850_m1) using different total RNA amounts (0.1–100 ng) from whole-brain tissue and whole-blood samples of a transgenic mouse. For assay validation, we used standard curves for each assay (Gapdh, Snca, Eno2 and Syp) using a transgenic mouse whole-brain and whole-blood RNA samples. In addition, the slope of the relative efficiency plot for SNCA and each internal control was determined to validate the assays. The slope in the relative efficiency plot for mouse Syp and human SNCA in brain (0.045) and for mouse Gapdh and human SNCA in blood (0.1) were the smallest, showing a standard value required for the validation of the relative quantitative method (Figs 3A and 6A). Thus, for the extended study, we chose Syp and Gapdh as the internal control for the whole brain and blood, respectively. In addition, for a subset group of brain samples, we used the geometric mean of mouse Syp and Eno2 as a normalization control and confirmed the selection of Syp as a representative normalization control for the entire brain set.
Western blotting and antibodies
The expression of human SNCA protein in the brain tissues was determined by western blotting with the mouse mAb LB509 (Zymed) and mAb 211 that are specific for the human orthologue of SNCA, with mAb Syn-1 (Transduction Labs) and hSA3. The latter is a polyclonal Ab raised and affinity-purified against full-length, human SNCA, as described (33). Syn-1 and hSA3 Ab react with both the mouse and human orthologues of SNCA (31).
Whole brains from 2- to 3-month-old mice were homogenized in 10-fold volume of 50 mm Tris–HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, in the presence of a protease inhibitor cocktail (Sigma, St Louis, MO, USA) using a hand-held motorized pestle. After centrifugation to remove particulates at 100 000g (30 min; 4°C), total protein concentrations were determined by the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA, USA), and 20 µg of each sample was run on 15% Tris–glycine SDS–PAGE. Proteins were transferred to PVDF membranes (Immobilon-P, Millipore, Bedford, MA, USA), and blots were blocked with 2% casein in 0.1 m Tris, 0.2 m NaCl, pH 9.5. Secondary antibody was horseradish peroxidase-coupled goat anti-mouse IgG (Amersham, Chicago, IL, USA), used at 1:10 000. The immunoreactive proteins were detected using the ECL system (Amersham) according to manufacturer's instructions. For the analysis of F1 generation mouse lines, transgene expression was determined by western blots in comparison with recombinant human SNCA (50 ng).
Enzyme-linked immunoadsorbent assays
SNCA protein levels were assessed using a 384-well-based sandwich ELISA system as described recently in detail (33, 34). For specific quantification of the human orthologue, we employed the Ab sandwich [hSA3/211-B]; for the quantification of total SNCA (i.e. mouse and human combined), we employed the sandwich pair [hSA3/Syn1-B], where ‘B’ stands for biotinylation). Recombinant protein was used in standard dilutions for the interpolation of SNCA concentration, as described (33). ELISA signals were monitored at 405 nm every 5 min for up to 60 min. Saturation kinetics were examined for the identification of time point(s) where standards and sample dilutions were in the log phase (33). To control inter-assay variability, all samples were run using two different dilutions in triplicate on the same day with the same lot of standards. Also, a mouse WT versus knock-out Snca curve was generated and included as positive and negative control for the [hSA3/Syn1-B] assay (33); both WT and knock-out snca mouse tissue served as negative controls for the human SNCA-specific [hSA3/211-B] assay. The relative concentration estimates of human SNCA in transgenic mouse brain and blood specimens were calculated according to the standard curve. Frozen mouse brains were thawed, weighed (mg), individually homogenized in three volumes (µl) of Triton X-100-containing lysis buffer and centrifuged at 100 000g during 30 min at 4°C, as described (54). Note that each round of homogenization included at least one representative from each of the two mouse lines of the three genotypes. Protein concentrations were calculated and equalized prior to loading. Frozen whole-blood specimens from decapitated mice (drawn in EDTA-containing tubes) were thawed, diluted with normal saline-containing Triton X-100 (1%) and protease inhibitors (Roche) and centrifuged before ELISA analysis.
Statistical analysis
SNCA-mRNA fold expression value of each sample was analyzed repetitively and the results of all replicates were averaged. The average values for each sample (12 samples per line) were used to calculate the average fold expression of each of the six lines. All average values were expressed as mean ± SEM. The significance of the difference between the mRNA measurements of the three strains carrying the different SNCA-Rep1 alleles was analyzed by the one-sided Wilcoxon rank-sum test. Correlations were assessed by linear regression analyses. The general linear model (GLM) method was used to evaluate the effect of the primary explained variable (SNCA-Rep1 genotype) as well as other secondary variable (sex) on the RNA levels. The GLM is a procedure unifying the ordinary linear regression and ANOVA as well as other procedures based on the least square computation such as ANCOVA. Since gender may also show an effect on the RNA levels, it was included in the model as a factor. Where the P-value of the maximal model remains significant, an effect of each single term was estimated calculating the type III sum of squares and the corresponding F-value and its P-value. Protein concentrations of all mice from each line were used to calculate the average protein concentration of human SNCA of each of the six lines, which were expressed as mean mean ± SEM. All analyses were carried out using STATA/IC10.0 statistical software (StataCorp, College Station, TX, USA).
AUTHOR CONTRIBUTIONS
K.D.C. performed all the experiments and analyses for the DNA dosage and RNA levels; D.G. did the statistical analyses; P.M. performed the western and all ELISA experiments for the protein analyses; C.L. contributed to the genotyping analysis; A.R. and D.J.B. maintained, breed the mice and carried out tissue harvesting; B.M.O. generated, sequenced and analyzed the PAC recombinant clones; O.M.A.E. co-developed the human-specific ELISA; M.G.S. designed and supervised the protein experiments and analyses; O.C.-F. designed and supervised the DNA and RNA experiments and analyses; R.L.N. and O.C.-F. conceived and designed the study; M.G.S., R.L.N. and O.C.-F. drafted and finalized the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported in part by the Institute for Genome Sciences and Policy at Duke University (to O.C.-F.); the Intramural Research Program of the National Human Genome Research Institute (to R.L.N); the National Institutes of Health/National Institute for Neurological Disorders and Stroke (grant number 5P50-NS038375) and the Parkinson Research Consortium at the University of Ottawa (to M.G.S.). Funding to pay the Open Access publication charges for this article was provided by the Institute for Genome Sciences and Policy at Duke University (to O.C.-F.).
REFERENCES
- 1.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 2.Kruger R., Kuhn W., Muller T., Woitalla D., Graeber M., Kosel S., Przuntek H., Epplen J.T., Schols L., Riess O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat. Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 3.Zarranz J.J., Alegre J., Gomez-Esteban J.C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atares B., et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 4.Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., et al. Alpha-synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs J., Nilsson C., Kachergus J., Munz M., Larsson E.M., Schule B., Langston J.W., Middleton F.A., Ross O.A., Hulihan M., et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 6.Ross O.A., Braithwaite A.T., Skipper L.M., Kachergus J., Hulihan M.M., Middleton F.A., Nishioka K., Fuchs J., Gasser T., Maraganore D.M., et al. Genomic investigation of alpha-synuclein multiplication and parkinsonism. Ann. Neurol. 2008;63:743–750. doi: 10.1002/ana.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 8.Ibanez P., Bonnet A.M., Debarges B., Lohmann E., Tison F., Pollak P., Agid Y., Durr A., Brice A. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 9.Pankratz N., Wilk J.B., Latourelle J.C., Destefano A.L., Halter C., Pugh E.W., Doheny K.F., Gusella J.F., Nichols W.C., Foroud T., et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum. Genet. 2008;124:593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrer M., Kachergus J., Forno L., Lincoln S., Wang D.S., Hulihan M., Maraganore D., Gwinn-Hardy K., Wszolek Z., Dickson D., et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann. Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 11.Miller D.W., Hague S.M., Clarimon J., Baptista M., Gwinn-Hardy K., Cookson M.R., Singleton A.B. Alpha-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62:1835–1838. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- 12.Spillantini M.G., Schmidt M.L., Lee V.M., Trojanowski J.Q., Jakes R., Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 13.Gai W.P., Power J.H., Blumbergs P.C., Blessing W.W. Multiple-system atrophy: a new alpha-synuclein disease? Lancet. 1998;352:547–548. doi: 10.1016/s0140-6736(05)79256-4. [DOI] [PubMed] [Google Scholar]
- 14.Chiba-Falek O., Lopez G.J., Nussbaum R.L. Levels of alpha-synuclein mRNA in sporadic Parkinson disease patients. Mov. Disord. 2006;21:1703–1708. doi: 10.1002/mds.21007. [DOI] [PubMed] [Google Scholar]
- 15.Grundemann J., Schlaudraff F., Haeckel O., Liss B. Elevated alpha-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson's disease. Nucleic Acids Res. 2008;36:e38. doi: 10.1093/nar/gkn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pals P., Lincoln S., Manning J., Heckman M., Skipper L., Hulihan M., Van den Broeck M., De Pooter T., Cras P., Crook J., et al. Alpha-synuclein promoter confers susceptibility to Parkinson's disease. Ann. Neurol. 2004;56:591–595. doi: 10.1002/ana.20268. [DOI] [PubMed] [Google Scholar]
- 17.Mueller J.C., Fuchs J., Hofer A., Zimprich A., Lichtner P., Illig T., Berg D., Wullner U., Meitinger T., Gasser T. Multiple regions of alpha-synuclein are associated with Parkinson's disease. Ann. Neurol. 2005;57:535–541. doi: 10.1002/ana.20438. [DOI] [PubMed] [Google Scholar]
- 18.Mizuta I., Satake W., Nakabayashi Y., Ito C., Suzuki S., Momose Y., Nagai Y., Oka A., Inoko H., Fukae J., et al. Multiple candidate gene analysis identifies alpha-synuclein as a susceptibility gene for sporadic Parkinson's disease. Hum. Mol. Genet. 2006;15:1151–1158. doi: 10.1093/hmg/ddl030. [DOI] [PubMed] [Google Scholar]
- 19.Winkler S., Hagenah J., Lincoln S., Heckman M., Haugarvoll K., Lohmann-Hedrich K., Kostic V., Farrer M., Klein C. {alpha}-Synuclein and Parkinson disease susceptibility. Neurology. 2007;69:1745–1750. doi: 10.1212/01.wnl.0000275524.15125.f4. [DOI] [PubMed] [Google Scholar]
- 20.Xia Y., Rohan de Silva H.A., Rosi B.L., Yamaoka L.H., Rimmler J.B., Pericak-Vance M.A., Roses A.D., Chen X., Masliah E., DeTeresa R., et al. Genetic studies in Alzheimer's disease with an NACP/alpha-synuclein polymorphism. Ann. Neurol. 1996;40:207–215. doi: 10.1002/ana.410400212. [DOI] [PubMed] [Google Scholar]
- 21.Touchman J.W., Dehejia A., Chiba-Falek O., Cabin D.E., Schwartz J.R., Orrison B.M., Polymeropoulos M.H., Nussbaum R.L. Human and mouse alpha-synuclein genes: comparative genomic sequence analysis and identification of a novel gene regulatory element. Genome Res. 2001;11:78–86. doi: 10.1101/gr.165801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrer M., Maraganore D.M., Lockhart P., Singleton A., Lesnick T.G., de Andrade M., West A., de Silva R., Hardy J., Hernandez D. Alpha-synuclein gene haplotypes are associated with Parkinson's disease. Hum. Mol. Genet. 2001;10:1847–1851. doi: 10.1093/hmg/10.17.1847. [DOI] [PubMed] [Google Scholar]
- 23.Mizuta I., Nishimura M., Mizuta E., Yamasaki S., Ohta M., Kuno S. Meta-analysis of alpha synuclein/NACP polymorphism in Parkinson's disease in Japan. J. Neurol. Neurosurg. Psychiatr. 2002;73:350. doi: 10.1136/jnnp.73.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellick G.D., Maraganore D.M., Silburn P.A. Australian data and meta-analysis lend support for alpha-synuclein (NACP-Rep1) as a risk factor for Parkinson's disease. Neurosci. Lett. 2005;375:112–116. doi: 10.1016/j.neulet.2004.10.078. [DOI] [PubMed] [Google Scholar]
- 25.Maraganore D.M., de Andrade M., Elbaz A., Farrer M.J., Ioannidis J.P., Kruger R., Rocca W.A., Schneider N.K., Lesnick T.G., Lincoln S.J., et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 26.Chiba-Falek O., Touchman J.W., Nussbaum R.L. Functional analysis of intra-allelic variation at NACP-Rep1 in the alpha-synuclein gene. Hum. Genet. 2003;113:426–431. doi: 10.1007/s00439-003-1002-9. [DOI] [PubMed] [Google Scholar]
- 27.Chiba-Falek O., Nussbaum R.L. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum. Mol. Genet. 2001;10:3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs J., Tichopad A., Golub Y., Munz M., Schweitzer K.J., Wolf B., Berg D., Mueller J.C., Gasser T. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 29.Williams-Gray C.H., Goris A., Foltynie T., Brown J., Maranian M., Walton A., Compston D.A., Sawcer S.J., Barker R.A. No alterations in alpha-synuclein gene dosage observed in sporadic Parkinson's disease. Mov. Disord. 2006;21:731–732. doi: 10.1002/mds.20863. [DOI] [PubMed] [Google Scholar]
- 30.Anderson J.P., Walker D.E., Goldstein J.M., de Laat R., Banducci K., Caccavello R.J., Barbour R., Huang J., Kling K., Lee M., et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J. Biol. Chem. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 31.Cullen V., Lindfors M., Ng J., Paetau A., Swinton E., Kolodziej P., Boston H., Saftig P., Woulfe J., Feany M.B., et al. Cathepsin D expression level affects alpha-synuclein processing, aggregation, and toxicity in vivo. Mol. Brain. 2009;2:5. doi: 10.1186/1756-6606-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Agnaf O.M., Salem S.A., Paleologou K.E., Curran M.D., Gibson M.J., Court J.A., Schlossmacher M.G., Allsop D. Detection of oligomeric forms of alpha-synuclein protein in human plasma as a potential biomarker for Parkinson's disease. FASEB J. 2006;20:419–425. doi: 10.1096/fj.03-1449com. [DOI] [PubMed] [Google Scholar]
- 33.Mollenhauer B., Cullen V., Kahn I., Krastins B., Outeiro T.F., Pepivani I., Ng J., Schulz-Schaeffer W., Kretzschmar H.A., McLean P.J., et al. Direct quantification of CSF alpha-synuclein by ELISA and first cross-sectional study in patients with neurodegeneration. Exp. Neurol. 2008;213:315–325. doi: 10.1016/j.expneurol.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Scherzer C.R., Grass J.A., Liao Z., Pepivani I., Zheng B., Eklund A.C., Ney P.A., Ng J., McGoldrick M., Mollenhauer B., et al. GATA transcription factors directly regulate the Parkinson's disease-linked gene alpha-synuclein. Proc. Natl Acad. Sci. USA. 2008;105:10907–10912. doi: 10.1073/pnas.0802437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbour R., Kling K., Anderson J.P., Banducci K., Cole T., Diep L., Fox M., Goldstein J.M., Soriano F., Seubert P., et al. Red blood cells are the major source of alpha-synuclein in blood. Neurodegener. Dis. 2008;5:55–59. doi: 10.1159/000112832. [DOI] [PubMed] [Google Scholar]
- 36.Stan A.D., Ghose S., Gao X.M., Roberts R.C., Lewis-Amezcua K., Hatanpaa K.J., Tamminga C.A. Human postmortem tissue: what quality markers matter? Brain Res. 2006;1123:1–11. doi: 10.1016/j.brainres.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weis S., Llenos I.C., Dulay J.R., Elashoff M., Martinez-Murillo F., Miller C.L. Quality control for microarray analysis of human brain samples: the impact of postmortem factors, RNA characteristics, and histopathology. J. Neurosci. Methods. 2007;165:198–209. doi: 10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Ross O.A., Gosal D., Stone J.T., Lincoln S.J., Heckman M.G., Irvine G.B., Johnston J.A., Gibson J.M., Farrer M.J., Lynch T. Familial genes in sporadic disease: common variants of alpha-synuclein gene associate with Parkinson's disease. Mech. Ageing Dev. 2007;128:378–382. doi: 10.1016/j.mad.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonsch D., Lederer T., Reulbach U., Hothorn T., Kornhuber J., Bleich S. Joint analysis of the NACP-REP1 marker within the alpha synuclein gene concludes association with alcohol dependence. Hum. Mol. Genet. 2005;14:967–971. doi: 10.1093/hmg/ddi090. [DOI] [PubMed] [Google Scholar]
- 40.Brighina L., Schneider N.K., Lesnick T.G., de Andrade M., Cunningham J.M., Mrazek D., Rocca W.A., Maraganore D.M. Alpha-synuclein, alcohol use disorders, and Parkinson disease: a case–control study. Parkinsonism Relat. Disord. 2009 doi: 10.1016/j.parkreldis.2008.11.011. Epub ahead of print February, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scherzer C.R., Eklund A.C., Morse L.J., Liao Z., Locascio J.J., Fefer D., Schwarzschild M.A., Schlossmacher M.G., Hauser M.A., Vance J.M., et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc. Natl Acad. Sci. USA. 2007;104:955–960. doi: 10.1073/pnas.0610204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masliah E., Rockenstein E., Veinbergs I., Mallory M., Hashimoto M., Takeda A., Sagara Y., Sisk A., Mucke L. Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287:1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- 43.van der Putten H., Wiederhold K.H., Probst A., Barbieri S., Mistl C., Danner S., Kauffmann S., Hofele K., Spooren W.P., Ruegg M.A., et al. Neuropathology in mice expressing human alpha-synuclein. J. Neurosci. 2000;20:6021–6029. doi: 10.1523/JNEUROSCI.20-16-06021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiba-Falek O., Kowalak J.A., Smulson M.E., Nussbaum R.L. Regulation of alpha-synuclein expression by poly (ADP ribose) polymerase-1 (PARP-1) binding to the NACP-Rep1 polymorphic site upstream of the SNCA gene. Am. J. Hum. Genet. 2005;76:478–492. doi: 10.1086/428655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCulloch C.C., Kay D.M., Factor S.A., Samii A., Nutt J.G., Higgins D.S., Griffith A., Roberts J.W., Leis B.C., Montimurro J.S., et al. Exploring gene–environment interactions in Parkinson's disease. Hum. Genet. 2008;123:257–265. doi: 10.1007/s00439-008-0466-z. [DOI] [PubMed] [Google Scholar]
- 46.Morozova N., O'Reilly E.J., Ascherio A. Variations in gender ratios support the connection between smoking and Parkinson's disease. Mov. Disord. 2008;23:1414–1419. doi: 10.1002/mds.22045. [DOI] [PubMed] [Google Scholar]
- 47.Lewis J., Melrose H., Bumcrot D., Hope A., Zehr C., Lincoln S., Braithwaite A., He Z., Ogholikhan S., Hinkle K., et al. In vivo silencing of alpha-synuclein using naked siRNA. Mol. Neurodegener. 2008;3:19. doi: 10.1186/1750-1326-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiao L., Hamamichi S., Caldwell K.A., Caldwell G.A., Yacoubian T.A., Wilson S., Xie Z.L., Speake L.D., Parks R., Crabtree D., et al. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol. Brain. 2008;1:17. doi: 10.1186/1756-6606-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feany M.B., Bender W.W. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 50.Maries E., Dass B., Collier T.J., Kordower J.H., Steece-Collier K. The role of alpha-synuclein in Parkinson's disease: insights from animal models. Nat. Rev. Neurosci. 2003;4:727–738. doi: 10.1038/nrn1199. [DOI] [PubMed] [Google Scholar]
- 51.Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 52.Dutra A.S., Mignot E., Puck J.M. Gene localization and syntenic mapping by FISH in the dog. Cytogenet. Cell Genet. 1996;74:113–117. doi: 10.1159/000134395. [DOI] [PubMed] [Google Scholar]
- 53.Lichter P., Cremer T., Borden J., Manuelidis L., Ward D.C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- 54.Schlossmacher M.G., Shimura H. Parkinson's disease: assays for the ubiquitin ligase activity of neural Parkin. Methods Mol. Biol. 2005;301:351–369. doi: 10.1385/1-59259-895-1:351. [DOI] [PubMed] [Google Scholar]
- 55.Gispert S., Del Turco D., Garrett L., Chen A., Bernard D.J., Hamm-Clement J., Korf H.W., Deller T., Braak H., Auburger G., et al. Transgenic mice expressing mutant A53T human alpha-synuclein show neuronal dysfunction in the absence of aggregate formation. Mol. Cell. Neurosci. 2003;24:419–429. doi: 10.1016/s1044-7431(03)00198-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.