Abstract
Dysregulation of the mammalian target of rapamycin (mTOR) kinase pathway is centrally involved in a wide variety of cancers and human genetic diseases. In mammalian cells, mTOR is part of two different kinase complexes: mTORC1 composed of mTOR, raptor and mLST8, and mTORC2 containing mTOR, rictor, sin1 and mLST8. Whereas, mTORC1 is known to be a pivotal regulator of cell size and cell cycle control, the question whether the recently discovered mTORC2 complex is involved in these processes remains elusive. We report here that the mTORC1-mediated consequences on cell cycle and cell size are separable and do not involve effects on mTORC2 activity. However, we show that mTORC2 itself is a potent regulator of mammalian cell size and cell cycle via a mechanism involving the Akt/TSC2/Rheb cascade. Our data are of relevance for the understanding of the molecular development of the many human diseases caused by deregulation of upstream and downstream effectors of mTOR.
INTRODUCTION
The serine/threonine protein kinase mTOR (mammalian target of rapamycin) is the central player within a signalling cascade regulating a wide variety of different cellular functions. In mammalian cells two structurally and functionally distinct mTOR-containing complexes have been identified. mTORC1 (also known as the mTOR/raptor complex) contains raptor (regulatory associated protein of mTOR) and mLST8 (also known as GβL). Whereas the function of mLST8 is not really clarified, raptor regulates mTORC1 functioning as a scaffold for recruiting mTORC1 substrates. The major substrates of mTORC1 known so far are 4EBP1 (eukaryotic initiation factor 4E binding protein-1) and p70S6K (ribosomal p70S6 kinase), both regulators of mRNA translation. mTORC1 phosphorylates and activates p70S6K at T389 to activate the ribosomal protein S6 via phosphorylation at S240/244 (1,2).
The only recently discovered kinase complex mTORC2 (mTOR/rictor complex) also contains the mLST8 protein, but is marked by the rictor (rapamycin-insensitive companion of mTOR), sin1 (stress-activated protein kinase-interacting protein) and protor subunits in place of raptor. Rictor and sin1 appear to stabilize each other through binding, building the structural foundation for mTORC2. mTORC2 phosphorylates the oncogenic kinase Akt (also known as protein kinase B) at S473. Downstream of the phosphatidylinositol-3-kinase (PI3K), PDK1 (phosphoinositide-dependent kinase-1) phosphorylates Akt at T308, what in conjunction with the mTORC2-mediated phosphorylation of Akt drives full activation of this kinase (3,4). Over 100 putative Akt substrates have been reported, although only a few of them meet the criteria suggested for the definition of a bonafide Akt substrate (5).
Akt phosphorylates and blocks the inhibitory effects of the complex of the two tuberous sclerosis gene products TSC1/TSC2 towards Rheb (Ras homolog enriched in brain), which is a potent regulator of mTORC1. In addition, PRAS40 (proline-rich Akt substrate 40 kDa) is phosphorylated by Akt at T246 releasing its inhibitory effects on mTORC1. In summary, Akt stimulates mTORC1 signalling by inhibiting the function of tuberin and PRAS40 (3,6).
The growth of an organ or a whole organism is regulated by a coordinated increase in cell number and in cell size. To remain constant during proliferative conditions, both DNA content and cell mass must double in the course of each cell division cycle. Accordingly, although separable, cell cycle progression and cell growth (increase in cell size and cell mass) are coordinated. In 2002 and 2004, Fingar et al. (7,8) published their pioneering work showing that mTOR controls mammalian cell size and cell cycle progression via its downstream targets p70S6K and 4EBP1. Since these studies were performed before the discovery of two mTOR-containing kinase complexes with different functions and using the inhibitor rapamycin to block mTORC1 activity, two major questions must be answered to clarify mTOR's role in cell cycle and cell size control: (i) Is the regulation of mTORC2 an essential component for mTORC1-mediated cell cycle and/or cell size control? (ii) Is mTORC2 itself capable of regulating mammalian cell cycle and/or cell size?
Making use of non-transformed, non-immortalized primary human fibroblasts we found that (i) the mTORC1-mediated consequences on cell cycle and cell size are separable and do not involve effects on mTORC2 activity; (ii) In addition, we report here for the first time that mTORC2 is a potent regulator of mammalian cell size and cell cycle via a mechanism involving Akt/TSC2/Rheb.
RESULTS
Rapamycin-mediated effects on mTORC1 on cell cycle and cell size are separable and do not involve effects on mTORC2 activity
Many of the signalling components of the mTOR cascade are frequently deregulated in a wide variety of cancers and other human diseases (9). Accordingly, it is not surprising that the regulation of mTOR was reported to be different depending on the transformation status of the used cell system (10). Another problem of many mTOR studies is that they have been performed with ectopically overexpressed components of the mTOR cascade or using in vitro assay systems. All experiments presented in this report have been performed using primary, non-transformed, non-immortalized, diploid human IMR-90 fibroblasts (11) studying endogenous proteins. Modulation of endogenous activities has exclusively been obtained either via the widely used mTORC1 inhibitor rapamycin or via specific short-interfering RNAs (siRNAs). Endogenous mTORC1 and mTORC2 activities have been studied by analysing their endogenous functional readouts phospho-T389 p70S6K and phospho-S473 Akt, respectively.
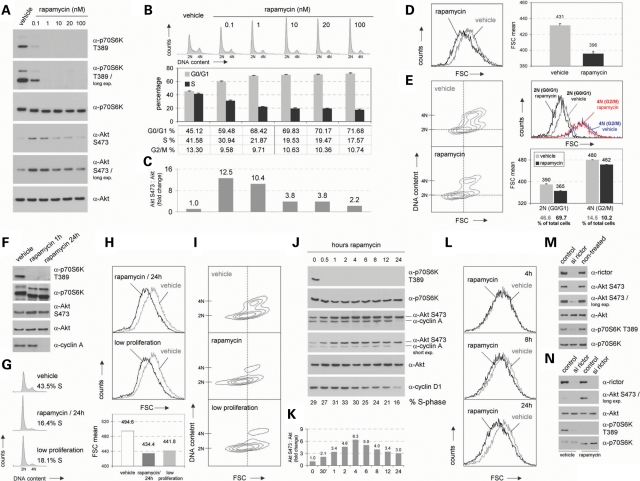
First, we determined the optimal concentration of rapamycin to block mTORC1 in our cell system. Titration experiments revealed that low rapamycin concentrations blocked mTORC1 but also induced endogenous Akt S473 phosphorylation. The latter phenomenon, which we observed in several experiments in the study performed here (see below), is very likely owing to the well known earlier described mTORC1-induced feedback mechanism involving the inhibiting phosphorylation of insulin receptor substrate 1 via p70S6K (1–4). For the experiments in this study we decided to use 100 nm, because at this concentration mTORC1 is efficiently blocked and rapamycin mediates its well-known cell cycle effects with only minor effects on Akt phosphorylation (Fig. 1A–C).
Figure 1.
mTORC1-mediated consequences on cell cycle and cell size are separable and do not involve effects on mTORC2 activity. (A) Logarithmically growing non-transformed, non-immortalized IMR-90 fibroblasts, derived from human fetal lung tissue, were treated with the mTOR inhibitor rapamycin at final concentrations ranging from 0.1 nm to 100 nm for 24 h. Control cells were treated with an equal volume of DMSO (vehicle). Total lysates were prepared and analysed for the expression level of p70S6K and Akt as well as for the phosphorylation status at T389 and S473, respectively. (B) In addition, so treated cells were stained with propidium iodide and cytofluorometrically analysed for DNA distribution. Representative DNA profiles are presented (upper panel). The percentage of cells in G0/G1, S and G2/M is indicated (lower panel). (C) Phospho-Akt S473 signals detected in (A) were densitometrically scanned and normalized to total Akt levels. (D) 100 nm rapamycin-treated IMR-90 cells derived from the same pool of cells as analysed in (A, B and C) were cytofluorometrically investigated for overall cell size via FSC (Forward Scatter) analyses. Overlays of FSC histograms are presented to enable direct comparison of control cells versus rapamycin-treated cells (left panel). Mean FSC values obtained from these analyses are presented (right panel). (E) In addition, cell size according to different cell cycle phases was examined via two-dimensional contour blot analyses of FSC versus DNA content. To visualize FSC shifts between different samples, a crosshair has been set into the center of the G0/G1 population of control cells (left panel). For details regarding different types of FSC analyses compare the Methods section in the text. Furthermore, G0/G1-phase cells (2 N) and G2/M-phase cells (4 N) were selectively gated and analysed for cell size distribution via FSC. Mean FSC values and the percentage of gated cells are presented (right panel). (F) Lysates of IMR-90 cells treated with 100 nm rapamycin for the indicated times were analysed for the expression level of total p70S6K (on long exposures the used α-p70S6K antibody recognizes also lower migrating p85S6K) and total Akt as well as for their phosphorylated forms as indicated. In addition, same lysates were examined for cyclin A levels. (G) Logarithmically growing IMR-90 cells (vehicle) were either treated with 100 nm rapamycin for 24 h as described in (A) or were grown under low proliferation conditions. So treated cells were cytofluorometrically analysed for DNA distribution. The percentage of cells in S-phase is indicated. (H) In parallel, overall cell size was cytofluorometrically investigated via FSC (upper panel). Mean FSC values obtained from these analyses are presented (lower panel). (I) In addition, cell size according to different cell cycle phases was examined via two-dimensional contour blot analyses of FSC versus DNA content. (J) IMR-90 fibroblasts were treated with 100 nm rapamycin for the indicated times and total cell lysates were analysed for the expression levels of p70S6K T389, total p70S6K, Akt S473, total Akt, cyclin A and cyclin D1. In addition, the percentage of cells in S-phase was determined on the flow cytometer. (K) Phospho-Akt S473 signals detected in (J) were densitometrically scanned and normalized to total Akt levels. (L) In parallel, overall cell size distribution of so treated cells was cytofluorometrically analysed via FSC at the indicated time points. (M) Logarithmically growing IMR-90 fibroblasts were transfected with pooled short-interfering RNAs (siRNAs) targeting human rictor (si rictor). Cells treated with non-targeting siRNAs (control) or cells which were left entirely untreated were analysed in parallel. About 72 h after initial transfection, lysates were prepared and analysed for indicated proteins via immunoblotting. (N) Experiments were basically performed as described in (M) with the exception that cells were treated with 100 nm rapamycin or DMSO for the final 24 h of incubation. So treated cells were lysed and examined for the expression level of rictor, S473 phosphorylated Akt, total Akt, T389 phosphorylated p70S6K and total p70S6K.
At this concentration rapamycin triggered a very efficient block of mTORC1 activity (detected by downregulation of phospho-T389 p70S6K and by faster migration of p70S6K protein) (Fig. 1A and F), but was without such effects on mTORC2 activity (compare phospho-S473 Akt levels and Akt protein migration) (Fig. 1A, C and F), strongly decreased the amount of S-phase cells (Fig. 1B and G; P < 0.05) and reduced cell size demonstrated by a shift detected on Forward scatter (FSC) histograms (Fig. 1D and H; P < 0.05). Here, it is important to note that whenever such a cell size phenotype is detected, two questions must be answered. (i) Does the cell size reduction occur in all cell cycle phases? (ii) Is the observed cell size reduction really a result of altered cell growth or just of an altered rate of cell cycle progression? To allow investigation of the first question DNA was stained and two-dimensional contour blot analyses of FSC versus DNA content were performed (12) (Fig. 1E and I). Furthermore, from the cell populations analysed in the two-dimensional contour blots, G0/G1 cells (2 N) and G2/M cells (4 N) have been selectively gated and analysed via FSC histograms (Fig. 1E, upper right panel), FSC mean values (Fig. 1E, lower right panel) and the % of the gated cells (Fig. 1E, under the lower right panel) are presented. These data demonstrate that rapamycin's effects on cell size regulation are independent of the cell cycle. In addition, it is proven that the two-dimensional contour blot analyses used in this study provide a suitable approach to study cell size effects over the cell cycle. With regard to the second question one has to consider that FSC analyses of cells cultivated under low proliferation conditions also suggest decreased cell size. However, in these cases the decreased FSC is because of an increased amount of G0/G1 cells within the cell population rather than to real cell size reduction. To clarify this issue we cultivated IMR-90 cells under low proliferation conditions without refreshing the growth medium for 48 h. Two-dimensional contour blot cell size analyses revealed that low proliferation conditions only increased the amount of G0/G1 cells without any effects on cell size, whereas rapamycin triggers a real size reduction of cells in all cell cycle phases (Fig. 1I).
To further investigate the kinetics of rapamycin's effects on cell size and cell cycle progression, we performed a time-course experiment. Whereas diminished mTORC1 activity (phospho-T389 p70S6K) occurred already after half an hour and the effects on cell size (FSC) started already early being good visible after 8 h, the effects on cell cycle regulation (detected by decreasing cyclin A and cyclin D1 protein levels and by % S-phase cells) were hardly detectable before 24 h. Interestingly, in this experiment no negative effects on the phosphorylation of Akt S473 were detected (Fig. 1J–L).
In this study, we performed the widely used analyses of endogenous phosphorylated Akt S473 as functional readout for mTORC2 activity. Especially for the here presented study, important reasons for the usage of this approach instead of in vitro kinase assays exist. (i) We wanted to really study the endogenous activites in primary non-immortalized non-transformed cells. (ii) Since it is known that the intracellular localization of both, p70S6K and Akt, is regulated, one cannot expect the in vitro usage of ectopic p70S6K and Akt as substrates to really reflect the in vivo endogenous availability of these substrates. (iii) Since it could be that the availability of both, p70S6K and Akt, is regulated throughout the cell cycle, one cannot expect the in vitro usage of constitutive ectopic p70S6K and Akt as substrates to really reflect the in vivo endogenous availability of these substrates in all the cell cycle experiments. We performed additional experiments to prove that analysing endogenous phosphorylated Akt really reflects endogenous mTORC2 activity under our experimental conditions in the cell system used here. Compared with both non-treated cells and cells transfected with non-targeting siRNAs, siRNAs specific for the mTORC2 component rictor trigger downregulation of endogenous Akt S473 phosphorylation (Fig. 1M). This is even true in cells, in which mTORC1 activity has been blocked via rapamycin (Fig. 1N). These findings demonstrate that analysing endogenous levels of Akt S473 phosphorylation reflects a reliable and specific approach to study endogenous mTORC2 activity in our cells.
Taken together, these data demonstrate that blocking mTORC1 mediates cell size reduction in the cells of all cell cycle phases and affects cell cycle regulation separable from its cell size effects. Most importantly, all that occurs without downregulation of mTORC2-mediated Akt S473 phosphorylation.
Blocking mTORC1 activity delays S-phase induction with smaller G1 cells entering S but without effects on mTORC2 activity
Cell growth in size is a requirement for cell proliferation, as cells must double in size before dividing (13,14). From the data obtained so far three questions arose: (i) Does blocking of mTORC1 delay S-phase induction in primary non-transformed, non-immortalized human cells? (ii) Is mTORC2 activity affected at any time point during the transition of rapamycin-treated G1 cells into S phase? (iii) Do such cells with blocked mTORC1 activity pass the G1–S phase transition to start replication at smaller size?
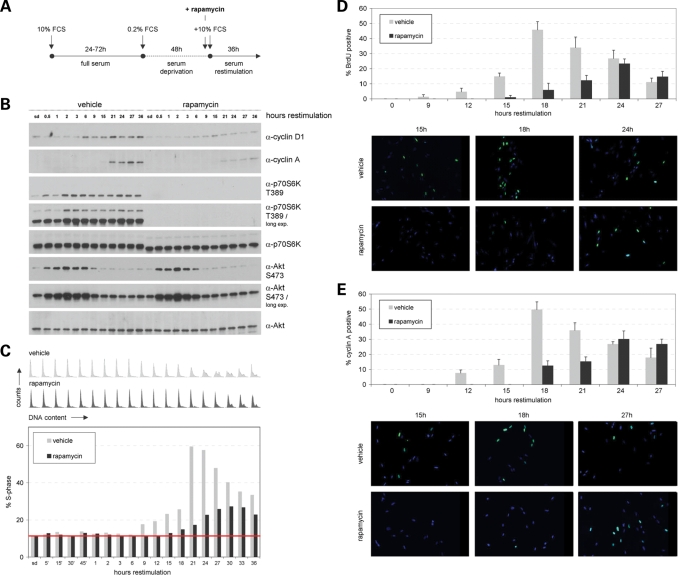
To investigate these issues, IMR-90 cells were serum arrested and restimulated with and without rapamycin (Fig. 2A). The discovery that mTOR exists in two structurally and functionally distinct kinase complexes and the observation that under certain circumstances a rapamycin-mediated block of mTORC1 is accompanied by effects on mTORC2-mediated Akt phosphorylation (depending on the transformation status of the analysed cell) (10,11) made it essential to also investigate the role of mTORC2. Western blot analyses studying the restimulated cells described above revealed that the rapamycin-triggered block of mTORC1-mediated p70S6K T389 phosphorylation leads to delayed S-phase induction without any effects on mTORC2-mediated Akt S473 phosphorylation (Fig. 2B and C).
Figure 2.
Blocking mTORC1 activity delays S-phase induction without effects on mTORC2 activity. IMR-90 fibroblasts, synchronized in G0/G1 via serum deprivation, were serum restimulated to re-enter the cell cycle in the presence or absence of the mTOR inhibitor rapamycin. (A) Schematic outline of the synchronization procedure. Briefly, cells growing under full serum (10% FCS) conditions for 24–72 h were serum-deprived in medium containing 0.2% serum (0.2% FCS) for 48 h and serum restimulated (10% FCS) for additional 36 h. Rapamycin was added 30 min prior to restimulation at a concentration of 100 nm. Control cells were treated with an equal volume of DMSO and analysed in parallel. (B) Cells treated as described in (A) were lysed and examined for the expression level of cyclin D1, cyclin A, p70S6K T389, p70S6K, Akt S473 and Akt at the indicated time points via immunoblotting. (C) In addition, cells were cytofluorometrically analysed for DNA content at the indicated time points. DNA profiles are presented (upper panel). In addition, the percentage of cells in S-phase was determined for DMSO (vehicle)- and rapamycin-treated cells. To facilitate interpretation of S-phase induction, a baseline corresponding to the S-phase values of serum-deprived vehicle- and rapamycin-treated cells was inserted into the graph. (D) Experiments as described in (A, B and C) were repeated and the percentage of cells actively undergoing DNA replication was analysed by immunocytochemical BrdU incorporation assay at specific time points as indicated (at least 250 cells were scored for each timepoint). (E) In addition, cells derived from the same pool of cells as described in (D) were immunocytocemically stained for cyclin A expression (at least 250 cells were scored for each timepoint).
Four independent approaches—western blot analyses of cyclin D1 and A, analysing DNA distribution, BrdU incorporation assays and cytochemical cyclin A analyses—revealed that blocking mTORC1 activity via rapamycin indeed causes a delay in S-phase entry for some hours (Fig. 2B–E).
FSC analyses and two-dimensional contour blot studies of FSC versus DNA content demonstrated that rapamycin-treated cells start to replicate at a smaller size compared with non-treated counterparts (Fig. 3A and B). To additionally confirm this observation, we compared two time points during restimulation from the experiment described in Figure 2B and C with and without rapamycin with similar G0/G1–S–G2/M phase distributions. Flowcytometric FSC analyses of gated G0/G1 and of gated S+G2/M phase cells revealed a pronounced difference in cell size at the G1/S-phase transition between rapamycin-treated and non-treated cells (Fig. 3C). In addition, we also compared two time points during restimulation from the experiment described in Figure 2D and E representing the first strong BrdU incorporation in cells with and without rapamycin. Comparable to the experiment described above, FSC analyses of gated G0/G1 and of gated S + G2/M phase cells again revealed a pronounced difference in cell size at the G1/S-phase transition between rapamycin-treated and non-treated cells (Fig. 3D; P < 0.05). In summary, these findings prove that cells with blocked mTORC1 activity indeed start to replicate later and at a smaller cell size.
Figure 3.
Blocking mTORC1 activity causes G1 cells to enter S-phase at smaller size. IMR-90 fibroblasts, synchronized and restimulated in the presence or absence of 100 nm rapamycin as described in Figure 2, were analysed on a flowcytometer. At specific time points as indicated, cells from the experiment presented in Figure 2B and C were examined for overall cell size via FSC analyses (A) (in addition, mean FSC values for each time point are presented in numbers) and for cell size according to different cell cycle phases via two-dimensional analyses of FSC versus DNA content (B) (in addition, the percentage of cells in S-phase is presented for each time point). (C) Cells from the experiment presented in Figure 2B and C were analysed in more detail. DMSO-treated cells (vehicle) at 15 h restimulation were compared with rapamycin-treated cells at 27 h restimulation via two-dimensional FSC analysis (left panel). In addition, G0/G1-phase cells and S+G2/M-phase cells were selectively gated and analysed for cell size distribution via FSC (upper right panel). Mean FSC values are presented (lower right panel). (D) Rapamycin or vehicle-treated cells derived from the same experiment presented in Figure 2D and E were analysed as outlined in (C) with the exception that DMSO-treated cells (vehicle) at 15 h restimulation were compared with rapamycin-treated cells at 24 h restimulation.
Taken together, our findings show for the first time that mTORC1 can control cell cycle progression and cell size in cells without any effects on mTORC2 activity.
mTORC2 regulates mammalian cell size and cell cycle progression
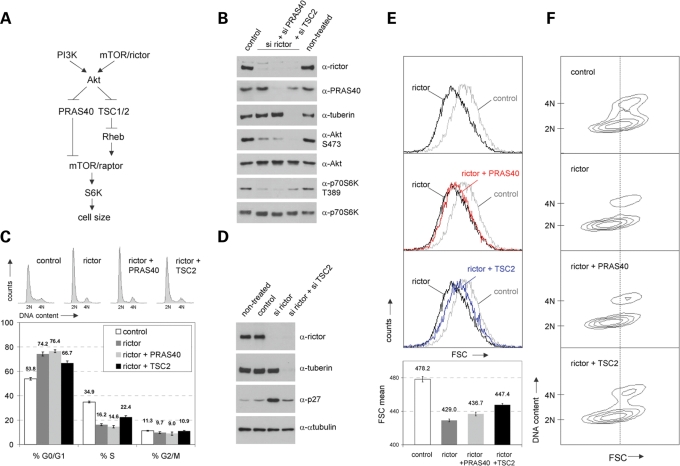
Next, we investigated the question whether the kinase complex mTORC2 itself is capable of regulating mammalian cell size and/or cell cycle. We made use of rictor- and raptor-specific siRNAs to downregulate endogenous mTORC2 and mTORC1 activities in primary non-transformed, non-immortalized human fibroblasts. As expected, knockdown of rictor triggered downregulation of endogenous phospho-S473 Akt levels and the same is true for the effects of raptor-specific siRNAs on endogenous phospho-T389 p70S6K levels (Fig. 4A). Once again, these results prove that analysing the phosphorylation status of these two substrates as endogenous functional readouts is a specific and reliable approach to study the endogenous activities of the two mTOR kinase complexes. In agreement with earlier observations using rapamycin to block mTORC1 activity (7,8) (Figs 1–3), downregulation of endogenous mTORC1 activity via raptor-specific siRNAs mediated deregulation of cell cycle progression (from 56.2% to 83.3% G0/G1 cells and from 35.4% to 12.6% S phase cells) (Fig. 4B; P < 0.05) and of size of cells in all cell cycle phases (Fig. 4C and D; P < 0.05).
Figure 4.
mTORC2 regulates mammalian cell size and cell cycle progression. Logarithmically growing IMR-90 fibroblasts were transfected with short-interfering RNAs (siRNAs) targeting human raptor and rictor, respectively. Cells treated with non-targeting siRNA were analysed in parallel and served as a negative control (control). Forty-eight hours after transfection, cells were replated at low density and were grown for another 20 h. (A) Cells as described above were lysed and examined for the expression level of raptor, rictor, S473 phosphorylated Akt, total Akt, T389 phosphorylated p70S6K and total p70S6K. α-Tubulin was co-analysed as an additional loading control. (B) Cells derived from the same pool of cells described in (A) were cytofluorometrically analysed for DNA distribution. Representative DNA profiles (upper panel) and the percentage of cells in G0/G1, S and G2/M (lower panel) are presented. Apart from the quantification of DNA distribution, cells were cytofluorometrically examined for overall cell size via FSC analyses (C) and for cell size according to different cell cycle phases via two-dimensional blots of FSC versus DNA content (D).
Interestingly, siRNA-mediated knockdown of rictor also mediated strong effects on both, cell cycle progression (from 56.2% to 76.6% G0/G1 cells and from 35.4% to 18.8% S phase cells) (Fig. 4B; P < 0.05) and cell size regulation (Fig. 4C; P < 0.05). Two-dimensional contour blot analyses of FSC versus DNA content revealed that mTORC2-induced cell size regulation is not a consequence of its effects on cell cycle progression since knockdown of rictor triggers a size reduction in all cell cycle phases (Fig. 4D).
These experiments demonstrate mTORC2 (i) to be a potent regulator of mammalian cell cycle progression; (ii) to be a potent cell size regulator; and (iii) to be able to control size of cells in all phases of the mammalian cell cycle.
mTORC2 regulates cell size and cell cycle via a mechanism involving Akt/TSC2/Rheb
Next we wanted to obtain insights into the underlying molecular mechanism of how mTORC2 controls cell cycle progression and cell size in mammalian cells. The major function of mTORC2 is to phosphorylate and thereby activate Akt (3,4). So far, although many putative Akt substrates have been reported, only very few of them meet all the criteria, which have been defined to be necessary to characterize a bonafide Akt substrate (5). Since mTORC1 is very well known to be a pivotal regulator of both, cell cycle and cell size control (1), Akt substrates, which are involved in mTORC1 regulation were of special interest.
One Akt substrate playing a major role in mTORC1 regulation is TSC2 (tuberin). TSC2 (in a complex with TSC1) is phosphorylated by Akt on S939 and T1462 and this phosphorylation inhibits its ability to act as a GTPase-activating protein against Rheb, which in turn regulates mTORC1's potential to activate p70S6K (15–20) (Fig. 5A). Accordingly, one could speculate that blocking mTORC2 activity could trigger downregulation of mTORC1 via activation of TSC2. In agreement with this model we found that modulation of mTORC2 via rictor-specific siRNAs really, as a consequence, triggers downregulation of mTORC1 activity in primary human cells (Figs 1M, N, 4A and 5B). To clarify the role of this pathway for mTORC2-mediated cell size and cell cycle regulation, we first wanted to investigate the effects of rictor-specific siRNA treatment in cells with knockdown of TSC2. Knockdown of TSC2 indeed reactivated mTORC1 activity (represented by endogenous phospho-T389 p70S6K levels), but not mTORC2 activity (endogenous phospho-S473 Akt levels), in cells treated with rictor-specific siRNAs (Fig. 5B). These results prove that the effects of downregulated mTORC2 activity on mTORC1 are indeed mediated via the TSC2-involving cascade. And strikingly, both mTORC2-mediated effects on cell cycle (DNA analyses and investigation of the cell cycle inhibitor p27) and cell size were significantly diminished upon TSC2 knockdown (Fig. 5C–F; P < 0.05). Taken together, these findings provide strong evidence that in mammalian cells mTORC2 controls both, cell cycle progression and cell size, via its potential to regulate TSC2.
Figure 5.
mTORC2 regulates cell size and cell cycle via a mechanism involving TSC2. (A) Simplified schematic presentation of the potential functional interaction of mTORC2 and mTORC1 in cell size regulation. Both, PI3K and mTORC2 (mTOR/rictor) are necessary to drive full activation of the serine-threonine kinase Akt, which in turn negatively regulates PRAS40 and the TSC1/2 complex to activate mTORC1 (mTOR/raptor) known to exert positive effects on cell size regulation via its effector S6K. (B–F) Logarithmically growing IMR-90 fibroblasts were either transfected with rictor siRNA alone or were co-transfected with siRNAs targeting rictor and PRAS40 or rictor and TSC2. Non-targeting siRNA (control) was analysed in parallel. Sixty hours post transfection, cells were replated into fresh growth medium and incubated for additional 20 h. For experiments presented in (D), cells were transfected as described above but were grown for a total of 96 h without replating. (B) siRNA-treated cells were harvested, lysed and protein levels of rictor, PRAS40 and tuberin were examined via immunoblotting to prove efficient knockdown of target proteins. In addition, the expression level of Akt and p70S6K as well as the corresponding phosphorylation status at S473 and T389, respectively, were analysed. Lysates of non-treated cells, which were co-analysed in parallel, were included to prove the here used conditions of siRNA treatment to be physiologic and technically sound (compare control versus non-treated). (C) siRNA-treated cells derived from the same pool of cells described in (B) were stained with propidium iodide and cytofluorometrically analysed for cell cycle distribution. Representative DNA profiles (upper panel) and the percentage of cells in G0/G1, S and G2/M (lower panel) are presented. (D) Lysates of cells treated as indicated were examined for the expression levels of rictor, tuberin, p27 and α-tubulin. In addition, cells were examined for overall cell size via FSC analyses (E) and for cell size according to different cell cycle phases via two-dimensional blots of FSC versus DNA-content (F) on the flow cytometer.
The notion that mTORC1's potential to regulate p70S6K activity is involved in the mechanism of mTORC2's effects on cell size and cell cycle control was additionally supported by studying the effects of knockdown of PRAS40. PRAS40 is another Akt substrate being involved in mTORC1 regulation. PRAS40 was identified as a raptor-binding protein, which does not interact with mTORC2. It is also phosphorylated directly by mTORC1, but not mTORC2, predominantly at S183. Akt-mediated phosphorylation of PRAS40 at T246 prevents its inhibition of mTORC1 (Fig. 5A). Importantly, PRAS40 regulates mTORC1 via a totally different mechanism compared with TSC2. PRAS40 functions as a direct inhibitor of mTORC1 substrate binding (21–26). Accordingly, in a cell in which mTORC1 kinase activity is downregulated via the mTORC2/Akt/TSC2/Rheb cascade (presented in Fig. 5A), one would not expect knockdown of PRAS40 to reactivate mTORC1-mediated phosphorylation of p70S6K at T389. Knockdown of PRAS40 might increase mTORC1's potential to bind to its substrates but would not reactivate mTORC1-mediated p70S6K phosphorylation in a cell, in which rictor-specific siRNA treatment triggers downregulation of mTORC2-mediated Akt activity and activation of TSC2 to block mTORC1's kinase activity. In agreement with this notion, we found knockdown of PRAS40 not be able to reactivate mTORC1-induced T389 p70S6K phosphorylation in cells treated with rictor-specific siRNAs (Fig. 5B). Our findings that modulating PRAS40 was without consequences for mTORC2-dependent effects on cell cycle and cell size regulation (Fig. 5C, E and F), additionally support the here suggested model of mTORC2 controlling both, cell cycle progression and cell size, by regulating TSC2.
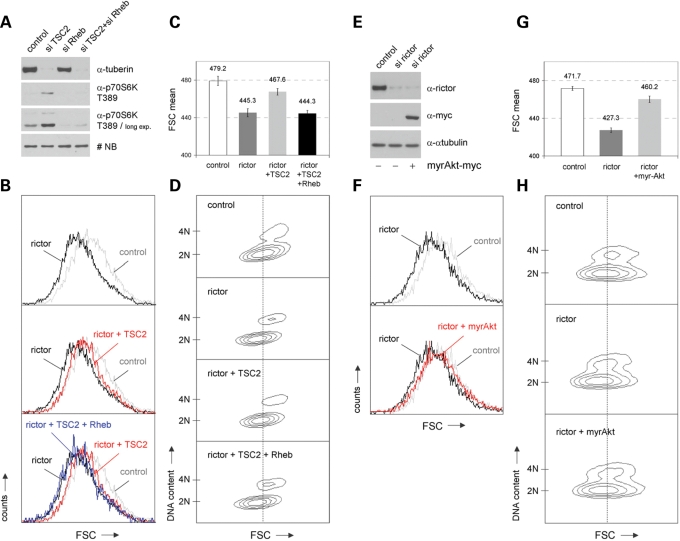
Our data presented so far provide strong evidence that mTORC2 affects cell cycle and cell size regulation via modulating the potential of TSC2 to regulate mTORC1. This potential of TSC2 is mediated via Rheb (compare also Fig. 5A). Our findings suggest that knockdown of endogenous mTORC2 activity via rictor siRNAs allows TSC2 to efficiently block Rheb's activity to control mTORC1. In line with this assumption, TSC2 should not be able to revert the effects of rictor siRNA treatment on cell size in a Rheb-negative (-diminished) background. First, transfection experiments proved that the activating effects of TSC2-specific siRNAs on p70S6K activity depend on functional Rheb (Fig. 6A). Furthermore, we observed again that TSC2 siRNA treatment counteracts the negative effects of rictor-specific knockdown on cell size regulation. Cotransfection with Rheb-specific siRNAs demonstrated that these TSC2-mediated effects also depend on functional Rheb (Fig. 6A–D; P < 0.05).
Figure 6.
The role of Rheb and Akt for mTORC2-mediated cell size effects. (A) IMR-90 cells were transfected with siRNAs targeting either TSC2, Rheb or both. Lysates of so treated cells were examined for the expression levels of tuberin, phospho-p70S6K T389 and total p70S6K. A non-specific band (NB) serves as a loading control. (B–D) IMR-90 fibroblasts were either treated with rictor siRNA alone or were co-transfected with siRNAs targeting rictor and TSC2 or rictor, TSC2 and Rheb, respectively. Cells were replated into fresh growth medium 60 h post transfection and incubated for additional 20 h. (B) So treated cells were stained with propidium iodide and cytofluorometrically analysed for overall cell size via FSC analyses. (C) Mean FSC values obtained from these analyses are presented. (D) In addition, cell size according to different cell cycle phases was examined via two-dimensional blots of FSC versus DNA-content. (E–H) Rictor- and control siRNA-treated cells were grown for 72 h, replated and transiently transfected with an empty expression vector or with an expression vector containing myristoylated (constitutively active) Akt (myrAkt) together with GFP-spectrin expression plasmids as reporter. Another 36 h later, cells were harvested and analysed via immunoblotting and flowcytometry. (E) The expression levels of rictor, ectopic Akt (via myc-antibody) and α-tubulin were analysed. (F) In addition, GFP-positive cells were examined for overall cell size via FSC analyses. (G) Corresponding mean FSC values obtained from these analyses are indicated. (H) Cell size according to different cell cycle phases was examined via two-dimensional blots of FSC versus DNA content.
As already described above, we assume that the effects of mTORC2 on cell size depend on its potential to regulate the Akt kinase: downregulating endogenous rictor leads to diminshed Akt activity, what induces the cascade presented in Figure 5A. Last but not least, to test whether Akt regulation indeed represents the initial relevant step for the mTORC2-mediated cell size control, we tested the effects of rictor-specific siRNA treatment in cells expressing myristoylated Akt. These experiments demonstrated that downregulation of mTORC2 via rictor siRNAs cannot control mammalian cell size in cells harboring constitutive Akt kinase activity (Fig. 6E–H; P < 0.05).
In summary, our here described findings show that mTORC2 regulates mammalian cell size via a mechanism involving the Akt/TSC2/Rheb cascade.
DISCUSSION
In mammalian cells, the mTOR pathway has been shown to be a pivotal regulator of both cell cycle progression and cell size. Accordingly, studies have been performed before the discovery that mTOR is also part of a second kinase complex named mTORC2. In addition, in many of the underlying experiments mTOR activity has been modulated via rapamycin (see e.g. 7,8). Although originally believed to exclusively block mTORC1, rapamycin has been shown to also affect mTORC2 activity at least under specific experimental conditions (10,11). Accordingly, with regard to mTOR's role in cell size and cell cycle control in mammalian cells, two major question remained elusive: (i) Is the regulation of mTORC2 essentially involved in mTORC1-mediated cell cycle and/or cell size control? (ii) Is mTORC2 itself capable of regulating mammalian cell cycle and/or cell size?
To answer these questions we felt it to be essential to use an optimal biological cell system. In the past, many data on mTOR regulation were obtained from ectopic overexpression experiments in transformed cells or using in vitro kinase assays. Varying results on mTOR regulation have been reported in many different immortalized or transformed cell lines, what is mainly because of the fact that a wide variety of upstream regulators of mTOR are deregulated in many different types of diseases and tumors (3,9–11,27). All experiments presented in this study have been performed using non-transformed, non-immortalized primary human diploid IMR-90 fibroblasts harboring a finite lifetime and a normal karyotype (11). In addition, with the exception of ectopic expression of myristoylated Akt no overexpression experiment is included, but we exclusively modulated endogenous activities.
It was already reported earlier that mTORC1's effects on cell cycle and cell size are separable (7,8). We have confirmed and expanded this knowledge: (i) The approach of two-dimensional contour blot analyses of FSC versus DNA content allowed the demonstration that blocking endogenous mTORC1 via rapamycin reduces the size of cells in all cell cycle phases. (ii) Identical results were obtained in cells, in which endogenous mTORC1 was diminished via raptor-specific siRNAs. (iii) Rapamycin time-course experiments revealed that the effects of downregulated mTORC1 on cell size occur far before the cell cycle deregulation. (iv) Re-stimulation experiments with and without rapamycin showed that blocking mTORC1 activity delays S-phase induction with smaller G1 cells starting to replicate.
Four independent experimental data support our conclusion that mTORC1-mediated cell cycle regulation does not need to involve effects on mTORC2 activity. (i) Rapamycin treatment of logarithmically growing cells triggers accumulation of G0/G1 cells accompanied by a downregulation of the amount of S-phase cells without effects on endogenous mTORC2 activity. (ii) A rapamycin time-course experiment revealed that after 24 h blocking mTORC1 activity cyclin A levels, cyclin D1 levels and % S-phase cells decreased, but no effects could be detected on mTORC2 activity. (iii) Serum re-stimulation approaches revealed that a rapamycin-mediated block of mTORC1 significantly delays S-phase entry without any effects on mTORC2 activity. (iv) Downregulation of endogenous mTORC1 activity via raptor-specific siRNAs in logarithmically growing cells also triggered accumulation of G0/G1 without negatively affecting mTORC2. That mTORC2 deregulation is also not playing an essential role for mTORC1-triggered cell size control was demonstrated (i) by showing that rapamycin triggers size reduction in all cell cycle phases of cycling cells without affecting mTORC2; (ii) by obtaining the same results upon blocking of mTORC1 via raptor-specific siRNAs; (iii) by detecting deregulated cell size control in a rapamycin time-course experiment without effects on endogenous mTORC2 activity; (iv) by demonstrating that rapamycin-treated cells enter the replicative phase at smaller size but do not exhibit any modulation of endogenous mTORC2 activity. In summary, these findings allow the conclusion that in mammalian cells mTORC1 can regulate cell cycle progression and cell size control independent of any effects on endogenous mTORC2 activity.
Next, we found that downregulating endogenous mTORC2 activity via rictor-specific siRNAs causes an accumulation of G0/G1 cells accompanied by a decrease of S-phase cells. Our here obtained data using primary non-transformed, non-immortalized human fibroblasts are in perfect agreement with a recent study reporting the same cell cycle phenotype upon knockdown of rictor in the MCF7 breast cancer and PC3 prostate cancer cell lines (28), suggesting that this potential of mTORC2 might be independent of the transformation status of the cell. Still, that mTORC2 itself is involved in tumor development is suggested by the recent finding that many gliomas overexpress rictor accompanied with elevated mTORC2 activities (29).
Drosophila mutants removing the critical TORC2 components, rictor and sin1, showed minor growth impairment (30). The aspect whether mTORC2 is involved in mammalian cell size regulation remained elusive so far. We report here that siRNA-induced knockdown of endogenous mTORC2 activity triggers strong effects on cell size regulation of non-transformed primary human cells. Several independent experiments revealed that rictor-siRNA-mediated size reduction to be comparable to the effects of a raptor-specific knockdown. Two-dimensional contour blot analyses of FSC versus DNA content demonstrated that mTORC2-mediated cell size regulation is not a consequence of its effects on cell cycle progression described above. We found that knockdown of rictor to trigger a size reduction in all cell cycle phases very comparable to the effects of raptor-specific siRNAs. Accordingly, we conclude mTORC2 to be a potent cell size regulator and to be able to control cell size of cells in all phases of the mammalian cell cycle.
mTORC2 is a major regulator of Akt kinase activity. One Akt substrate that has been reported to be a pivotal regulator of mammalian cell cycle and cell size is the tuberous sclerosis gene product TSC2. Akt is known to regulate growth by directly phosphorylating TSC2 (1,4,15–20). To investigate whether TSC2 is essentially involved in the mTORC2-mediated cell cycle and/or cell size regulation, we analysed the effects of TSC2-specific siRNA treatment on the rictor-siRNA-induced deregulations described above. Several independent experiments revealed that the rictor-siRNA-triggered accumulation of G0/G1 cells heavily depends on the endogenous levels of TSC2. The same was found to be true for the effects of rictor knockdown on the size of cells in all different cell cycle phases.
In the past, different observations were reported regarding the question whether mTORC2 affects the mTORC1-induced phosphorylation of p70S6K. Very likely, owing to the usage of different approaches to modulate mTORC2 and of different transformed immortalized cells, modulating mTORC2 was described to be either without any effects on p70S6K phosphorylation, or to trigger its up- or downregulation (31–33). However, whenever we downregulated endogenous mTORC2 activity via rictor-specific siRNAs in non-transformed, non-immortalized primary human cells, we observed a very pronounced downregulation of endogenous mTORC1-mediated p70S6K phosphorylation on T389. This is one observation that provides evidence for the mTORC1/p70S6K cascade to be involved mTORC2-induced cell size and cell cycle regulation. As discussed above, we proved TSC2 to be of essential relevance for these mTORC2-dependent regulations and TSC2 is very well known to exert its cell size effects via regulation of mTORC1/p70S6K. Knockdown of TSC2 reverted the cell cycle and cell size effects of rictor-specific siRNAs accompanied with a reactivation of endogenous T389 p70S6K phosphorylation. In addition, this finding shows that the effects of downregulated mTORC2 activity on mTORC1 are indeed mediated via the TSC2-involving cascade. Knockdown of PRAS40 could not reactivate p70S6K phosphorylation and also did not have any influence on the mTORC2-triggered cell size and cell cycle effects.
All these findings provide strong evidence that mTORC2 affects cell size regulation via modulating the potential of TSC2 to regulate mTORC1. This potential of TSC2 is known to be mediated via Rheb (1–4). We found that TSC2's potential to counteract mTORC2-mediated cell size control indeed depends on functional Rheb. Taken together with our here reported observation that downregulation of mTORC2 via rictor siRNAs cannot control mammalian cell size in cells harboring constitutive Akt kinase activity, these findings allow the conclusion that mTORC2 is a potent regulator of mammalian cell size via a mechanism involving the Akt/TSC2/Rheb cascade.
MATERIALS AND METHODS
Cell culture
IMR-90 (ATCC no. CCL-186) are non-transformed, non-immortalized human diploid fibroblasts from fetal lung and have a finite lifetime being capable of attaining around 58 population doublings before the onset of senescence. They were obtained from the American Type Culture Collection at passage number 10 (population doubling 26) and were grown in Dulbecco's modified Eagle's medium (DMEM) at 4.5 g/l glucose, supplemented with 10% calf serum and antibiotics (30 mg/l penicillin, 50 mg/l streptomycin sulphate) at 37°C and 5% CO2. In the course of experiments, IMR-90 cells were grown for no more than eight additional passages (≤47 total population doublings) and were regularly analysed by standard karyotyping to confirm a normal diploid karyotype. For synchronization in G0/G1, IMR-90 were used at passage number 15 and deprived of serum in medium containing 0.2% FCS for 48 h. Cells were stimulated to re-enter the cell cycle by the addition of 10% serum for another 36 h. Experiments involving the mTOR inhibitor rapamycin (Calbiochem) were performed in the absence (DMSO vehicle control) or presence of the drug at final concentrations as indicated.
Transfections
Pooled short interfering RNAs (siRNAs) specifically targeting human raptor, rictor, PRAS40, TSC2 or Rheb were purchased from Dharmacon (ON-TARGETplus SMART pool reagents) and were delivered to the cells at a final range of 50–100 nm using Lipofectamine 2000 reagent (Invitrogen) following the transfection protocol provided by the manufacturer. A pool of four non-targeting siRNAs was used as a control for non-sequence-specific effects. In experiments where simultaneous knockdown of two or three genes was performed, the overall amount of siRNA for each reaction was kept constant by the addition of non-targeting siRNAs. Cells were trypsinized, replated into fresh growth medium 48–60 hours post-transfection and grown for another 20 h until harvest. In some experiments siRNA-treated cells were grown for 72 h, replated and retransfected with DNA using the following plasmids: pUSE-myr-Akt1 (activated Akt, N-terminal myristoylation, myc-tagged), pUSE empty vector (both purchased from Millipore), GFP-spectrin expression vector. Transfections with pUSE expression plasmids and GFP-spectrin plasmids were performed with Lipofectamine 2000 at a final ratio of 1:5. Cells were harvested for analysis 36 h later.
Flow cytometry
For cytofluorometric DNA and cell size analyses, cells were harvested by gentle trypsinization and fixed by rapid submersion in ice-cold 85% ethanol. After overnight fixation at −20°C, DNA was stained in an appropriate volume of staining solution containing 0.25 mg/ml propidium iodide, 0.05 mg/ml RNase and 0.1% Triton X-100 in citrate buffer, pH 7.8. A total of 1.5×104 to 3×104 cells per sample (in case of DNA transfections 1×104 to 1.5×104 GFP-spectrin-positive cells per sample) were collected in linear amplification mode and analysed on a Beckton Dickinson FACScan using CELLQUEST and MODFIT software. Cell size analyses via FSC were basically performed in two different ways as previously described (12): overall cell size distribution, regardless of the different cell cycle phases, was investigated via one-dimensional FSC histograms. To enable direct comparison of different samples, overlays of FSC histograms were performed. In addition, mean FSC values, representing the average FSC distribution of the analysed cell pool, were obtained using CELLQUEST histogram statistical tools. Cell size distribution according to the different cell cycle phases was analysed via two-dimensional, so called contour blots of FSC versus DNA content. Contour blots, representing a particular, density-related presentation of two parameters in relation to cell numbers, were generated in probability density mode at 20%, means the outermost contour line represents 10% of the total number of events, the next contour represents 30%, then 50%, 70% and 90%. Except contour blots shown in Figure 2E, left panel, which are presented in linear density mode and served as the template to gate G0/G1 and S + G2/M cells, all blots have been generated in the same way. To visualize FSC shifts between blots of different samples, a bar, or alternatively a crosshair, was set into the center of the G0/G1 (2 N) population of control cells. Alternatively, FSC overlays of G0/G1, S and/or G2/M gated populations are presented.
Protein extraction and immunoblotting
Extracts of cellular total protein were prepared by physical disruption of cell membranes by repeated freeze-and-thaw cycles. Briefly, cells were washed with PBS and harvested by scraping. Pellets were lysed in buffer A containing 20 mm Hepes, pH 7.9, 0.4 m NaCl, 2.5% glycerol, 1 mm EDTA, 0.5 mm DTT, 1 mm PMSF, 0.5 mm NaF, 0.5 mm Na3VO4 supplemented with 2 mg/ml aprotinin, 2 mg/ml leupeptin, 0.3 mg/ml benzamidinchlorid, 10 mg/ml trypsin inhibitor by freezing and thawing. Supernatants were collected by centrifugation at 15 000 rpm for 20 min at 4°C and stored at −80°C. Proteins were resolved using 10% SDS–PAGE and transferred to nitrocellulose. Blots were stained with Ponceau-S to visualize the amount of loaded protein (11). For immunodetection antibodies specific for the following proteins were used: tuberin C-20 (Santa Cruz, no. sc-893), raptor clone 24C12 (Cell Signaling, no. 2280), rictor clone 53A2 (Cell Signaling, no. 2114), PRAS40 (Cell Signaling, no. 2610), cyclin D1 M-20 (Santa Cruz, no. sc-718), cyclin A H-432 (Santa Cruz, no. sc-751), phospho-p70S6 kinase T389 clone 108D2 (Cell Signaling, no. 9234), p70S6 kinase clone 49D7 (Cell Signaling, no. 2708), phospho-Akt S473 clone D9E (Cell Signaling, no. 4060), Akt (pan) clone C67E7 (Cell Signaling, no. 4691), Kip1/p27 clone 57 (Transduction Laboratories, no. K25020), c-myc clone 9E10 (BD Pharmingen, no. 554205) and α-tubulin (DM1A, Calbiochem, no. CP06). Rabbit polyclonal and monoclonal antibodies were detected using anti-rabbit IgG, a HRP-linked heavy and light chain antibody from goat (no. A120-101P, Bethyl Laboratories); mouse monoclonal antibodies were detected using anti-mouse IgG, a HRP-linked heavy and light chain antibody from goat (no. A90-116P, Bethyl Laboratories). Signals were detected using the enhanced chemiluminescence method (Pierce). Some of the presented western blots were densitometrically analysed using LabWorks image acquisition and analysis software. The results are given in relation to the according control value set as 1.
Brdu incorporation assay and immunofluorescence
The DNA Replication Assay Kit (Millipore) was performed according to the manufacturer's instructions. Briefly, cells were grown on coverslips, starved and restimulated as described above, pulse-labeled for 60 min with 10 µm BrdU and fixed in 95% ethanol/5% glacial acetic acid. BrdU-positive cells were detected using a monoclonal antibody against BrdU and an Alexa Fluor 488 goat anti-mouse secondary antibody (no. A11029, Molecular Probes). For immunodetection of cyclin A, cells were fixed in 4% paraformaldehyde for 10 min at room temperature, treated with 0.1% Triton X-100 for 15 min and blocked for non-specific binding in PBS containing 3% BSA for another 30 min. After 1 h incubation with anti-cyclin A antibody (H-432, Santa Cruz, no. sc-751) cyclin A-positive cells were visualized by incubation with Alexa Fluor 488 goat anti-rabbit IgG (no. A11034, Molecular Probes). Cell nuclei were counterstained with DAPI. Fluorescence was analysed on a conventional microscope equipped with a video camera and a fluorescence attachment (model Axioplan 2 imaging with FluoArc, Zeiss) using a Plan-Neofluor 40x oil immersion (NA 1.3) objective. MetaSystems software was used for image acquisition and Adobe Photoshop 7.0 was used for further processing. At least 250 cells were scored for each time point.
Statisical analyses
The significance of the observed differences was determined by Student's t-test (paired, two-tailed) using Graph-Pad INSTAT software. P-values > 0.05 are defined as not significant.
FUNDING
Research in our laboratory is supported by the FWF Austrian Science Fund (P18894-B12), by the Herzfeldeŕsche Familienstiftung and by the Marie Curie Research Network of the European Community as part of the Framework program 6 (FP6 036097-2). Funding to pay the Open Access publication charges for this article was provided by Funding to pay the Open Access publication charges for this article was provided by the FWF Austrian Science Fund.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Wullschleger S., Loewith R., Hall M. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Dann S.G., Selvaraj A., Thomas G. mTOR complex 1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol. Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Guertin D.A., Sabatini D.M. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Yang Q., Guan L. Expanding mTOR signalling. Cell Res. 2007;17:666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 5.Manning B.D., Cantley L.C. AKT/PKB signalling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosner M., Hanneder M., Siegel N., Valli A., Hengstschläger M. The tuberous sclerosis gene products hamartin and tuberin are multifunctional proteins with a wide spectrum of interacting proteins. Rev. Mutat. Res. 2008;658:234–246. doi: 10.1016/j.mrrev.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Fingar D.C., Salama S., Tsou C., Harlow E., Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fingar D.C., Richardson C.J., Tee A.R., Cheatham L., Tsou C., Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosner M., Hanneder M., Siegel N., Valli A., Fuchs C., Hengstschläger M. The mTOR pathway and its role in human genetic diseases. Rev. Mutat. Res. 2008;659:284–292. doi: 10.1016/j.mrrev.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Sarbassov D.D., Ali S.M., Sengupta S., Sheen J-H., Hsu P.P., Bagley A.F., Markhard A.L., Sabatini D.M. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Rosner M., Hengstschläger M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Hum. Mol. Genet. 2008;17:2934–2948. doi: 10.1093/hmg/ddn192. [DOI] [PubMed] [Google Scholar]
- 12.Rosner M., Hofer K., Kubista M., Hengstschläger M. Cell size regulation by the human TSC tumor suppression proteins depends on PI3K and FKBP38. Oncogene. 2003;22:4786–4798. doi: 10.1038/sj.onc.1206776. [DOI] [PubMed] [Google Scholar]
- 13.Baserga R. Is cell size important? Cell Cycle. 2007;6:814–816. doi: 10.4161/cc.6.7.4049. [DOI] [PubMed] [Google Scholar]
- 14.Blagosklonny M.V., Pardee A.B. The restriction point of the cell cycle. Cell Cycle. 2002;1:103–110. [PubMed] [Google Scholar]
- 15.Dan H.C., Sun M., Yang L., Feldman R.I., Sui X-M., Ou C.C., Nellist M., Yeung R.S., Halley D.J.J., Nicosia S.V., Pledger W.J., Cheng J.Q. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J. Biol. Chem. 2002;277:35364–35370. doi: 10.1074/jbc.M205838200. [DOI] [PubMed] [Google Scholar]
- 16.Goncharova E.A., Goncharov D.A., Eszterhas A., Hunter D.S., Glassberg M.K., Yeung R.S., Walker C.L., Noonan D., Kwiatkowski D.J., Chou M.M., Panettieri R.A., Krymskaya V. Tuberin regulates p70S6Kinase activation and ribosomal protein S6 phosphorylation. J. Biol. Chem. 2002;277:30958–30967. doi: 10.1074/jbc.M202678200. [DOI] [PubMed] [Google Scholar]
- 17.Inoki K., Li Y., Zhu T., Wu J., Guan L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 18.Manning B.D., Tee A.R., Logsdon M.N., Blenis J., Cantley L.C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 19.Potter C.J., Pedraza L.G., Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell. Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 20.Tee A.R., Fingar D.C., Manning B.D., Kwiatkowski D.J., Antley L.C., Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signalling. Proc. Natl Acad. Sci. USA. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonseca B.D., Smith E.Q., Lee V., MacKintosh C., Proud C.G. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signalling downstream of this complex. J. Biol. Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 22.Vander Haar E., Lee S-I., Bandhakavi S., Griffin T.J., Kim H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell. Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 23.Oshiro N., Takahashi R., Yoshino K-C., Tanimura K., Nakashima A., Eguchi S., Miyamoto T., Hara K., Takehana K., Avruch J., Kikkawa U., Yonezawa K. The proline-rich Akt substrate of 40kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thedieck K., Polak P., Kim M.L., Molle K.D., Cohen A., Jeno P., Arrieumerlou C., Hall M.N. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE. 2007;2:e1214. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y., Thoreen C.C., Peterson T.R., Lindquist R.A., Kang S.A., Spooner E., Carr S.A., Sabatini D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Wang L., Harris T.E., Roth R.A., Lawrence J.C. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X., Shu L., Hosoi H., Murit K.G., Houghton P.J. Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J. Biol. Chem. 2002;277:28127–28134. doi: 10.1074/jbc.M202625200. [DOI] [PubMed] [Google Scholar]
- 28.Hietakangas V., Cohen S.M. Tor complex 2 is needed for cell cycle progression and anchorage-independent growth of MCF7 and PC3 tumor cells. BMC Cancer. 2008;8:282. doi: 10.1186/1471-2407-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masri J., Bernath A., Martin J., Jo O.D., Vartanian R., Funk A., Gera J. mTORC2 activity is elevated in gliomas and promotes growth and cell motility via overexpression of rictor. Cancer Res. 2007;67:11712–11720. doi: 10.1158/0008-5472.CAN-07-2223. [DOI] [PubMed] [Google Scholar]
- 30.Hietakangas V., Cohen S.M. Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev. 2007;21:632–637. doi: 10.1101/gad.416307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarbassov D.D., Ali S.M., Kim D-H., Guertin D.A., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 32.Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signalling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Yue P., Kim Y.A., Fu H., Khuri F.R., Sun Y. Enhancing mammalian target of rapamycin (mTOR)-targeted cancer therapy by preventing mTOR/raptor inhibition-initiated, mTOR/rictor-independent Akt activation. Cancer Res. 2008;68:09–7418. doi: 10.1158/0008-5472.CAN-08-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]