Abstract
Summary: The EMBRACE Registry is a web portal that collects and monitors web services according to test scripts provided by the their administrators. Users are able to search for, rank and annotate services, enabling them to select the most appropriate working service for inclusion in their bioinformatics analysis tasks.
Availability and implementation: Web site implemented with PHP, Python, MySQL and Apache, with all major browsers supported. (www.embraceregistry.net)
Contact: steve.pettifer@manchester.ac.uk
1 INTRODUCTION
‘Web services’ have become important tools in bioinformatics, allowing databases and algorithms to be accessed programmatically as computational components in programs, workflows and interactive analysis tools. Although these services are becoming common, with an growing adoption of standard protocols and technologies, the mechanisms for collecting and publicizing them are less mature. Service providers commonly resort to advertising their tools informally by email, or by listing them on institutional web pages and project wikis. This rather ad hoc approach has been sufficient to get web services into the mainstream of bioinformatics research and development; however, it comes with a number of limitations that are now being recognized by the community: finding suitable web services is difficult unless you know where to look; determining whether a web service is still operational is a matter of trying the service and seeing whether it appears to work correctly; and using a web service in the first instance requires a considerable amount of expertise—a problem that can be compounded by not knowing whether the service is in fact working as advertised.
A number of mechanisms for finding services have emerged over recent years (Goble et al., 2008). Particularly notable in this field are BioMoby Central (Wilkinson and Links, 2002), and the DAS Registry (Prlic et al., 2007), which provide single points of contact for finding biological services based on those specific technologies. The more general SeekDa (seekda.com) search engine indexes many thousands of SOAP-based services found by automated ‘crawling’ of the web, including several hundred that are relevant to biology or bioinformatics. Of these, only the DAS registry (which is restricted to recording DAS services) actively monitors the behaviour and status of its contents. Though tools exist to test the validity or presence of web service interfaces (e.g. www.soapui.org), these are unable to determine whether or not a the logic of service is functioning. It is still commonplace, therefore, to find services that are broken or no longer maintained.
The EMBRACE Network of Excellence has produced a web service registry that attempts to tackle these problems. Inspired by the project's own need to collect and advertise the growing number of databases and tools developed by project partners, and by the need within the consortium to share experiences about the provision and use of web services, the registry allows users to register, annotate, monitor and search for services, and acts as a ‘web2.0’-style community server, putting users and providers in touch with one another. Unlike ‘passive’ mechanisms for recording the existence of web services, this registry actively monitors the registration and ongoing behaviour of a service, giving providers and consumers up-to-date status notifications by email or via Twitter (www.twitter.com), if a service is behaving unexpectedly. The existence of a formal registration mechanism raises the question of what exactly constitutes a web service, and debates on this matter continue in the bioinformatics community and beyond (Stockinger et al., 2008). The approach of the this project, embodied in its registry, is to recommend a set of industry-standard technologies defined by the Web Service Interoperability organization and to provide tools that help developers move towards adopting these, while at the same time recognizing that a wide variety of other approaches exist for pragmatic or historical reasons. The registry therefore allows all manner of services to be added, and aims to provide documentation and support for users wishing to bring their services in line with standard practices. The registry thus supports WSDL/SOAP, REST, DAS and ‘home grown’ service types, with the dual intention of lowering the barriers to adoption and actively encouraging best practice.
2 FUNCTIONALITY AND ARCHITECTURE
One of the effects of the loosely coupled environment afforded by web services is that automated tasks rely on tools and resources provided by institutions located all around the world. The unexpected failure of one of these tools can have dire consequences for an analysis task. Even with today's comparatively reliable network connectivity, the heterogeneous nature of the back-end infrastructure that drives most bioinformatics web services means that occasional interruptions to services are inevitable. Determining reliably whether a service is working correctly at any given moment has been a real problem. The EMBRACE registry addresses this problem by combining automated monitoring mechanisms with high-level application-specific tests deposited by the service providers. The results from these tests are used to generate easy-to-read reports about the availability and reliability of a service over time. The registry itself is programmatically accessible as a web service, allowing other tools to automatically register and modify services and tests, as well as querying content.
Figure 1 shows the registry's architecture. The Drupal community server (www.drupal.org) acts both as a readily customizable web2.0-style foundation for the rest of the registry (including user registration and management, forums, blogs, tagging, rating and search facilities), and as a content management framework. Custom Drupal modules provide web service-specific functionality, with MySQL acting as a back-end database server. A separate Python harness, executing in an isolated virtual machine to limit damage caused by rogue code, executes whatever tests exist for a particular service and reports their status to the database.
Fig. 1.
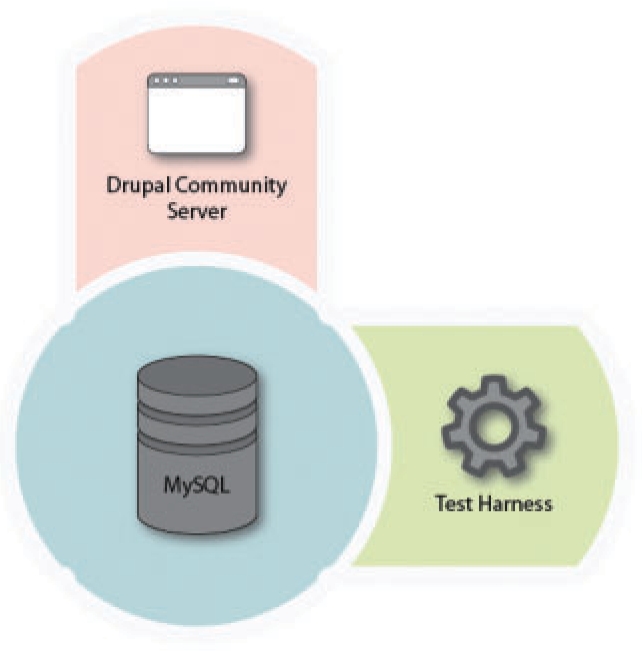
The architecture of the EMBRACE registry.
Some of the reasons for service failure are impossible to differentiate from one another from a client's point of view, and simply result in the service being unavailable. These errors can be detected by the generic tests run by the registry, (e.g. is the server currently accessible via the internet? If it is a DAS service, is it still returning an XML document that conforms to the DAS schema? If it is a WSDL service, are messages valid according to its WSDL description?). Other, perhaps more pernicious, problems can occur when the service is ostensibly working but is in fact returning plausible, but erroneous results. To detect these, the registry relies on the service-specific tests uploaded by the tool's curator (e.g. does this service correctly predict region X on protein Y?). The results of all these tests are combined to give an overall health status for each service, which is represented by one of the status indicators shown in Table 1. Although, in many ways, a gross simplification of the reality, these indicators provide a useful overview of a service's state for both providers and consumers; users can register an interest in particular services, and be notified by email, RSS or Twitter when their status changes.
Table 1.
Service status icons and their meanings
 |
3 RESULTS AND DISCUSSION
The registry has been active since October 2008; at the time of writing, it has accumulated ∼700 services from around 60 distinct users, principally from within the EMBRACE, BioSapiens and Enfin projects. Initially developed to collect the output of these projects, its monitoring and testing facility has already been of real use to service providers, identifying numerous service outages before they have become problems to the consumers. In several cases, it has also spotted significant but intermittent problems with what were considered to be ‘production quality’ services that had been thought to be running reliably for some considerable time. Based on the notifications generated by the registry, these services have now all been fixed, and have been running reliably, with confidence in their behaviour added by continued registry testing. We are now working closely with the BioCatalogue project (www.biocatalogue.org; Goble et al., 2008), funded by the UK Biotechnology and Biological Sciences Research Council, with a view to migrating the registry's content. This will ensure that its functionality and accumulated data will be secure beyond the end of the EMBRACE network, in early 2010.
Funding: The EMBRACE project is funded by the European Commission within its 6th Framework Programme, under the thematic area ‘Life sciences, genomics and biotechnology for health’, contract number LHSG-CT-2004-512092.
Conflict of Interest: none declared.
REFERENCES
- Goble C, et al. Data curation+process curation=data integration+science. Brief. Bioinform. 2008;9:506–517. doi: 10.1093/bib/bbn034. [DOI] [PubMed] [Google Scholar]
- Prlic A, et al. Integrating sequence and structural biology with DAS. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger H, et al. Experience using web services for biological sequence analysis. Brief. Bioinform. 2008;9:493–505. doi: 10.1093/bib/bbn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Links M. BioMOBY: an open-source biological web services proposal. Brief. Bioinform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]


