Abstract
Summary: We describe ChromA, a web-based alignment tool for chromatography–mass spectrometry data from the metabolomics and proteomics domains. Users can supply their data in open and standardized file formats for retention time alignment using dynamic time warping with different configurable local distance and similarity functions. Additionally, user-defined anchors can be used to constrain and speedup the alignment. A neighborhood around each anchor can be added to increase the flexibility of the constrained alignment. ChromA offers different visualizations of the alignment for easier qualitative interpretation and comparison of the data. For the multiple alignment of more than two data files, the center-star approximation is applied to select a reference among input files to align to.
Availability: ChromA is available at http://bibiserv.techfak.uni-bielefeld.de/chroma. Executables and source code under the L-GPL v3 license are provided for download at the same location.
Contact: stoye@techfak.uni-bielefeld.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Modern analytical methods in biology and chemistry use separation techniques coupled to sensitive detectors, such as gas chromatography–mass spectrometry (GC-MS) and liquid chromatography–mass spectrometry (LC-MS). These hyphenated methods provide high-dimensional data. Comparing such data manually to find corresponding signals is a tedious task, as each experiment usually consists of thousands of individual scans, each containing hundreds or even thousands of distinct signals.
In order to allow successful identification of metabolites or proteins within such data, especially in the context of metabolomics and proteomics, an accurate alignment and matching of corresponding features between two or more experiments is required. Such a matching algorithm should capture fluctuations in the chromatographic system which lead to non-linear distortions on the time axis (Strehmel et al., 2008).
Many different algorithms for the alignment of GC-MS/LC-MS data have been proposed and published, but only some of them are easily accessible or contained in publicly available toolkits (De Vos et al., 2007; Jonsson et al., 2005, 2006; Kohlbacher et al., 2007; Smith et al., 2006; Sturm et al., 2008). The tool presented here, ChromA, is immediately accessible for pairwise alignment and easy to use via the web frontend (see Supplementary Fig. 1) and as a web service. It provides different visual representations of the alignment, focusing on differences and similarities between the chromatograms. We additionally offer ChromA as an immediately deployable JAVAtm Web Start application and for download as a platform-independent command-line tool. These allow alignment of more than two chromatograms, using the center-star approximation to select a reference chromatogram among all input files to align to.
To compute the pairwise alignment, we use dynamic time warping (DTW) due to its applicability to data with non-linear time scale distortions (Itakura, 1975; Kruskal and Liberman, 1999; Sakoe and Chiba, 1978). It is suitable to globally align chromatograms, which are sequences of mass spectra. Every mass spectrum is preprocessed to nominal mass bin accuracy. In contrast to other methods (Robinson et al., 2007), there is no need for a priori selection of peaks for alignment, but a priori knowledge can be used to improve and speedup the alignment.
2 DATA MANAGEMENT AND METHODS
Currently, netcdf files (Rew and Davis, 1990) following the ASTM/AIA/ANDI-MS standard (Matthews and Miller, 2000) and xml files following the mzXML format (Pedrioli et al., 2004) can be read. Aligned chromatograms are stored in netcdf files, whereas general processing results, statistics and status information are saved in tab-separated value text format for easier access. All files generated during a run of ChromA, their creator and their designation (preprocessing, alignment, visualization, etc.), are stored in an xml file to allow an easy integration with data curation and analysis platforms for metabolomic experiments, for example, MeltDB (Neuweger et al., 2008).
In our software, we included different local distance and similarity functions between mass spectral intensity vectors, like the Euclidean distance, cosine similarity and linear correlation (Prince and Marcotte, 2006), to calculate a retention time alignment of chromatograms with DTW. Additionally, we included the Hamming distance on binarized vectors and a very fast function based on squared difference of total ion current (TIC) (Reiner et al., 1979), which is available for quick evaluation. Depending on the local function used, we apply different weights to provide a smooth warping. ChromA allows the user to define a number of optional configuration choices. As a preprocessing step, intensities contained in user-defined mass bins may be removed from consideration by the alignment. Additionally, manually or automatically matched peaks (Robinson et al., 2007; Styczynski et al., 2007) may be included as anchors to constrain the alignment to pass through their positions (see Supplementary Fig. 2).
Even though the worst case complexity of DTW is still of order 𝒪(m2) in space and time, where m is the number of scans in a chromatogram, we can achieve large speedups in practice. An alignment of two chromatograms with about 5400 scans each, 500 nominal mass bins, 38 defined anchors and a maximum scan deviation of 10% (about 540 scans to the left and right of the diagonal) using the cosine score as local similarity was calculated in 12 s on a MacBook with 2.4 GHz Core2 Duo processor, using around 500 MB of memory. Without any constraints, the same alignment was calculated in 7 min. The multiple alignment of 20 chromatograms using the center-star approximation required computation of 190 pairwise alignments. Using the aforementioned constraints, it was calculated within 40 min, without constraints in <24 h.
With the introduction of anchors to DTW, we address one major issue of peak-alignment algorithms, namely the problem of prior peak detection, by allowing strong peak candidates, such as reference compounds with unique mass traces (LC-MS) or characteristic fragmentation patterns (GC-MS), to be included, but at the same time allowing an alignment of weaker peaks. To allow the alignment additional flexibility, a neighborhood of radius n can be defined for all anchors.
3 VISUALIZATIONS
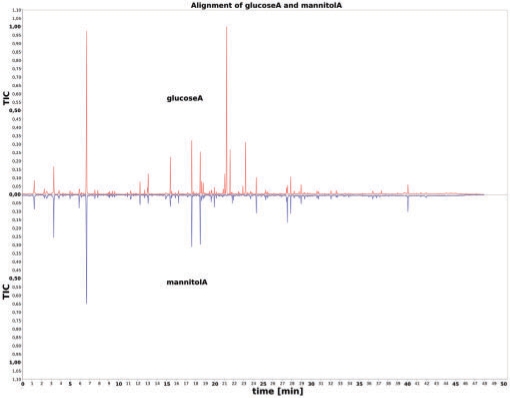
ChromA provides a number of visualizations for alignments, variable data and chromatograms, which are generated using the open source library JFreeChart (Gilbert and Morgner, 2009). In order to visualize alignments, we implemented different chart types. Figure 1 shows a plot of the TIC of the second chromatogram below the first chromatogram's TIC after alignment. Corresponding peaks are easily spotted with this visualization, as well as peaks that are only present in one of the chromatograms. We additionally provide visualizations of a multiple alignment of TICs before and after the alignment using the Web Start version of ChromA (Supplementary Figs 3 and 4), as well as an exemplary mass sensitive visualization of nominal mass 73 (silylation agent) before and after the alignment (Supplementary Figs 5 and 6).
Fig. 1.
Visualization of TICs after DTW alignment with ChromA. The TIC of file glucoseA is displayed above the TIC of file mannitolA. Files were obtained from experiments with Xanthomonas campestris pv. campestris B100 raised on different carbon sources (Neuweger et al., 2008). Chromatograms were aligned based on cosine similiarity between nominal mass-spectral intensity vectors.
4 CONCLUSION
ChromA is an easily accessible tool for retention time alignment of GC-MS and LC-MS chromatograms. Integration of the positions of matched peaks or of already identified compounds as anchors speeds up alignment calculation, yet still provides enough flexibility for it. The visualizations provided allow easy qualitative comparison of both unaligned and aligned replicate and non-replicate chromatograms. The framework used to develop ChromA, Maltcms (modular application toolkit for chromatography–mass spectrometry), available at http://maltcms.sourceforge.net, published under the GNU L-GPL v3 license, will be extended in the future, so we would like to encourage other researchers to join the project and contribute to it.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Tony Watt for providing the example GC-MS data available for evaluation on the web site.
Conflict of Interest: none declared.
REFERENCES
- De Vos RC, et al. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protocols. 2007;2:778–791. doi: 10.1038/nprot.2007.95. [DOI] [PubMed] [Google Scholar]
- Gilbert D, Morgner T. JFree Chart. 2009 Available at http://www.jfree.org/ (last accessed date April 30, 2009) [Google Scholar]
- Itakura F. Minimum prediction residual principle applied to speech recognition. IEEE Trans. Acoust. Speech. Signal. Process. 1975;23:67–72. [Google Scholar]
- Jonsson P, et al. High-Throughput data analysis for detecting and identifying differences between samples in GC/MS-based metabolomic analyses. Anal. Chem. 2005;77:5635–5642. doi: 10.1021/ac050601e. [DOI] [PubMed] [Google Scholar]
- Jonsson P, et al. Predictive metabolite profiling applying hierarchical multivariate curve resolution to GC-MS data a potential tool for multi-parametric diagnosis. J. Proteome Res. 2006;5:1407–1414. doi: 10.1021/pr0600071. [DOI] [PubMed] [Google Scholar]
- Kohlbacher O, et al. TOPP–the OpenMS proteomics pipeline. Bioinformatics. 2007;23:191–197. doi: 10.1093/bioinformatics/btl299. [DOI] [PubMed] [Google Scholar]
- Kruskal JB, Liberman M. The symmetric time warping problem: from continuous to discrete Ch. 4. In: Sankoff D, Kruskal J, editors. Time Warps, String Edits, and Macromolecules. Stanford, CA: CSLI Publications, Stanford University; pp. 125–161. [Google Scholar]
- Matthews L, Miller T. ASTM protocols for analytical data interchange. J. Assoc. Lab. Autom. 2000;5:60–61. [Google Scholar]
- Neuweger H, et al. MeltDB: a software platform for the analysis and integration of metabolomics experiment data. Bioinformatics. 2008;24:2726–2732. doi: 10.1093/bioinformatics/btn452. [DOI] [PubMed] [Google Scholar]
- Pedrioli PGA, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- Prince JT, Marcotte EM. Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Anal. Chem. 2006;78:6140–6152. doi: 10.1021/ac0605344. [DOI] [PubMed] [Google Scholar]
- Reiner E, et al. Characterization of normal human cells by pyrolysis gas chromatography mass spectrometry. Biomed. Mass. Spectrom. 1979;6:491–498. doi: 10.1002/bms.1200061107. [DOI] [PubMed] [Google Scholar]
- Rew RK, Davis GP. NetCDF: an interface for scientific data access. IEEE Comput. Graph. Appl. Mag. 1990;10:76–82. [Google Scholar]
- Robinson M, et al. A dynamic programming approach for the alignment of signal peaks in multiple gas chromatography-mass spectrometry experiments. BMC Bioinformatics. 2007;8:419. doi: 10.1186/1471-2105-8-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoe H, Chiba S. Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans. Acoust. Speech. Signal. Process. 1978;26:43–49. [Google Scholar]
- Smith CA, et al. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Strehmel N, et al. Retention index thresholds for compound matching in GC–MS metabolite profiling. J. Chromatogr. B. 2008;871:182–190. doi: 10.1016/j.jchromb.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Sturm M, et al. OpenMS – an open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styczynski MP, et al. Systematic identification of conserved metabolites in GC/MS data for metabolomics and biomarker discovery. Anal. Chem. 2007;79:966–973. doi: 10.1021/ac0614846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.