Abstract
Summary: PINE-SPARKY supports the rapid, user-friendly and efficient visualization of probabilistic assignments of NMR chemical shifts to specific atoms in the covalent structure of a protein in the context of experimental NMR spectra. PINE-SPARKY is based on the very popular SPARKY package for visualizing multidimensional NMR spectra (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). PINE-SPARKY consists of a converter (PINE2SPARKY), which takes the output from an automated PINE-NMR analysis and transforms it into SPARKY input, plus a number of SPARKY extensions. Assignments and their probabilities obtained in the PINE-NMR step are visualized as labels in SPARKY's spectrum view. Three SPARKY extensions (PINE Assigner, PINE Graph Assigner, and Assign the Best by PINE) serve to manipulate the labels that signify the assignments and their probabilities. PINE Assigner lists all possible assignments for a peak selected in the dialog box and enables the user to choose among these. A window in PINE Graph Assigner shows all atoms in a selected residue along with all atoms in its adjacent residues; in addition, it displays a ranked list of PINE-derived connectivity assignments to any selected atom. Assign the Best-by-PINE allows the user to choose a probability threshold and to automatically accept as “fixed” all assignments above that threshold; following this operation, only the less certain assignments need to be examined visually. Once assignments are fixed, the output files generated by PINE-SPARKY can be used as input to PINE-NMR for further refinements.
Availability: The program, in the form of source code and binary code along with tutorials and reference manuals, is available at http://pine.nmrfam.wisc.edu/PINE-SPARKY.
Contact: whlee@nmrfam.wisc.edu; markley@nmrfam.wisc.edu
1 INTRODUCTION
Despite rapid progress toward automating many facets of research in structural biology, visualization and expert verification of computational results continue to be required. PINE-NMR (Bahrami et al., 2009) is an automated protein NMR assignment package that accepts, as input, the amino acid sequence of a protein and peak lists associated with defined NMR experiments and provides, as output, probabilistic backbone and side chain assignments and an analysis of the secondary structure. PINE-NMR can accommodate prior information about assignments or stable isotope labeling schemes. PINE-NMR achieves robust and consistent results that have been shown to be effective in subsequent steps of NMR structure determination. In cases where the input data do not support unequivocal assignments (because of weak signals or too many missing signals) PINE-NMR provides multiple ranked possibilities that need to be evaluated. The PINE-SPARKY software package described here provides a graphical interface for reviewing possible assignments in the context of their experimental basis (peaks in multidimensional NMR spectra) and for choosing among them. The software enables the expert to inject additional knowledge into the assignment process in an efficient and straightforward manner. The functionality of PINE-SPARKY is different from the SPARKY spin graphs extension, which shows connectivities between assigned peaks, but will not handle PINE-NMR results.
2 IMPLEMENTATION
We selected SPARKY as the viewing and verification tool, because currently it is the most popular NMR visualization and assignment program according to software citations in BMRB (Ulrich et al., 2007). Another benefit is that SPARKY enables programmers to utilize its internal classes to write Python extensions. PINE-SPARKY consists of two parts: (i) PINE2SPARKY, which converts PINE-NMR assignments and their associated probabilities to SPARKY inputs and (ii) PINE. SPARKY extensions, which support intuitive interfaces that enable various visualization and assignment tasks.
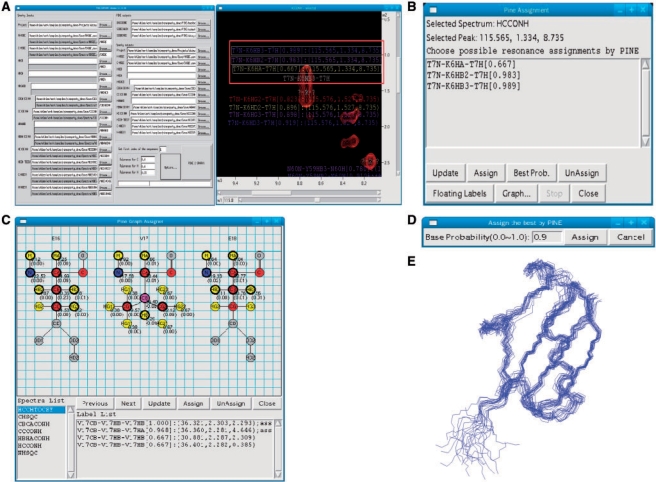
PINE2SPARKY converter: Multiple assignments and their probabilities (output from PINE-NMR) are converted into labeled objects, and these objects are incorporated into SPARKY save files by the PINE2SPARKY converter (Fig. 1A). After the user chooses which assignment is correct, the incorrect labels can be removed. Colors of the labels are associated with the level probability. These can be configured by the user, but the default spectrum is blue for the highest probability and red for the lowest. We developed PINE2SPARKY under Lazarus, an IDE of Free Pascal, and the software is compatible with multiple operating systems (MS Windows, MacOSX and Linux).
Fig. 1.
PINE-SPARKY user interface and its use in 3D structures determination. (A) PINE2SPARKY converter and SPARKY labels. (B) PINE Assigner box. (C) PINE Graph Assigner box. (D) Assign the Best by PINE box. (E) Three-dimensional structure of ubiquitin determined from PINE-SPARKY assignments.
PINE Assigner is a dialog box. The peak to be analyzed is selected prior to opening the dialog box. The dialog box lists all possible assignments for that peak (Fig. 1B) and contains buttons that simplify the assignment selection process. Each button is labeled with its function (Update, Assign, Best probability, Unassign, Floating labels, Graph, Stop, Close).
PINE Graph Assigner is a graphical window consisting of four parts: the covalent structural representation of a tripeptide, a list of spectra associated with different NMR experiments that PINE-NMR used for the assignment (Fig. 1C), buttons with defined functions (Previous residue, Next residue, Update, Assign, Unassign, Close), and list of labels. When the user chooses a residue from the protein sequence, the graphical window displays all the atoms in that residue as well as the atoms in the residues sequentially to either side. Atoms with assignments are color coded (yellow for 1H, red for 13C, blue for 15N); gray denotes atoms that PINE-NMR was unable to assign. Chemical shifts and their standard deviations associated with the assignments are displayed below and to the right of each assigned atom. When the user clicks on an individual atom and a spectrum, PINE Graph Assigner displays a ranked list of PINE-derived assignment connectivities to that atom from that spectrum. By going to the spectrum view, the user sees a list of available peak labels associated with the chosen atom. One can assign or unassign peaks with a few mouse clicks. The list of spectra includes only those currently loaded into PINE-SPARKY.
Assign the Best by PINE enables the user to bypass the manual steps needed to fix assignments. The user can choose a threshold, such as 90%, and Assign the Best by PINE will fix all assignments with probabilities greater than or equal to this value (Fig. 1D).
3 RESULTS AND CONCLUSION
We used NMR data from the 76-residue protein, human ubiquitin, to illustrate the use of PINE-SPARKY in a structure determination project. 1H–15N HSQC, 1 H–13C HSQC, CBCA(CO)NH, HNCACB and HBHA(CO)NH data sets were collected to support backbone assignments, and (H)CC(CO)NH, H(CC)(CO)NH and HCCH-TOCSY data sets were collected to support sidechain assignments. 15N-edited NOESY 13C-edited NOESY data sets were used in a subsequent structure determination. NMRpipe (Delaglio et al., 1999) was used to process all NMR spectra, and NMRdraw (Delaglio et al., 1999) was used to pick peaks in all but the NOESY data sets. ATNOS (Herrmann et al., 2002) was used to pick NOESY peaks. We generated a SPARKY project and save files with the processed spectra. PINE-NMR was used to generate probabilistic assignments, and these were uploaded via the PINE2SPARKY converter. Tolerances for 13C 15 N were set at 0.4 ppm, and that for 1H was set to 0.03 ppm. Assign the Best by PINE was performed with a threshold of 0.9 (90%) with all (non NOESY) NMR spectra. Peaks that remained unassigned after that process were assigned with PINE Graph Assigner and PINE Assigner. Assign the Best by PINE with 0.9 threshold assigned more than 90% of the peaks automatically. After this the procedure, it was possible to quickly assign the remaining peaks with small number of clicks using PINE Graph Assigner. TALOS (Cornilescu et al., 1999) was used to determine torsion angle constraints from the assigned chemical shifts: 106 torsion angles involving 53 residues were judged to be ‘good’ by TALOS, and these were used constraints along with the NOESY data in 3D structure calculations by CYANA (Güntert, 2004). In the resulting 20 best structures, the root mean standard deviation was 0.46 Å for backbone atoms and 1.22 Å for all heavy atoms in the structured regions (Fig. 1E). The following is an analysis of the time required to determine the structure following initial data collection: PINE-NMR run (∼1 h), PINE-SPARKY analysis (30 min), TALOS analysis (20 min), CYANA structure determination (7 min) with 16 CPUs.
Funding: National Institutes of Health [grant numbers P41 RR02301 and 1U54 GM074901].
Conflict of Interest: none declared.
REFERENCES
- Bahrami A, et al. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 2009;5:1–12. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, et al. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Bax A. Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 1994;4:738–744. [Google Scholar]
- Cornilescu G, et al. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Herrmann T, et al. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- Pervushin K, et al. Attenuated T2relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich E, et al. BioMagResBank. Nucleic Acids Res. 2007;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K. Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 1990;265:22059–22062. [PubMed] [Google Scholar]
- Yang D, Kay LE. TROSY triple-resonance four-dimensional NMR spectroscopy of a 46 ns tumbling protein. J. Am. Chem. Soc. 1999;121:2571–2575. [Google Scholar]
- Zhang L, Yang D. SCAssign: a sparky extension for the NMR resonance assignment of aliphatic side-chains of uniformly13C15N-labeled large proteins. Bioinformatics. 2006;22:2833–2834. doi: 10.1093/bioinformatics/btl477. [DOI] [PubMed] [Google Scholar]