Abstract
Motivation: Recent advances in high-throughput technologies have made it possible to investigate not only individual protein interactions, but also the association of these proteins in complexes. So far the focus has been on the prediction of complexes as sets of proteins from the experimental results. The modular substructure and the physical interactions within the protein complexes have been mostly ignored.
Results: We present an approach for identifying the direct physical interactions and the subcomponent structure of protein complexes predicted from affinity purification assays. Our algorithm calculates the union of all maximum spanning trees from scoring networks for each protein complex to extract relevant interactions. In a subsequent step this network is extended to interactions which are not accounted for by alternative indirect paths. We show that the interactions identified with this approach are more accurate in predicting experimentally derived physical interactions than baseline approaches. Based on these networks, the subcomponent structure of the complexes can be resolved more satisfactorily and subcomplexes can be identified. The usefulness of our method is illustrated on the RNA polymerases for which the modular substructure can be successfully reconstructed.
Availability: A Java implementation of the prediction methods and supplementary material are available at http://www.bio.ifi.lmu.de/Complexes/Substructures/.
Contact: caroline.friedel@bio.ifi.lmu.de
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Cellular processes of all sorts are shaped by proteins associated in complexes. Thus, the identification of such complexes and the interactions within the complexes have become a major experimental focus. While direct, physical interactions can be identified by the yeast two-hybrid (Y2H) approach (Fields and Song, 1989), affinity purification methods followed by mass spectrometry, such as tandem affinity purification (TAP) (Rigaut et al., 1999), can also identify indirect interactions via other proteins in complexes. Recently, the TAP systems was applied by Gavin et al. (2006) and Krogan et al. (2006) to identify protein complexes in the yeast Saccharomyces cerevisiae on a genome scale.
In the TAP system, epitope-tagged proteins (baits) are expressed and purified in consecutive affinity columns (Rigaut et al., 1999). Proteins interacting directly or indirectly with the bait, so-called preys, are then co-purified with the bait and identified by mass spectrometry. Ideally, the purification of one bait would yield the complete protein complex the bait is involved in. However, proteins may be co-purified which bind unspecifically to the bait (false positives), while proteins from the same protein complex may fail to bind tightly enough and be missed in the screen (false negatives). Due to these measurement errors and the large size of these datasets, sophisticated methods are necessary to predict the actual complexes from the purification results.
The first predictions methods were developed by the groups of Gavin et al. and Krogan et al. themselves. Since the resulting complexes showed only relatively little agreement, advanced methods have been developed recently (Collins et al., 2007; Friedel et al., 2008; Hart et al., 2007; Pu et al., 2007), which improved predictive performance significantly. Here, most approaches use a two-step approach by first calculating interaction scores and then predicting the complexes from those scores. Thus, complexes are predicted as sets of associated proteins and the substructure of the complexes or the physical interactions within these is not considered. As the predictions of the best approaches differ significantly although the overall prediction quality is the same, a more detailed analysis of the complex structure is necessary for the individual complexes.
So far, few computational methods have been developed for analyzing the substructure of protein complexes. Aloy et al. (2004) used homology modeling and electron microscopy to at least partially resolve interactions between subunits of 54 experimentally derived complexes. The method of Hollunder et al. (2007) identifies subsets of proteins which occur more frequently in different complexes than expected at random. Gavin et al. (2006) distinguished between core elements and modules or attachments in their protein complex predictions, but did not predict direct interactions.
Scholtens et al. (2005) and Bernard et al. (2007) developed approaches to model the physical topology of protein complexes from affinity purification results as well as physical interactions from Y2H experiments. However, Scholtens et al. used this only as an intermediate step in predicting protein complexes and did not evaluate the actual interactions they predicted. Bernard et al. showed that accurate predictions can be obtained by applying their approach to combined affinity purification and Y2H results, but did not evaluate to what degree their results depend on the Y2H interactions used additionally.
Here, we investigated whether the topology of protein complexes can be predicted from the affinity purification results alone. The topology of a protein complex describes both the direct physical interactions within the complex (the complex scaffold) and its hierarchical substructure, i.e. the subdivision of the complex into smaller components. Since most methods for predicting protein complexes from affinity purification results calculate interaction scores as an intermediate step, we developed a method to extract the complex scaffolds from these densely connected scoring networks.
Our algorithm calculates the union of all maximum spanning trees (MSTs) from the interaction scores for each complex. The MSTs are then extended by interactions which are not accounted for by indirect interactions via other proteins. We applied our method to confidence scores and protein complexes calculated from the yeast affinity purification experiments of Gavin et al. and Krogan et al. with the Bootstrap method (Friedel et al., 2008). Our approach predicts physical interactions with superior accuracy than baseline approaches and the method by Bernard et al. (2007). Furthermore, the distance in the resulting network between two proteins reflects the similarity of their subcomponent annotations. Accordingly, the substructure of the protein complexes can be resolved and subcomplexes can be identified.
2 METHODS
In the following, let C={C1,…, Cn} be a set of protein complexes with Ci a set of proteins and G=(V, E) a weighted network of interaction scores. Here, V is the set of all proteins and E the set of all interactions between them. We assume that all scores are confidence values in the range of 0 to 1. The function w : E→[0, 1] defines the weight, i.e. the confidence score, of each edge. Interactions not contained in the network are given a weight of 0. If the scoring method calculates general scores from −∞ (or 0) to ∞, edge weights are scaled to [0, 1].
We assume that each complex is connected in the network of actual physical interactions. This means that each protein can be reached from every other protein in the same complex by a path of physical (direct) interactions. This network of direct interactions is denoted as the scaffold of the complex and the scaffold is predicted separately for each complex.
2.1 Maximum spanning trees
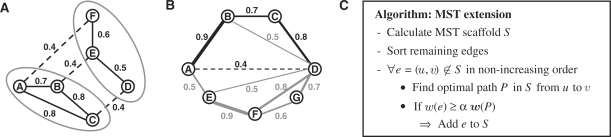
For each complex, our algorithm takes as input the network of interaction scores for all protein pairs in this complex. From this interaction network, we predict interactions for the complex scaffold by calculating MSTs. A spanning tree is a tree which connects all vertices in the network. The MST is the spanning tree with the maximum sum of edge weights. As several different MSTs can exist in a network, we determine the union of all possible MSTs to predict the physical interactions in the complex scaffold. To calculate the union of all MSTs, we used a modification of the Kruskal algorithm (Cormen et al., 2000) described by Bandelt et al. (1999) (Fig. 1A). The runtime of this algorithm is in O(|E|log|V|) as for the original Kruskal algorithm and its correctness follows from the work of Carroll (1995).
Fig. 1.
(A) The algorithm for determining the union of all MSTs (Bandelt et al., 1999). Edge weights are processed in decreasing order. For each edge weight δ (in the example δ=0.4), all edges with weight δ (dashed lines) which connect different connected components (gray ellipses) are added to the current MST network (solid lines). (B) Illustration of how MSTs are extended (for α = 1). The current scaffold is indicated by solid lines. To determine if the interaction between A and D (dashed line) should be added to the network, we first find the optimal path with maximum probability between the two nodes. In this case, this is A → B → C → D which has a weight of 0.9 · 0.7 · 0.8=0.504. Since the weight of edge (A, D) is < 0.504, the edge is discarded. If the weight of (A, D) were > 0.504, it would be added to the scaffold network. (C) Algorithm for calculating the extended MST networks (eMSTα).
2.2 Extending the MSTs
Although the combination of all MSTs is no longer necessarily a tree, the resulting networks are extremely sparse and many physical protein interactions are missed. As a consequence, we extend this network by interactions which cannot be explained by an indirect interaction via other proteins in the MST scaffold. For this purpose, we compare an interaction (u, v) in the original network to the best indirect interaction between u and v in the current scaffold network. If the edge weight is at least as high as a factor α times the weight of the best indirect interaction, the interaction is added to the MST network. The resulting network is denoted as eMSTα and the parameter α tunes the density of the resulting scaffold network. By default, α is set to 1.
For calculating the best indirect interaction, we use the fact that all edge weights are confidence values in [0, 1]. As a consequence, the weight of an edge is interpreted as the probability that this edge is a physical interaction. Here, we assume independence between the edge probabilities. The probability of an indirect interaction between two proteins u and v is then defined as the maximum probability of any path between them in the current scaffold [without the edge (u, v)] (Fig. 1B).
The probability of a path P is calculated as the product of the edge probabilities on this path (w(P)). By taking the absolute values of the logarithms of the edges weights, the path with maximum probability can be efficiently calculated as the path with the smallest sum of transformed edge weights. This optimal path between a pair of nodes can then be efficiently calculated using Dijkstra's algorithm for shortest paths (Cormen et al., 2000). Thus, the worst-case runtime of the algorithm is O(|E|2log|V|) using binary heaps. Since the scaffold networks are relatively small and sparse, this is sufficiently fast for practical purposes.
To identify interactions which cannot be explained by a sequence of sufficiently strong interactions via other proteins, we process candidate interactions in the order of non-increasing edge weights (Fig. 1C). For each interaction e, we calculate the optimal alternative path P with maximum probability w(P) between the corresponding proteins in the current scaffold. The interaction e is added to the scaffold if w(e)≥αw(P) and the scaffold is updated whenever a new interaction is identified (Fig. 1C). Since we never remove any interaction and consequently any path from the scaffold, no interaction is missed in this way. In the following, we show for α≤1 that no interaction is added with a better alternative path in the final scaffold.
Lemma 1. —
∀e=(u, v) in the final scaffold network S, we have that w(e)≥αw(P) for all alternative paths P between u and v in S if α≤1.
Proof. —
By contradiction: assume, there exists a path P for an edge e such that w(e)<αw(P). Since the weight of each edge is ≤1 and edge weights are multiplied to get w(P), we have for each edge f ∈ P that w(P)≤w(f). Thus, w(e)<αw(f) ∀f∈P and w(e)<w(f)∀f∈P if α≤1. As a consequence, all edges on this path have been processed before e and this path was already contained in the scaffold S when e was added. For the best alternative path Popt between u and v at this time, we have that w(Popt)≥w(P). As a consequence, αw(Popt)≥αw(P)>w(e). This is a contradiction to the construction of the scaffold network. ▪
2.3 Identification of subcomplexes
To characterize the substructure of the complexes and identify subcomplexes, we apply a simple partitioning approach to the scaffold network determined with the MST or extended MST approach. For this purpose, interactions are processed in the order of non-increasing edge weights. If the current interaction connects a singleton protein (= a protein not contained in any subcomplex yet) either to a subcomplex or another singleton protein, the protein is included in this subcomplex or combined with the singleton to create a new subcomplex. As we never merge two subcomplexes which both contain more than one protein, this results in a disjoint partitioning of the network without having to define cutoff parameters.
3 RESULTS
The MST and extended MST approaches were applied to interaction scores and complex predictions calculated from the combined results of the genome-scale TAP experiments of Gavin et al. (2006) and Krogan et al. (2006) in yeast. Here, we used confidence scores and protein complexes (409 complexes, BT-409) predicted with the unsupervised Bootstrap approach that we presented recently (Friedel et al., 2008). The Bootstrap confidence scores have been shown to be more accurate than any other scoring method and the Bootstrap complexes have the same quality as the best supervised predictions.
All Bootstrap confidence scores are between 0 and 1 and the original network contains 62 876 interactions. By restricting this to interactions within the BT-409 complexes, we obtained 9918 interactions (15.8% of the original set). The MST approach extracted 1658 interactions and the extended MST approach (with α=1) 3085 interactions. As baseline classifier a simple algorithm was used which calculates the connected network for each complex. The connected network is defined as the network Gτ within each complex where w(e)≥τ ∀e∈Eτ and τ is the largest value such that Gτ is connected. This baseline approach predicted 5404 interactions.
3.1 Reference interactions
To compile a reference set of physical interactions, we extracted all yeast protein–protein interactions from the BioGRID database (Breitkreutz et al., 2008) (release 2.0.45, October 1, 2008) determined with the Y2H method. Furthermore, Y2H interactions from the recently published study by Yu et al. (2008) were included as they were not yet contained in this BioGRID release. From the complete set of Y2H interactions, we extracted a second set of interactions from small-scale experiments in which ≤100 interactions were determined. We used only Y2H interactions for the reference sets, since other experimental methods, such as co-immunoprecipitation or pull-down assays, do not only detect physical but also indirect interactions via other proteins.
Additionally, we inferred physical interactions from large-scale Y2H studies for other species and known domain–domain interactions extracted from 3D structures of protein complexes. Y2H interactions were predicted for yeast from large-scale studies for other species (Giot et al., 2003; Li et al., 2004; Rual et al., 2005; Stelzl et al., 2005) using orthology assignments from the Inparanoid database (Berglund et al., 2008). Interactions were predicted if both interaction partners had orthologs in yeast. Domain–domain interactions were taken from the iPfam (Finn et al., 2005) and 3DID (Stein et al., 2005) databases and mapped to the yeast proteome. Only interactions between different protein chains in the crystal structure were considered.
Since the Y2H system is prone to measurement errors, we also analyzed separately the core (806 interactions) and non-core (3669 interactions) Y2H interactions determined in yeast by Ito et al. (2001) to investigate the influence of measurement errors in the benchmark set. Note that the non-core interactions were not included in the complete Y2H reference set. Yu et al. (2008) recently showed that the non-core Ito interactions have tremendously lower confirmation rates than the core interactions and contain mostly false positive interactions. This allowed us to compare the evaluation results for two datasets with large differences in accuracy.
3.2 Evaluation of predictive accuracy
To evaluate the predictive accuracy of the presented methods, true positive rate (TPR) and false positive rate (FPR) with regard to the reference networks were calculated. TPR is defined as the fraction of reference interactions within the BT-409 complexes recovered by the prediction methods. FPR is the fraction of interactions within the BT-409 complexes not contained in the reference network but predicted to be in the scaffold. To compare prediction accuracy of different methods, we calculated the TPR/FPR ratio for each method. Analytical results (see Supplementary Material) showed that this measure allows to determine a correct ranking of the performance of the different prediction methods despite measurement errors in the reference networks.
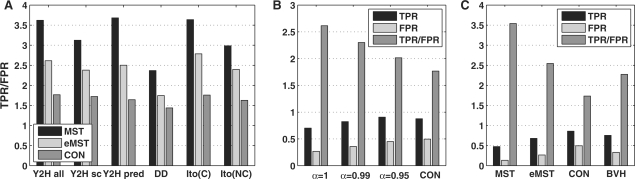
For all reference networks significant improvements in predictive accuracy were obtained with the MST approach (Fig. 2A) compared with the baseline approach. Generally, TPR/FPR ratios are almost twice as high as for the connected networks. For instance, on the complete Y2H network only 13.6% false positives are identified by the MST approach at a TPR of 49.1% compared with 49.6% false positives for the connected approach. Although the connected approach recovers 87.6% of the true positives, the TPR/FPR ratio of the MST approach cannot be reached even if only the most confident interactions in the connected network are considered (see Supplementary Fig. 1). As the accuracy of the non-core Ito interactions within complexes is comparable with the core network (see Supplementary Material), it showed the same ranking of the methods as the other networks.
Fig. 2.
(A) Ratio of TPR to FPR for the MST, extended MST (eMST, α = 1) and connected (CON) approach on the complete Y2H network (Y2H all), the small-scale Y2H interactions (Y2H sc), the predicted interactions from Y2H experiments for other species (Y2H pred), the domain–domain interactions from the 3DID and iPfam databases (DD) and the core [Ito(C)] and non-core [Ito(NC)] interactions from the study by Ito et al. (2001). (B) TPR and FPR for decreasing values of α and the connected networks on the complete Y2H network. (C) Comparison of prediction accuracy of the MST, eMST and connected approach to the predictions by Bernard et al. (2007) (BVH) on the complete Y2H network. For this purpose, Y2H interactions from the studies of Uetz et al. (2000) and Ito et al. (2001) were not included in the calculation of TPR and FPR as they were used for training by Bernard et al. (2007).
The higher specificity of the MST approach results in a significantly lower sensitivity which can be increased by extending the MSTs. Although the FPR consequently increases as well, the overall performance of the extended MSTs is still significantly better than for the baseline predictions. Figure 2B illustrates TPR and FPR on the complete Y2H network in yeast for decreasing values of α used for extending the MSTs. The more conditions are relaxed for extending the networks, the more interactions are added. Thus, more true interactions are recovered, but also more wrong predictions are made. Even so more true positives can be recovered with the extended MST approach for α=0.95 at a lower FPR than for the connected network.
We also compared our approach against the predictions by Bernard et al. (2007) (Fig. 2C). The predictions of Scholtens et al. (2005) were not evaluated as they are used only as an intermediate step in complex prediction and cannot be obtained from the R implementation of the algorithm. Since Bernard et al. used Y2H interactions from the studies of Uetz et al. (2000) and Ito et al. (2001) as training data, these interactions were excluded in the calculation of TPR and FPR to obtain unbiased estimates. The resulting TPR/FPR ratios show that although the Bernard et al. approach is superior to the connected baseline classifier, it has a lower accuracy than the extended MST and in particular the MST approach. Since the focus of the Scholtens et al. method is on modeling co-complex membership and not specifically physical interactions, we expect that its prediction accuracy is also lower.
Apart from prediction accuracy, our method improves significantly on runtime. Applying the extended MST approach to the complete Bootstrap network took < 3 min on one processor of an Intel Core2Duo with 2.4 GHz. Even if we include the runtime of the Bootstrap algorithm (∼1.5 h), this is one order of magnitude less than the 12.5–15 h reported by Bernard et al. (2007). Due to the large memory requirements of the R implementation by Scholtens et al. (>8 GB for both the Gavin et al. and Krogan et al. dataset), runtime of their algorithm could not be evaluated.
3.3 Substructure resolution in the scaffold network
An analysis of the 195 complexes with more than two proteins showed a negative correlation between the density of the complex scaffolds predicted by the extended MST approach and the size of the complexes (Spearman correlation coefficient: −0.34, P-value: 1.3×10−6). Eighty-three (42.6%) of these complexes are fully connected, but they have an average size of only 3.8. Thus, for small complexes a globular structure is predicted in most cases where the majority of proteins interact physically. For the remaining larger complexes (average size 9.5) sparser networks are predicted (∼28.4% of interactions not contained in the MST networks) and, as a consequence, more complex substructures.
Proteins in the same subcomplexes are more closely associated in the network of physical interactions than proteins in different parts of the complex. Thus, we investigated whether the distance of two proteins in the scaffold network, i.e. the number of interactions on the shortest (unweighted) path between them, accurately reflects the similarity of the subcomplexes they are part of. Similarity of the subcomplexes of two proteins was calculated as the fraction of Gene Ontology (GO) (Ashburner et al., 2000) cellular component annotations they have in common. For this purpose, only GO terms corresponding to protein complexes were considered. This simple overlap measure allows for a more fine-grained analysis of the subcomponents within complexes than more sophisticated between measures and which take into account the hierarchical structure of the GO and yield similar results for a complex and its subcomplexes.
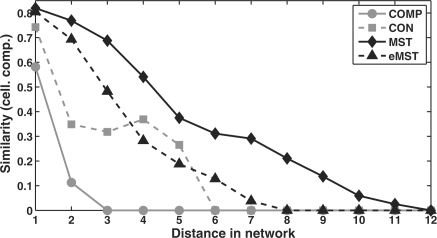
Our results showed that the similarity of subcomponents of two proteins decreased with the distance between the corresponding proteins (Fig. 3). It decreased less rapidly for the MST and extended MST networks since these networks are more sparse. Thus, proteins involved in different subcomponents of a complex are separated from each other by many interactions in the predicted scaffolds, whereas proteins involved in the same subcomponents are close to each other. As the small distances in the baseline predictions make it difficult or even impossible in many cases to identify a substructure in the networks, resolution of the subcomponent structure is significantly improved by the MST and extended MST approaches.
Fig. 3.
Correlation of the distance between a protein pair in the complete (COMP), connected (CON), MST and eMST networks to the fraction of GO cellular compartment annotations the two proteins have in common. Here, only GO terms corresponding to protein complexes were considered. Averages are taken over all protein pairs with the same distance. Subcomponent similarity increases at a distance of 4 for the connected network due to outliers among the few protein pairs with this distance.
3.4 Evaluation of subcomplex identification
Our results showed that the scaffold network determined with the extended MST network reflects the subcomponent structure of the complexes. To determine the actual subcomplexes, we applied our subcomplex identification algorithm described in Section 2.3 to this network. For 50 of the 195 complexes containing more than two subunits at least two subcomplexes were identified. The majority of these complexes (44/50 = 88%) were not fully connected in the extended MST network and their average size (13.7) was significantly larger than for complexes which were not subdivided (4.8). For 46 of these complexes with available GO annotations, we compared subcomponent similarity within the complete complex with the predicted subcomplexes using the same measure as before. On average, the similarity within the subcomplexes identified was increased by ∼20% compared with the complete complex. Thus, the subcomponent structure of the complexes could be better characterized by identifying subcomplexes in the scaffold network with our method.
Subcomplex predictions were analyzed in more detail for the 21 complexes with at least 10 subunits, at least two predicted subcomplexes and available protein complex annotations in the GO (see http://www.bio.ifi.lmu.de/Complexes/Substructures/). For three complexes, which consisted of partially overlapping complexes clustered together due to a few shared proteins (DNA-directed RNA polymerases; SWI/SNF and RSC chromatin remodelers; INO80, SWR1 and NuA4 complexes), this allowed us to identify the different complexes. Here, physical interactions were only predicted between proteins in the same complex. Shared proteins were found at the interface between the subcomplexes and were strongly associated with more than one subcomplex. Another two complexes also corresponded to overlapping complexes (SAGA and TFIID complexes; Rpd3L and Rpd3S complexes), but the overlaps were too large to separate the complexes. Nevertheless, meaningful subsets of proteins were identified which are contained together in some but not all of the overlapping complexes. For the SAGA complex, for instance, we could distinguish two proteins which are contained in the SAGA complex but not (SPT8) or only in a C-terminally truncated form (SPT7) in the SAGA-like complex (Pray-Grant et al., 2002).
For six complexes (spliceosome; the vacuolar ATPase; the 90S preribosome in two overlapping forms; a splicing factor complex; and the kinetochore), known subcomplexes were correctly identified to a large extend. For another five complexes (Proteasome, regulatory particle; mitochondrial large ribosomal subunit; small subunit of the ribosome; RNase MRP; and vesicle transport complexes), subsets of proteins were identified for which it was not quite clear how they corresponded to the subcomponent structure of the complex as it was not sufficiently described in the literature.
For the 16 complexes described above, similarity of the subcomponents as described in the GO was increased in the subcomplexes compared with the complete complex (15 complexes) or stayed the same (1 complex). For the remaining five of the 21 analyzed complexes, similarity of subcomponents was slightly lower (by ∼2.4%) in the subcomplexes we identified than in the complete complex as no detailed subcomplex structure has been described in the GO. However, we could recover several subcomplexes that have previously been described in the literature such as the RIX1 and YTM1-ERB1-NOP7 subcomplexes of the preribosome (Krogan et al., 2004) (see http://www.bio.ifi.lmu.de/Complexes/Substructures/ for detailed results). In the following, we illustrate with the example of the DNA-directed RNA polymerase complexes how a detailed analysis of the complex scaffold and the predicted subcomplexes can lead to a better understanding of the complex structure.
3.5 Analysis of the DNA-directed RNA polymerase
The DNA-directed RNA polymerase complex is one of the largest predicted complexes in the BT-409 set and contains 46 proteins. It effectively consists of three separate RNA polymerase complexes (RNA polymerase I, II and III), which have been clustered into one complex since they have many proteins in common. Such complexes which overlap to a large degree are a general problem for complex prediction algorithms and other complex prediction approaches also cluster the three polymerases together. The crystal structure of polymerase II is known, whereas only little structural information is available for polymerases I and III (Cramer et al., 2008).
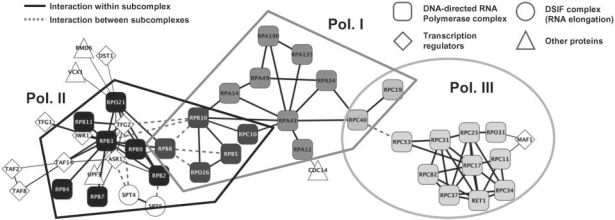
In the extended MST network (Fig. 4), the subdivision of the complex into polymerase complexes I, II and III is clearly visible which is not the case for the complete or connected networks. Due to the clear separation of the three complexes in the extended MST network, they could be successfully determined using our subcomplex identification approach. Physical interactions were only predicted between proteins contained in the same polymerase and no interactions are observed between different polymerases. The polymerase III complex is connected by two shared proteins (RPC19, RPC40) to the polymerase I complex. The latter one is connected to the polymerase II complex via a group of five proteins (RPB5, RPO26, RPB8, RPB10 and RPC10) contained in all three RNA polymerase complexes. Since our complex identification algorithm predicts a disjoint partitioning of the complex, each of the shared proteins was assigned to only one of the subcomplexes. Nevertheless, as these shared proteins connect the different subcomplexes and are strongly associated to several subcomplexes, they can be identified in a straightforward way.
Fig. 4.
Predicted subnetworks for the DNA-directed RNA complex with the extended MST approach. Colors indicate the three subcomponents: Polymerase complexes I (gray), II (black) and III (light gray). Rectangles denote the actual polymerase proteins, diamonds transcription regulators and circles the DSIF transcription elongation factor complex. Proteins not previously reported to be involved in transcription are indicated by triangles. Interactions between predicted subcomplexes are dashed.
In the MST and extended MST network, the five proteins contained in all three polymerases are not directly connected to the other polymerase III proteins although they are subunits of this complex. If we relax the criterion for extending the MSTs (α = 0.99), the interaction between RPB10 and RPC40, which was reported previously (Flores et al., 1999), is added to the scaffold (see http://www.bio.ifi.lmu.de/Complexes/Substructures/). At first glance, this suggests that the interactions of the common proteins to polymerase III are mediated via this interaction. However, if we look at the crystal structure of polymerase II and the model for polymerase III (Cramer et al., 2008), we find that none of the common proteins are actually in physical contact in the complexes (possibly with the exception of RPB10 and RPC10).
Going back to the original purification experiments, we find that of the seven interactions predicted between the common proteins, six interactions are bait–prey interactions which have been found to be very reliable (Bader et al., 2004) and three of those are identified in both directions (bait–bait interactions). This indicates that the association between these proteins is very strong. Since they do not appear to physically interact, this is probably a consequence of the fact that they are contained together in three different complexes. This close association of the five proteins can be identified reliably from the extended MST network.
4 DISCUSSION
We presented an approach for predicting the topology of protein complexes, i.e. the scaffold of direct interactions which spans the complex. First, our method calculates the union of all MSTs in the interaction score network for a protein complex. In a subsequent step, this network is iteratively extended by interactions which cannot be explained by a path of alternative indirect interactions. The MST approach is applicable to all weighted interaction networks and in particular to interaction scores calculated from affinity purification assays with any of the recently published scoring methods. Confidence scores which are required for extending the MSTs in our algorithm, can be obtained by scaling any type of scores to [0, 1] or using our Bootstrap approach implemented in the ProCope software package (Krumsiek et al., 2008 available at http://www.bio.ifi.lmu.de/Complexes/ProCope/) to calculate scores from affinity purification experiments.
Predictive performance of subnetworks calculated from Bootstrap confidence scores was evaluated on experimentally determined physical interactions from Y2H experiments and domain–domain interactions from 3D structures. We could show that, despite measurement errors, these networks can provide an accurate ranking of the performance of different prediction algorithms. Furthermore, the accuracy of reference interactions within complexes is significantly higher than the overall accuracy of the Y2H system. The evaluation of our method on these reference networks showed a significantly higher predictive accuracy than for the baseline classifier and the method by Bernard et al. (2007). Thus, physical interactions can be identified with high accuracy from the purification results alone. Since < 50% of the complexes in the predicted complex set contain at least one Y2H interaction, and only 7% of the complexes are actually non-trivially connected (i.e. they are connected and contain more than two proteins) in the Y2H network, many of the direct interactions within complexes have not been identified yet. Here, the interactions predicted by our approach but not found in the Y2H network are promising starting points for experimental validation.
Protein complexes are not simply disordered clumps of proteins but they have an internal substructure and a well-defined spatial arrangement in which not all proteins interact physically. Our results show that the similarity of subcomponent annotations of two proteins is negatively correlated to their distance in the MST and extended MST network. As a consequence, these networks reflect the modular substructure of the corresponding complexes. Although the same negative correlation was also observed for the baseline prediction approaches, distances in these networks are very short and, thus, do not allow for a reasonable resolution of the complex structure.
The extended MST approach predicts a globular structure for the majority of small complexes, while sparser networks with more intricate structures are predicted for larger complexes. The corresponding subcomplexes can then be identified in the extended MST network using a straightforward partitioning approach. A detailed analysis of the 21 largest complexes showed that in this way known subcomplexes can be recovered and complexes can be determined which have been clustered together due to several shared proteins. These shared proteins can be easily identified as they connect the different subcomplexes and associate strongly with them. A further analysis of the subcomplexes we predicted, but which have not been described in the literature yet may lead to new insights into the structure and function of these complexes.
We illustrated this approach on the complex of DNA-directed RNA polymerases. While the substructure of the complex with three different RNA polymerases can only be partly observed in the baseline predictions, it is clearly evident in the network predicted with our approach and the subcomplexes can be easily identified. By relaxing the conditions for extending the MSTs slightly, the substructure of the complex can be further emphasized and important interactions can be identified. A further comparison of the predictions and the original purification experiments to the 3D structure revealed limitations of the TAP system in distinguishing between indirect interactions and the actual physical ones.
The approach we developed can be easily extended to include additional information in the form of known physical interactions or information from crystal structures on which proteins are too far apart to be physically interacting. In this way, interactions may be either enforced or forbidden when extending the MSTs and alternative indirect paths can either be created or removed.
5 CONCLUSIONS
In this article, we presented an approach for post-processing protein complex predictions to allow for a more detailed analysis and comparison of complexes predicted from affinity purification results. Based on MSTs, we infer physical interactions to identify and visualize the substructure of protein complexes in an intuitive way. We showed that physical interactions are enriched within the predicted networks and that the scaffold network reflects the subcomponent structure. Furthermore, individual subcomplexes can be identified from the scaffold network with a straightforward partitioning approach. This shows that the complex topology can be inferred from purification results alone despite the experimental limitations of purification assays in distinguishing the actual physical interactions. Accordingly, the algorithm presented here supports the in-depth analysis of the predicted protein complexes beyond the individual complex subunits.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Aloy P, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader JS, et al. Gaining confidence in high-throughput protein interaction networks. Nat. Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, et al. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Berglund A-C, et al. InParanoid 6: eukaryotic ortholog clusters with in paralogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, et al. Proceedings of the 11th Annual International Conference on Research in Computational Molecular Biology, RECOMB 2007. Oakland, CA, USA: 2007. Reconstructing the topology of protein complexes; pp. 32–46. April 21-25. [Google Scholar]
- Breitkreutz B-J, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JD. “Minimax length links” of a dissimilarity matrix and minimum spanning trees. Psychometrika. 1995;60:371–374. [Google Scholar]
- Collins SR, et al. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Cormen TH, et al. Introduction to Algorithms. 2. Cambridge, MA, Boston, MA: MIT Press, McGraw-Hill Book Company; 2000. [Google Scholar]
- Cramer P, et al. Structure of eukaryotic RNA polymerases. Ann. Rev. Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- Flores A, et al. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel CC, et al. Proceedings of the 12th Annual International Conference on Research in Computational Molecular Biology, RECOMB 2008. Singapore: 2008. Bootstrapping the interactome: unsupervised identification of protein complexes in yeast; pp. 3–16. March 30–April 2. [DOI] [PubMed] [Google Scholar]
- Gavin A-C, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Hart GT, et al. A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinformatics. 2007;8:236. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollunder J, et al. DASS: efficient discovery and p-value calculation of substructures in unordered data. Bioinformatics. 2007;23:77–83. doi: 10.1093/bioinformatics/btl511. [DOI] [PubMed] [Google Scholar]
- Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Krumsiek J, et al. ProCope–protein complex prediction and evaluation. Bioinformatics. 2008;24:2115–2116. doi: 10.1093/bioinformatics/btn376. [DOI] [PubMed] [Google Scholar]
- Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant MG, et al. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S, et al. Identifying functional modules in the physical interactome of Saccharomyces cerevisiae. Proteomics. 2007;7:944–960. doi: 10.1002/pmic.200600636. [DOI] [PubMed] [Google Scholar]
- Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rual J-F, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Scholtens D, et al. Local modeling of global interactome networks. Bioinformatics. 2005;21:3548–3557. doi: 10.1093/bioinformatics/bti567. [DOI] [PubMed] [Google Scholar]
- Stein A, et al. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.