Summary
LIM transcription factors bind to nuclear LIM interactor (Ldb/NLI/Clim) in specific ratios to form higher-order complexes that regulate gene expression. Here we examined how the dosage of LIM homeodomain proteins Isl1 and Isl2 and LIM-only protein Lmo4 influences the assembly and function of complexes involved in the generation of spinal motor neurons (MNs) and V2a interneurons (INs). Reducing the levels of Islet proteins using a graded series of mutations favored V2a IN differentiation at the expense of MN formation. Although LIM-only proteins (LMOs) are predicted to antagonize the function of Islet proteins, we found that the presence or absence of Lmo4 had little influence on MN or V2a IN specification. We did find, however, that the loss of MNs resulting from reduced Islet levels was rescued by eliminating Lmo4, unmasking a functional interaction between these proteins. Our findings demonstrate that MN and V2a IN fates are specified by distinct complexes that are sensitive to the relative stoichiometries of the constituent factors and we present a model to explain how LIM domain proteins modulate these complexes and, thereby, this binary-cell-fate decision.
Keywords: LIM transcription factor; Motor neuron; V2a interneuron; Cell-fate specification; Ldb (NLI, CLIM, Chip); Lmo4; Isl1; Isl2; Lhx3; Mouse; Chick
INTRODUCTION
Neuronal diversity in the developing central nervous system (CNS) depends on combinatorial transcription codes created by multiple transcription factors: however, our understanding of the nature of interactions among transcription factors and their in vivo relevance is still unclear (Jessell, 2000). LIM homeodomain factors represent a prototypical example of combinatorial regulation within the neural tube. Biochemical and genetic studies have shown that LIM proteins interact to generate heteromeric complexes, rather than each component functioning independently to control cell identity by additively regulating subsets of target genes (Jurata et al., 1998; Thaler et al., 2002; Thor et al., 1999). Although the protein interactions that mediate the formation of these higher-order transcription complexes have begun to be defined, the principles that govern the assembly, function and interrelationships between related LIM complexes have not been well characterized in mammals.
Motoneurons (MNs) and V2a interneurons (INs) are two adjacent populations located in the ventral spinal cord that exhibit a simple `LIM code': Isl1 and Lhx3 for MNs and Lhx3 alone for V2a INs (Thaler et al., 2002). V2a INs arise from p2 progenitors and extend axons ipsilaterally over multiple segments to control proper locomotion (Kimura et al., 2006; Lundfald et al., 2007). MNs, which arise from pMN cells ventral to p2 cells, are cholinergic cells that extend axons into the periphery and directly control muscle contractions. Indeed, MNs express two types of Islet proteins, Isl1 and Isl2, with similar but temporarily distinct expression patterns, implying the possible involvement of Isl2 in the LIM code together with Isl1 and Lhx3. Isl1 is expressed in all MNs during the time when MNs exit the cell cycle, and Isl2 subsequently appears in all MNs at least transiently (Ericson et al., 1992; Thaler et al., 2004; Tsuchida et al., 1994). Studies in mice demonstrated that Isl1 and Isl2 are essential for MN development. Elimination of Isl1 in Isl1 null mice triggered massive cell death in the ventral spinal cord, including MNs and INs, whereas MNs in Isl2 null mice survived but later exhibited defects in subsets of MNs (Pfaff et al., 1996; Thaler et al., 2004). Conversely, knockdown of both Isl1 and Isl2 in zebrafish resulted in cell-fate conversion without cell death (Hutchinson and Eisen, 2006). These studies indicate that LIM homeodomain factors Isl1 and Isl2 may serve redundant and/or distinct physiological roles during MN development.
Highlights for the assembly of LIM complex are protein-protein interactions between the LIM domain and the nuclear LIM interactor (Ldb/NLI/Clim) (Agulnick et al., 1996; Bach et al., 1997; Dawid et al., 1998; Jurata et al., 1996). NLI contains an N-terminal self-binding domain, which leads to the formation of NLI dimers that recruit multiple LIM factors into a single complex. The existence of LIM factor complex has been indicated by gain-of-function studies: a hexameric Isl1:Lhx3:NLI complex drives MN generation, whereas a tetrameric Lhx3:NLI complex drives V2a IN formation (Thaler et al., 2002). This raises two issues to be resolved to ensure proper assembly of LIM factor complex. First, NLI-based LIM complex requires a precise stoichiometric relationship between the constituent proteins within LIM complexes. In the context of MN-V2a generation, appropriate levels of Islet protein within the complex are likely to be a crucial determinant between MN and V2a IN. Therefore, identifying the existence and necessity of such complexes in vivo, as well as the contribution of Islet affecting the stoichiometry of the complex and its physiological consequences, need to be addressed. Second, assembly of LIM complexes is expected to be transient and dynamic, especially when LIM factors just appear during neural development. Thus, negative regulatory mechanisms might be necessary to deplete incomplete complexes when the proper ratio of LIM proteins is not yet established. Potential candidates for this negative regulation include LIM-only (LMO) proteins that lack DNA-binding domains (Bach et al., 1997; Sugihara et al., 1998; Visvader et al., 1997; Wadman et al., 1997). The binding property and minimal functional motif suggested that LMO may interfere with the formation of LIM transcription factor complexes (Lee et al., 2008; Milan et al., 1998; Zeng et al., 1998). Here we provide a new role for LMO proteins enriching stable MN-promoting complexes while inhibiting weak unstable complex formation.
Taken together, these observations lead to several predictions. First, the assembly of functional complexes for MN and V2a IN differentiation are based on precise stoichiometric relationships and therefore are highly dependent on maintaining the appropriate levels of the individual components. Second, the LIM homeodomain complexes implicated in V2a IN and MN specification are expected to be negatively regulated by LMOs. To examine the functional consequences of these predictions we genetically altered the dosage of LIM homeodomain proteins Isl1 and Isl2, and LIM-only protein Lmo4, and monitored the differentiation of MNs and V2a INs. Reduction of Islet protein concentration favored V2a IN differentiation at the expense of MN formation. This fate conversion could be shifted back toward MN generation by eliminating Lmo4. Our findings reveal the interrelationships between the factors for MN and V2a IN fates, and further suggest that the relative stoichiometries of the factors and the modulatory effects of Lmo4 are important determinants that influence the assembly of functional LIM complexes.
MATERIALS AND METHODS
Mouse lines
Generation of Isl1 null, Isl2 null, Lmo4 null, Isl1 hypo, Isl1fl/fl mice and transgenic mouse line Hb9::gfp have been reported previously (Lee et al., 2005; Pfaff et al., 1996; Sun et al., 2008; Thaler et al., 2004). The Isl1 hypo allele contains loxP sites encompassing exon4 of the mouse Isl1 gene with the neomycin-resistance gene. The neomycin-resistance gene was removed by crossing heterozygotes of Isl1 hypo mice to an FLPeR delete strain to generate Isl1fl/fl mice (Sun et al., 2008). Neural-deletion of Isl1 was achieved by using timed matings between heterozygous Isl1ko/+; Nestin::Cre+/- (Jackson Laboratory) mice and homozygous Isl1fl/fl animals. Embryos with the genotype: Isl1fl/ko; Nestin::Cre+/- were analyzed.
DNA constructs
Rat Isl1; mouse Isl2, Lhx3, NLI, Lmo4; DD-Isl1-Lhx3 are from Thaler et al. (Thaler et al., 2002) and Lee et al. (Lee et al., 2003). ΔNLI (aa 200-375) lacks the dimerization domain (DD).
TUNEL assays
TUNEL assays were performed using the ApopTag Fluorescein In Situ Apoptosis Detection Kit (Chemicon). Controls included wild-type sections treated with DNAse I and Isl1 mutant mice sections without TdT enzyme.
Immunohistochemistry
Embryos were obtained at embryonic day (E) 10.5-12.5 and fixed in 4% paraformaldehyde and cryosectioned for immunohistochemistry. Antibodies used in this study were as follows: guinea pig anti-Isl1 (Thaler et al., 2004), guinea pig anti-Isl2, rabbit anti-Isl1/2 (Ericson et al., 1992), rabbit and guinea pig anti-Hb9 (Thaler et al., 1999), rabbit anti-Lhx3 (Sharma et al., 1998), rabbit anti-NLI (Jurata et al., 1996), guinea pig anti-Chx10 (Thaler et al., 2002), rabbit anti-GFP (Invitrogen), monoclonal anti-HA (Babco), monoclonal anti-MNR2 (Developmental Studies Hybridoma Bank), rabbit anti-Nkx6-1 (Beta Cell Biology Consortium), rabbit anti-Irx3 (Dr T. M. Jessell), guinea pig anti-Lmo4 (Lee et al., 2005) antibodies. Fluorophore-conjugated species-specific secondary antibodies were used as recommended (Jackson Labs and Invitrogen).
In situ hybridization
Embryos were fixed in 4% paraformaldehyde, mounted and cryosectioned for in situ hybridization. Transverse sections were hybridized with digoxigenin-labeled probes specific for mouse Lmo4 amplified from mouse embryonic cDNA using the Advantage cDNA PCR Kit (Clontech) and cloned into pCRII vector using the TOPO cloning Kit (Invitrogen).
Chick in ovo electroporation
Chick eggs (Charles River and McIntyre Farms) were incubated in a humidified chamber, and DNA constructs were injected into the lumens of HH stage 10 to 12 chick embryonic spinal cords. Electroporation was performed using a square wave electroporator (BTX). Incubated chicks were harvested and analyzed at HH stage 20 to 25.
Cell culture and transient transfection experiment
Luciferase assays were performed by transfecting 293 cells with Hb9 motoneuron enhancer constructs together with CMV-β-galactosidase. After 48 hours, cell lysates were prepared to measure luciferase activity. The values were normalized to the β-galactosidase activity. For motoneuron induction assays, P19 mouse embryonic cells were cultured in α-minimum essential medium with 10% bovine fetal serum and retinoic acid. After transfection, cells were grown for 3 days and immunostained with anti-MNR2 antibody for analysis.
Protein interactions
293T cells were transfected with DNA constructs encoding FLAG-NLI, FLAG-ΔNLI, HA-Isl1 or HA-Lmo4 using Fugene6-HD (Roche). After 48 hours, cell lysates were prepared and incubated with FLAG-M2-conjugated agarose beads (Sigma) at 4°C, overnight. The immune complexes are visualized by SDS-PAGE and immunoblotting using anti-HA-HRP-conjugated antibody (Roche).
RESULTS
Transcription factors that specify MN and V2a IN differentiation
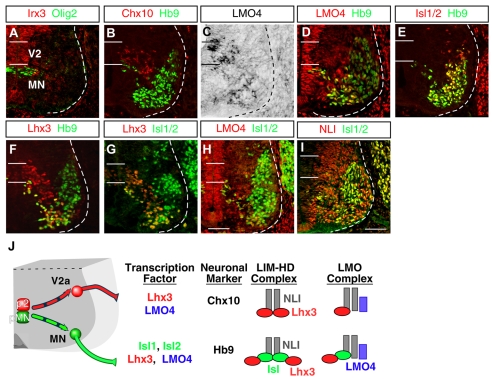
We first examined the expression of LIM factors present in the ventral spinal cord. The border between pMN and p2 domains was determined by a pMN marker Olig2 and Irx3 labeling p2 and above (Fig. 1A). Postmitotic V2a INs and MNs were traced by expressions of Chx10 (Vsx2 - Mouse Genome Informatics) and Hb9 (Mnx1 - Mouse Genome Informatics) (Fig. 1B). We found that Lmo4 is expressed in the region where V2a INs and MNs reside (Lee et al., 2005) (Fig. 1C,D,H). To carefully examine coexistence among LIM factors, we traced expression of these factors with various combinations (Fig. 1D-H) (Kenny et al., 1998; Pfaff et al., 1996). Most MNs co-expressed Isl1, Isl2 and Hb9 and subsets of MNs expressed Lhx3 and Lmo4. We also confirmed that NLI is widely expressed in the neural tube including developing V2a INs and MNs (Thaler et al., 2002) (Fig. 1I; data not shown). Thus, it is predicted that an array of NLI-based complexes containing LIM homeodomain and LMO proteins form in V2a INs and MNs (Ericson et al., 1992; Jurata et al., 1996; Thaler et al., 2002; Tsuchida et al., 1994) (Fig. 1J).
Fig. 1.
Combinatorial expression of transcription factors in ventral spinal neurons. (A-I) Expression of Irx3, Olig2, Chx10, Hb9, Lmo4, Isl1/2, Lhx3 and NLI in E11.5 mouse cervical spinal cord. MNs and V2a INs each express unique combinations of these factors. Expression of Lmo4 is assessed by both in situ hybridization (C) and immunohistochemistry (D,H). The locations of ventricular progenitor cells for MNs (pMN) and V2a INs (p2) are marked by two horizontal lines. Dashed lines indicate the boundary of the spinal cord. (J) The schematic diagram shows that the pMN domain generates motoneurons (MN, green), whereas p2 cells give rise to V2a INs (red). A summary of the combinatorial expression profile of transcription factors is shown along with neuronal-subtype markers and predicted higher-order regulatory complexes based on known protein-protein interactions. Scale bars: in I, 50 μm for A-C,E,I; in H, 40 μm for D,F-H.
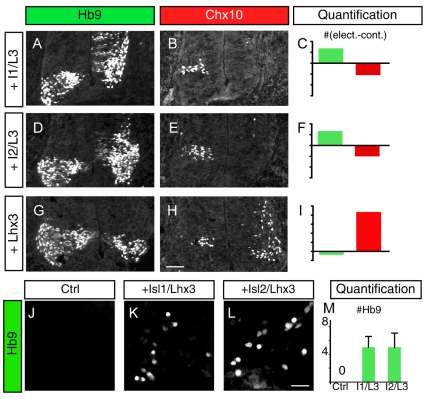
As Isl1 and Isl2 are highly similar proteins and are co-expressed during the early phases of motoneuron generation, we examined whether these proteins share similar activities, as found in zebrafish (Hutchinson and Eisen, 2006). Structure-function studies have shown that Isl1 binds to NLI and Lhx3, forming a hexameric DNA-binding complex (Fig. 1J) that promotes MN formation and suppresses V2a IN differentiation (Thaler et al., 2002). We electroporated Isl1 or Isl2 together with Lhx3 into chick neural tube, and monitored the number of MNs and V2a INs using Hb9 and Chx10 labeling. Because NLI is widely expressed, it was unnecessary to include expression constructs encoding this protein. We found that both Isl1 and Isl2 could generate MNs with similar efficiencies (Fig. 2A-F). Whereas Isl1+Lhx3 and Isl2+Lhx3 increased MN numbers, the ectopic expression of these transcription factors reduced the number of Chx10+ V2a INs. Similarly, both Isl1 and Isl2 in combination with Lhx3 induced Hb9, when transfected into P19 cells (Fig. 2J-M). Next we electroporated Lhx3 alone into the chick neural tube, which triggered the ectopic differentiation of V2a INs and suppressed endogenous MN production (Fig. 2G-I) (Thaler et al., 2002). Our findings indicate that, in the binary-cell-fate choices between MNs and V2a INs, Isl1 and Isl2 share the ability to drive MN differentiation, and the sum of Isl1 plus Isl2 probably defines the overall Islet-protein level available for the assembly of complexes.
Fig. 2.
Combinatorial regulation of MNs and V2a neuronal identity. (A,B) Misexpression of Isl1 and Lhx3 in the chick neural tube induces ectopic Hb9+ MNs at the expense of Chx10+ V2a INs. Chick MN nuclei are labeled using Mnr2 antibody, which detects both Mnr2 and Hb9. The electroporated side is on the right and the control side is on the left. (C) Differences in the number of MNs (green) and V2a INs (red) between the electroporated and control sides of the neural tube are plotted. For MNs each unit on the y-axis corresponds to 100 cells, and for V2a INs each unit is 10 cells. (D-F) Overexpression of Isl2 and Lhx3 causes an increase in MNs and a decrease in V2a INs, similar to Isl1 and Lhx3 overexpression. (G-I) Misexpression of Lhx3 triggers V2a IN development and inhibits MNs. (J-L) Transfection of Isl1+Lhx3 (K) or Isl2+Lhx3 (L) expression constructs into P19 cells induces Hb9. (M) Quantification of Hb9-expressing cells within the field of view (FOV). Each bar represents the average of at least five FOVs collected from two different experiments. Mean±s.e. is shown. Scale bars: in H, 50 μm for A,B,D,E,G,H; in L, 17 μm for J-L.
Characterization of Isl1 mutant mice
Mice heterozygous for a null mutation in Isl1 develop normally and produce viable adult animals. Despite the elimination of one copy of the Isl1 gene, Isl1 protein levels are comparable between heterozygous mutants and wild-type controls. Homozygous null mutants of Isl1 failed to develop beyond E9.5, probably because the cardiovascular system does not develop properly (Pfaff et al., 1996). Thus, null mutation of Isl1 has been difficult to use for characterizing the role of Isl1 in MN and V2a IN specification.
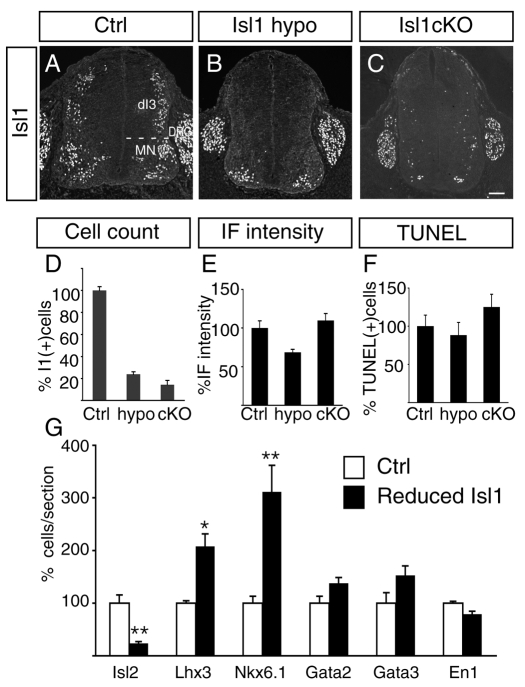
To circumvent this, we used two novel Isl1 mutations to attenuate gene expression: Isl1 hypomorph mice (Isl1 hypo) expressing low levels of Isl1 and Isl1 conditional mutant mice (Isl1 cKO) lacking Isl1 in neurons (Sun et al., 2008). Isl1 hypo mice contain a PGK-Neo cassette within an intron that reduces Isl1. For the Isl1 cKO, Isl1fl/fl mice were mated to Isl1ko/wt; Nestin::Cre animals (Betz et al., 1996; Pfaff et al., 1996; Sun et al., 2008). Embryos with the genotype Isl1fl/ko; Nestin::Cre (abbreviated Isl1 cKO below) were used for our analysis. We used Isl1 antibodies to monitor protein expression at cervical levels of E11.5 mutant embryos. Isl1 was detected in dorsal interneurons (dI3s), MNs and sensory neurons in the dorsal root ganglia (DRG) (Fig. 3A) (Ericson et al., 1992; Pfaff et al., 1996). In Isl1 hypo and cKO mutant embryos the number of Isl1+ cells in the cervical spinal cord was markedly reduced (Fig. 3A-E). The reduction of Isl1 was variable along the rostrocaudal axis of the spinal cord with the cervical level showing the highest degree of reduction. Isl1 was not detected in dI3 cells and the number of Isl+ MNs was reduced 76% in Isl1 hypo mutants and 86% in Isl1 cKO embryos (Fig. 3D). To estimate the Isl1 protein level in the remaining Isl1+ MNs, we quantified the fluorescence intensity of the immunolabeling from MN nuclei. In Isl1 hypo mutants, the labeling intensity was reduced ∼30% from controls, whereas the remaining Isl1+ cells in Isl1 cKO mice appeared to express normal levels of Isl1 (Fig. 3E). This difference is probably due to the nature of mutations. In Isl1 hypo mutants, protein expression from each Isl1 allele is attenuated and the residual Isl1 labeling could arise because feedback regulation compensates for this inefficiency with varying degrees of success. In Isl1 cKO, fewer cells expressed Isl1, although its expression level was comparable to the control. As Cre-mediated deletion of the floxed allele is an all-or-nothing event, the remnant Isl1+ cells in cKO mutants might be due to incomplete recombination in some cells.
Fig. 3.
Isl1 protein is reduced in Isl1 hypo and Isl1 cKO mutants. (A-C) Isl1 immunolabeling in E11.5 cervical spinal cord. In wild-type embryos (A), Isl1 is detected in MN nuclei, dI3s and sensory neurons in the DRG. In Isl1 hypo (B) and Isl1 cKO (C) mutants, Isl1 expression is significantly downregulated in MNs and absent in dI3 INs, but unchanged in DRG neurons. (D) Quantification of the number of Isl1+ cells per section in the E11.5 cervical spinal cord. Cell counts were restricted to the MN-containing region of the neural tube. (E) Measurement of Isl1 immunofluorescent intensity in E11.5 cervical MN nuclei. More than 20 cells were randomly selected. The average MN labeling intensity was normalized to the labeling of DRG neurons. (F) Quantification of TUNEL-positive cells in the ventral spinal cord. The amount of cell death in Isl1 hypo and Isl1 cKO mutants was comparable to controls. Each bar represents the average of at least eight sections from three embryos. Mean±s.e. is shown. (G) Analyses of ventral spinal neuron development in littermate controls (white bar) and Isl1 cKO mutants (black bar) assessed by marker expression. Asterisks indicate statistically significant differences compared with control (*P<0.05; **P<0.01, paired Student's t-test). Scale bar: in C, 100 μm for A-C.
The phenotype of the spinal cord in Isl1 hypo and cKO mutants differed significantly from Isl1 null mutants, which underwent massive cell death (Pfaff et al., 1996). The morphology was normal and minimal cell death occurred, similar to the level found in littermate controls (Fig. 3F). This suggests that low or even transient Isl1 in Isl1 mutant cells normally destined to become MNs is sufficient to spare them from the suicidal cell death found in Isl1 null mice. Next we examined whether p2 and pMN domains develop normally in Isl1 mutants. Judging from expressions of Irx3 and Olig2, it appears that both p2 and pMN domains were normally formed in Isl1 mutants (see Fig. S1 in the supplementary material). Nevertheless, the expression of markers that label specific neuronal subtypes was altered in Isl1 hypo and cKO mutants. Isl2 labeling in MNs was dramatically reduced, whereas Lhx3 and Nkx6-1 labeling was increased (Fig. 3G; see Fig. S1 in the supplementary material). As Isl2 is MN-specific whereas Lhx3 and Nkx6-1 are expressed in both MNs and V2a INs, our results suggest that cell-fate changes may have occurred between MNs and V2a INs in the Isl1 mutants.
Isl-protein levels influence the ratio of V2a INs to MNs
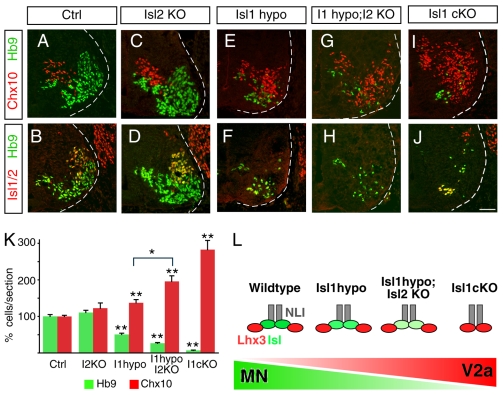
LIM homeodomain complexes are thought to form in precise stoichiometries: V2a IN-specifying complexes are tetramers with 2-Lhx3:2-NLI, and MN complexes are hexamers comprising 2-Islet: 2-Lhx3: 2-NLI (Fig. 1J) (Thaler et al., 2002). Thus, the level of Isl protein is predicted to influence the ratio of LIM complexes in MNs - as Isl levels decline, V2a IN complexes should become more prevalent (see Fig. 4L). To test the functional implications of this prediction in vivo, we assayed MN and V2a IN marker expression in Isl1 and Isl2 mutants. We found that the elimination of Isl2 alone had little influence on the overall numbers of Hb9+ MNs and Chx10+ V2a INs (Thaler et al., 2004) (Fig. 4C,D,K). Mutants homozygous for the Isl1 hypomorph allele displayed a reduction in MNs and a slight increase in V2a IN cells (Fig. 4E,F,K). Double labeling of Hb9 or Chx10 with Isl1 revealed that most MNs maintained Isl1 expression whereas all V2a INs lacked Isl1 in Isl1 hypo mice (see Fig. S1 in the supplementary material). Thus Isl1 mutant cells seem to express variable levels of Isl1, which determines cell fates between MNs and V2a INs.
Fig. 4.
The balance between MN and V2a IN development is sensitive to Isl protein levels. (A-J) Immunolabeling to detect Isl1/2+ and Hb9+ MNs, and Chx10+ V2a INs in E11.5 cervical spinal cords. Wild-type (Ctrl; A,B), Isl2 null (Isl2 KO; C,D), Isl1 hypo (E,F), Isl1 hypo; Isl2 KO (G,H) and Isl1 cKO (I,J) embryos were examined. (K) Quantification of Hb9+ MNs and Chx10+ V2a INs in transverse sections of E11.5 embryos. The loss of Isl labeling was found to correlate with an increase in V2a IN (Chx10) labeling. Each bar represents the average of eight sections collected from three different embryos. Mean±s.e. is shown. Asterisks indicate statistically significant differences compared with control or groups marked with bracket (*P<0.05; **P<0.01, paired Student's t-test). (L) Diagram depicting the hexameric 2-NLI: 2-Lhx3: 2-Isl1 transcription complex in wild-type and Isl mutant embryos. As Isl protein level drops, the formation of 2-NLI: 2-Lhx3 complexes for V2a IN development are likely to predominate. Scale bar: in J, 40 μm for A-J.
Next we intercrossed Isl1 hypo with Isl2 mutants to test whether further reduction of Islet levels influences MN-V2a IN specification. As a result, we observed a greater reduction of MN numbers and an increase in V2a IN cells (Fig. 4G,H,K). The elimination of Isl1 using the cKO mutation in combination with Nestin::Cre resulted in the most severe reduction in MN number and the greatest increase in V2a INs (Fig. 4I-K). Together, these results suggest that the extent of cell-fate conversion from MNs to V2a INs is determined by the levels of Isl1.
The conversion of MNs to V2a INs was further confirmed by crossing an Hb9::gfp transgene into the Isl1 hypo mice. Normally Hb9::gfp selectively marks MNs, which extend their axons to the periphery. In Isl1 hypo mice, ectopic Chx10+ V2a INs were also labeled by GFP, implying the conversion of cell fates from MNs to V2a INs (see Fig. S2 in the supplementary material). Furthermore, some GFP-labeled cell processes in Isl1 hypo mice extended along the rostrocaudal axis of the neural tube, mimicking the ipsilateral V2a IN trajectory over multiple segments of the neural tube. In addition, some GFP cells inappropriately crossed the midline as an indication of axon projection defects in Isl1 mutant cells. Thus, presumptive MNs labeled with the Hb9::gfp transgene displayed both molecular and cellular features similar to INs in Isl1 mutant embryos. These genetic studies provide in vivo evidence that the LIM complexes for V2a IN and MN specification are developmentally related and sensitive to the levels of Isl protein (Fig. 4L).
Lmo4 negatively regulates LIM homeodomain complexes for MN and V2a IN generation
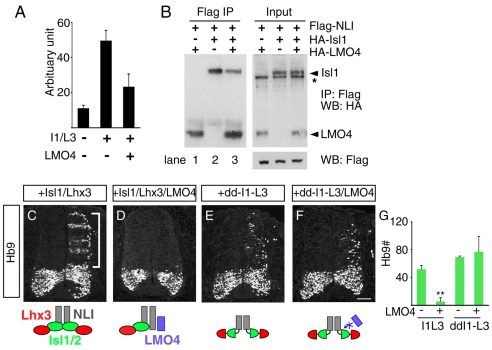
A potential function mediated by LIM-only factors such as Lmo4 is to interfere with the assembly and activity of LIM homeodomain complexes (see Fig. 1J). Consistent with this, we found that transcriptional activation of the motoneuron enhancer element M250 in the Hb9 gene by Isl1 plus Lhx3 was attenuated by Lmo4 (Fig. 5A) (Lee et al., 2004). If Lmo4 is a negative regulator of 2-NLI:2-Isl1:2-Lhx3 complexes in MNs, a simple prediction is that Lmo4 binding to the LIM interaction domain of NLI should be able to compete with Isl1 binding at this site. To test this we expressed NLI, Isl1 and Lmo4 in 293T cells and used immunoprecipitation to pull down protein complexes (Fig. 5B). The ratio of each construct is optimized to have Isl1 in excess to NLI, to ensure unbound NLI is not available to interact with Lmo4. In pairwise combinations, NLI was able to immunoprecipitate Lmo4 and Isl1 as expected (Deane et al., 2003; Jurata et al., 1998) (Fig. 5B, lanes 1 and 2). When NLI, Isl1 and Lmo4 were co-expressed, the presence of Lmo4 reduced the amount of Isl1 bound to NLI (Fig. 5B, compare lanes 2 and 3). Taken together, these results show that Lmo4 and Isl1 compete for NLI binding and support the notion that Lmo4 is a negative regulator of LIM homeodomain complexes.
Fig. 5.
Lmo4 competes with Isl1 for MN and V2a IN generation. (A) 293T cells were transiently transfected with the Hb9-derived M250 motoneuron enhancer linked to luciferase together with expression constructs encoding Isl1, Lhx3 and Lmo4. Enhancer activity is stimulated by Isl1+Lhx3. The addition of Lmo4 antagonizes the activity of Isl1+Lhx3. (B) In vitro co-immunoprecipitation assays using FLAG- and HA-epitope-tagged proteins. FLAG-NLI, HA-Isl1 and HA-Lmo4 were expressed in 293T cells as indicated and lysates were immunoprecipitated with anti-FLAG antibody-conjugated agarose beads. Co-precipitated HA-tagged proteins were identified by western blotting with anti-HA antibody. NLI interacts with Lmo4 and Isl1 (lanes 1 and 2). Isl1 binding to NLI is reduced when Lmo4 is present (lane 3). The asterisk marks a nonspecific band. (C-F)Lmo4 suppresses ectopic MN formation in chick embryos following electroporation with Isl1 and Lhx3 (C,D). The MN-inducing activity of chimera dd-I1-L3 is not blocked by Lmo4 (E,F). Diagrams depicting the transcription complexes are below. (G) Quantification of Hb9+ MNs in HH stage 20 embryos after electroporation. Asterisks indicate statistically significant differences compared with control (**P<0.005, paired Student's t-test). Scale bar: in F, 50 μm for C-F.
Given these considerations, we designed assays of cell differentiation to examine the function of Lmo4. The induction of ectopic MNs by misexpression of Isl1+Lhx3 in the chick neural tube was completely blocked when Lmo4 was included in the electroporation (Fig. 5C,D). To rule out the possibility that the negative effects of Lmo4 on MN generation were indirect, we examined the activity of a chimeric LIM homeodomain construct made by linking the homeodomains of Isl1 and Lhx3 to the dimerization domain from NLI (dd-Isl1-Lhx3). This construct is capable of inducing ectopic MNs similar to the native proteins (Fig. 5E) (Thaler et al., 2002), but Lmo4 is unable to displace Isl1 from NLI. As expected, Lmo4 failed to inhibit MN generation by dd-Isl1-Lhx3 (Fig. 5F). The ability of Lmo4 to inhibit MN generation by Isl1+Lhx3 but not dd-Isl1-Lhx3 (Fig. 5G) is further evidence that Lmo4 antagonizes the function of LIM homeodomain factors by competing for NLI binding. We also observed the similar results in the context of Lhx3 complex for V2a INs, supporting the LIM-specific competitive mechanisms of LMOs (see Fig. S3 in the supplementary material). Taken together, our findings indicate that Lmo4 functions as a negative regulator of transcriptional complexes for MN differentiation.
LMO4 attenuates MN differentiation
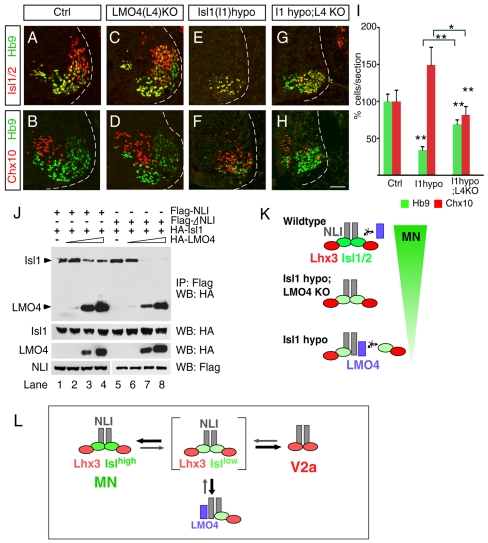
Although biochemical and misexpression experiments indicated that Lmo4 could antagonize the function of LIM complexes, it is unclear whether endogenous Lmo4 normally prevents MN and/or V2a IN generation, because Lmo4 overexpression in the chick neural tube did not obviously block the endogenous cell populations from differentiating (Fig. 5D). Therefore we examined neuronal specification in Lmo4 mutant mouse embryos (Lee et al., 2005). Previously, it was reported that Lmo4 mutant embryos display excencephaly (open brain) phenotypes without obvious developmental defects in the spinal cord (Hahm et al., 2004; Lee et al., 2005; Tse et al., 2004). We found that the normal numbers of Hb9+ MNs and Chx10+ V2a INs were present in Lmo4 mutant embryos at E11.5 (Fig. 6A-D). To further examine a possible role for Lmo4 in modulating the function of LIM homeodomain factors, we intercrossed Lmo4 heterozygotes and Isl1 hypo mutant mice. Strikingly, the MN-to-V2a IN conversion observed in Isl1 hypo mutants was shifted back toward a more normal distribution of cell types when Lmo4 function was eliminated (Fig. 6G-I). These findings provide genetic evidence that Lmo4 negatively regulates LIM homeodomain complexes for MN generation in vivo, but this activity seems to be restricted to cells that lack optimal levels of Isl1.
Fig. 6.
Lmo4 negatively regulates MN generation when Isl levels are reduced. (A-H) Immunocytochemical detection of Isl1/2, Hb9 and Chx10 in E11.5 cervical wild-type (A,B), Lmo4 null (C,D), Isl1 hypo (E,F) and Isl1 hypo; Lmo4 null (G,H) embryos. (I) Quantification of Hb9 and Chx10 expressions. The MN-to-V2a conversion in Isl1 hypo mutants is restored to a more normal cellular distribution in Isl1 Lmo4 double-mutant embryos. Each bar represents the average of at least eight sections from three embryos. Mean±s.e. is shown. Asterisks indicate statistically significant differences compared with control or groups marked with bracket (*P<0.05; **P<0.005, paired Student's t-test). (J) Co-immunoprecipitation assays to assess protein-protein interactions between FLAG-NLI, FLAG-ΔNLI, HA-Isl1 and HA-Lmo4. Lmo4 competes with Isl1 for binding to NLI in a dose-dependent manner (lanes 1-4). Isl1 is more easily displaced from NLI by Lmo4 when NLI-dimerization is prevented (ΔNLI; lanes 5-8). (K) Diagram depicting the putative Isl and Lmo4 complexes in each genetic background. (L) Models of transcription complexes and protein interactions for MN and V2a IN specification. NLI dimers are predicted to serve as the scaffold for building multimeric transcriptional complexes in MN and V2a IN development. NLI-Lmo4 interactions are likely to inhibit the function of complexes for MN specification when the LIM complex is suboptimal (brackets). Scale bar: in H, 40 μm for A-H.
Our observation that the antagonistic activity of Lmo4 was revealed only when the Isl1 level was low prompted us to reason that accumulation of incomplete LIM complex might happen due to the unavailability of Isl1 in Isl1 mutant mice and that Lmo4 might compete more efficiently with these intermediate complexes. To better understand the mechanistic basis for Lmo4-mediated antagonism of LIM complexes, we decided to compare the ability of Lmo4 to displace Isl1 from NLI dimers (when LIM complex is fully assembled) versus NLI monomers (when LIM complex is partial). For this, we used a dimerization domain deletion mutant of NLI, which only generates an Isl1-NLI monomer, a similar strategy proven to disrupt the LIM complex in Drosophila (Milan and Cohen, 1999; van Meyel et al., 1999). We expressed wild-type NLI or a dimerization domain deletion mutant of NLI with Isl1 and increasing levels of Lmo4 in 293T cells and performed immunoprecipitation to pull down protein complexes (Fig. 6J) (Jurata et al., 1996). Protein-interaction and crystal-structure studies have identified the LIM-interaction sites within the C-terminus of NLI, consistent with the finding that ΔNLI binds with similar efficiency to Isl1 as native NLI capable of self-dimerizing (Fig. 6J, lanes 1 and 5) (Deane et al., 2003; Jurata et al., 1996). Again, the assays were performed with Isl1 in excess of NLI, to ensure free NLI was not available for interactions with Lmo4. We found that the competition between Lmo4 and Isl1 for NLI binding was altered when higher-order complexes were prevented from assembling. We found that Lmo4 was much more effective at displacing Isl1 from NLI monomers (ΔNLI) than NLI dimers (Fig. 6J, compare lane 3 with 7). Taken together, our results suggest that Lmo4 is a negative regulator of LIM homeodomain complexes for MNs and that Lmo4 functions by displacing LIM homeodomain factors from NLI. Furthermore, the competitive interactions between LMO and LIM homeodomain factors appear to be influenced by the overall composition of the higher-order complexes, which are dependent upon expressing the appropriate ratios of stoichiometrically interacting proteins (Fig. 6K).
DISCUSSION
A variety of genetic studies have demonstrated that combinatorial transcription factor codes are employed to specify cell identity in the developing neural tube (Jessell, 2000). In some cases, such as with Hox, Ets and LIM factors, members of multi-gene families represent major components of the regulatory codes. However, even these gene families often synergize with unrelated factors to regulate gene expression in developmentally related but functionally distinct neuronal subtypes. Here we sought to understand the mechanistic underpinnings by which unique combinations of regulatory factors acquire specific activities for neuronal subtype specification by examining the function of multimeric LIM complexes. Our studies indicate that assembly of NLI-based transcription factor complexes are regulated at multiple levels during the development of MNs and V2a INs. We show that the Islet protein levels as well as the presence of Lmo4 contribute to the relative stoichiometries of the constituent components of LIM complexes in vivo. Below we discuss how MN and V2a IN development is regulated and speculate how Lmo4 might help to make cell-fate decisions more decisive when LIM protein concentrations are suboptimal.
Regulation of MN and V2a IN identity
Graded Shh signaling across the ventral neural tube induces different patterns of gene expression in the precursor cells for motoneurons (pMN cells) and V2a interneurons (pV2 cells) (Jessell, 2000). Both pMN and pV2 cells express NLI, Lhx3 and Lmo4; however, pMN progenitors express Olig2 and Ngn2 (Neurog2 - Mouse Genome Informatics), whereas pV2 cells express Irx3, Foxn4 and Mash1 (Ascl1 - Mouse Genome Informatics). Despite these `early' differences in gene expression patterns, the development of V2a INs and MNs is closely linked, and a variety of mutations leads to cell-fate inter-conversions among these neurons. For example, Olig2 and Notch1 mutants lack MNs but have increased V2a IN numbers, and Hb9 mutants exhibit cells with mixed identities expressing both MN and V2a IN markers (Muroyama et al., 2005; Thaler et al., 1999; Yang et al., 2006).
These initial diversities in gene expression are soon followed by the appearance of Isl1, the early postmitotic LIM homeodomain factor distinctively expressed in potential MNs around the cell cycle exit (Ericson et al., 1992). Likewise, we demonstrated that prospective MNs within Isl1 cKO and hypo mutant mice convert their identity to become V2a INs, supporting the conclusion that Isl1 plays a pivotal and ultimate role in segregating MN and V2a IN identities. In addition to our study to support the role of Isl1 in motoneuron identity, elimination of Isl1 in other cell types or tissues has revealed the broader role of Isl1 (Elshatory et al., 2007; Pan et al., 2008; Sun et al., 2008). As Isl1 continues to exist in postmitotic neurons with some restriction, it is most likely that Isl1 is necessary for subsequent differentiation processes as well.
Our observation is different from MN defects shown in Hb9 mutants with incomplete fate conversion resulting in mixed MN and V2a IN-like traits (Arber et al., 1999; Thaler et al., 1999). How do the Isl proteins act differently from Hb9 in promoting MN identity? Previous studies demonstrated that an Isl1-mediated mechanism occurs through direct interactions with Lhx3, leading to the formation of an MN-inducing hexameric complex at the expense of a V2a IN-inducing tetrameric complex (Fig. 6L) (Thaler et al., 2002). By contrast, Hb9, induced by Isl1, represses V2a IN genes such as Chx10 for the maintenance of MN identity in postmitotic MNs (Lee et al., 2008). Thus the timing of the appearance and the degree of fate conversion when it is absent indicate that Isl1 and Hb9 play a sequential role for MN development.
Stoichiometric regulation of LIM complexes by Islet protein level
Protein-protein interactions mediated by the LIM domains of LIM transcription factors form the basis for the assembly of higher-order regulatory complexes. The individual factors within these LIM complexes are bound to one another in specific stoichiometries, suggesting that the overall levels as well as the relative ratios of the binding partners dictate whether complete and functional complexes assemble. The strongest evidence for this stoichiometric model is from genetic studies in Drosophila, which have shown that the level of apterous (LIM homeodomain factor) and binding partner Chip (NLI ortholog) control dorsal wing development (Fernandez-Funez et al., 1998; Milan and Cohen, 1999; Milan et al., 1998; van Meyel et al., 1999). Recent studies using mammalian-derived cell lines have shown that single-stranded DNA-binding proteins (SSDP2/3) modulate interactions between the E3 ubiquitin ligase RLIM and NLI, and thereby contribute to the post-translational regulation of NLI levels (Gungor et al., 2007; Xu et al., 2007). In this report we tested whether the MN-V2a IN binary-cell-fate decision was influenced by LIM protein levels that comprise separate but related NLI-based transcription factor complexes (see Fig. 6L).
The LIM homeodomain factor Isl1 is expressed after pMN cells complete their final S-phase and become postmitotic motoneurons (Ericson et al., 1992; Thaler et al., 2004). Isl2, a paralog of Isl1, is expressed after Isl1, and therefore does not compensate for the loss of Isl1 in null mutants, but the two proteins are 73% identical and share the potential to perform the same functions. We found that both Isl1 and Isl2 cooperate with Lhx3 to specify MN identity. Likewise, studies in zebrafish have found that Isl1 and Isl2 have slightly different expression patterns but share similar activities (Hutchinson and Eisen, 2006). By combining the Isl1 hypo and Isl2 null mutations we found that the degree of MN-to-V2a IN inter-conversion was enhanced. This suggests that Isl1 and Isl2 share similar functions and that together the level of these two proteins influences which type of neuron is specified: Isllow favors the formation of V2a IN complexes, whereas Islhigh favors MN complexes (Fig. 6L).
Lmo4: a modulator of LIM homeodomain complexes
The strongest evidence that Lmo4 influences the activity of transcriptional complexes for cell-fate specification in the neural tube comes from Isl1 hypo mutants, in which the generation of motoneurons was restored without Lmo4. The genetic interaction as well as physical binding properties that we uncovered between Lmo4 and Isl1 suggest that Lmo4 is a negative regulator of MN generation. A similar action of Lmo4 has been suggested to inhibit Lhx3-driven complex within MNs in a later period of MN differentiation (Lee et al., 2008). Interestingly, Lmo4 plays a different role in V2b INs, another population generated from p2 progenitors (Joshi et al., 2009). In this case, Lmo4 acts as a linker to recruit other transcription factors lacking the LIM domain. Thus, it seems that Lmo4 plays a dual role to control assembly of transcription factor complexes by competing with LIM factors or recruiting non-LIM-domain-based transcription factors in the developing spinal cord.
Several observations suggest that the function of Lmo4 in MNs may be more subtle than simply acting as inhibitors by displacing LIM homeodomain factors from NLI. First, Lmo4 null mutants did not exhibit obvious defects in MN development (E9.5-13.5) unless Lmo4 null embryos were under extreme conditions in which Isl protein levels were greatly diminished. Second, overexpression of Lmo4 was not particularly effective at preventing endogenous MNs from developing. Third, the apparent effectiveness of Lmo4 to displace Isl1 from NLI was better with NLI-Isl1 dimers than 2-NLI: 2-Isl1 tetramers. This indicates that, under conditions in which NLI can dimerize with 2-Isl1 molecules, additional interactions may occur that further stabilize the complex, making it more resistant to Lmo4 competition.
Why might competitive interactions between Lmo4 and Isl1 be advantageous? During development neuroepithelial cells are confronted with the challenge of reading out small differences in graded signals such as Shh and ultimately making decisive fate decisions. Despite cross-inhibitory mechanisms to convert gradients into absolute readouts (Briscoe et al., 2000; Jessell, 2000), some cells, such as pMN cells and others, may transiently exhibit mixed identities (Ericson et al., 1996; Ruiz i Altaba, 1996). Perhaps Lmo4 may serve as an effective negative regulator under conditions in which Isl protein levels are not optimal relative to the proteins with which it interacts, whereas the endogenous MN cell population might be resistant to the negative effects of Lmo4 because the Isl levels are optimal for forming highly stable complexes. In this case, Lmo4 may represent an additional layer of regulation to ensure cell identity is accurately assigned. These predictions will best be tested through further characterization of the precise composition of LIM homeodomain complexes in vivo and careful measurement of the binding constants between NLI and its interacting factors under a variety of conditions.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/17/2923/DC1
Supplementary Material
We thank T. Jessell and S. Morton for anti-Irx3 antibody. We are grateful to members of the Pfaff lab for helpful discussions and encouragement, K. Lettieri for technical assistance. This study was supported by grants of the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs (A084135), Cell Dynamics Research Center (2009-0070938), KOSEF/MEST (2009-0059288), a fellowship from the Paralyzed Veterans of America (to M.-R.S.), NIH 1RO1 HL070867 (to S.E.) and NINDS R37NS037116 (to S.L.P.). S.L.P. is an investigator of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
References
- Agulnick, A. D., Taira, M., Breen, J. J., Tanaka, T., Dawid, I. B. and Westphal, H. (1996). Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature 384, 270-272. [DOI] [PubMed] [Google Scholar]
- Arber, S., Han, B., Mendelsohn, M., Smith, M., Jessell, T. M. and Sockanathan, S. (1999). Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 23, 659-674. [DOI] [PubMed] [Google Scholar]
- Bach, I., Carriere, C., Ostendorff, H. P., Andersen, B. and Rosenfeld, M. G. (1997). A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 11, 1370-1380. [DOI] [PubMed] [Google Scholar]
- Betz, U. A., Vosshenrich, C. A., Rajewsky, K. and Muller, W. (1996). Bypass of lethality with mosaic mice generated by Cre-loxP-mediated recombination. Curr. Biol. 6, 1307-1316. [DOI] [PubMed] [Google Scholar]
- Briscoe, J., Pierani, A., Jessell, T. M. and Ericson, J. (2000). A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435-445. [DOI] [PubMed] [Google Scholar]
- Dawid, I. B., Breen, J. J. and Toyama, R. (1998). LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14, 156-162. [DOI] [PubMed] [Google Scholar]
- Deane, J. E., Mackay, J. P., Kwan, A. H., Sum, E. Y., Visvader, J. E. and Matthews, J. M. (2003). Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4. EMBO J. 22, 2224-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory, Y., Deng, M., Xie, X. and Gan, L. (2007). Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J. Comp. Neurol. 503, 182-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson, J., Thor, S., Edlund, T., Jessell, T. M. and Yamada, T. (1992). Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science 256, 1555-1560. [DOI] [PubMed] [Google Scholar]
- Ericson, J., Morton, S., Kawakami, A., Roelink, H. and Jessell, T. M. (1996). Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell 87, 661-673. [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez, P., Lu, C. H., Rincon-Limas, D. E., Garcia-Bellido, A. and Botas, J. (1998). The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J. 17, 6846-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungor, C., Taniguchi-Ishigaki, N., Ma, H., Drung, A., Tursun, B., Ostendorff, H. P., Bossenz, M., Becker, C. G., Becker, T. and Bach, I. (2007). Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc. Natl. Acad. Sci. USA 104, 15000-15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm, K., Sum, E. Y., Fujiwara, Y., Lindeman, G. J., Visvader, J. E. and Orkin, S. H. (2004). Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol. Cell. Biol. 24, 2074-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, S. A. and Eisen, J. S. (2006). Islet1 and Islet2 have equivalent abilities to promote motoneuron formation and to specify motoneuron subtype identity. Development 133, 2137-2147. [DOI] [PubMed] [Google Scholar]
- Jessell, T. M. (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20-29. [DOI] [PubMed] [Google Scholar]
- Joshi, K., Lee, S., Lee, B., Lee, J. W. and Lee, S. K. (2009). LMO4 controls the balance between excitatory and inhibitory spinal V2 interneurons. Neuron 61, 839-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata, L. W., Kenny, D. A. and Gill, G. N. (1996). Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc. Natl. Acad. Sci. USA 93, 11693-11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata, L. W., Pfaff, S. L. and Gill, G. N. (1998). The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J. Biol. Chem. 273, 3152-3157. [DOI] [PubMed] [Google Scholar]
- Kenny, D. A., Jurata, L. W., Saga, Y. and Gill, G. N. (1998). Identification and characterization of LMO4, an LMO gene with a novel pattern of expression during embryogenesis. Proc. Natl. Acad. Sci. USA 95, 11257-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., Okamura, Y. and Higashijima, S. (2006). alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J. Neurosci. 26, 5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Lee, B., Joshi, K., Pfaff, S. L., Lee, J. W. and Lee, S. K. (2008). A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell 14, 877-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. K., Jurata, L. W., Funahashi, J., Ruiz, E. C. and Pfaff, S. L. (2004). Analysis of embryonic motoneuron gene regulation: derepression of general activators function in concert with enhancer factors. Development 131, 3295-3306. [DOI] [PubMed] [Google Scholar]
- Lee, S. K., Jurata, L. W., Nowak, R., Lettieri, K., Kenny, D. A., Pfaff, S. L. and Gill, G. N. (2005). The LIM domain-only protein LMO4 is required for neural tube closure. Mol. Cell. Neurosci. 28, 205-214. [DOI] [PubMed] [Google Scholar]
- Lundfald, L., Restrepo, C. E., Butt, S. J., Peng, C. Y., Droho, S., Endo, T., Zeilhofer, H. U., Sharma, K. and Kiehn, O. (2007). Phenotype of V2-derived interneurons and their relationship to the axon guidance molecule EphA4 in the developing mouse spinal cord. Eur. J. Neurosci. 26, 2989-3002. [DOI] [PubMed] [Google Scholar]
- Milan, M. and Cohen, S. M. (1999). Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol. Cell 4, 267-273. [DOI] [PubMed] [Google Scholar]
- Milan, M., Diaz-Benjumea, F. J. and Cohen, S. M. (1998). Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 12, 2912-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroyama, Y., Fujiwara, Y., Orkin, S. H. and Rowitch, D. H. (2005). Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature 438, 360-363. [DOI] [PubMed] [Google Scholar]
- Pan, L., Deng, M., Xie, X. and Gan, L. (2008). ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development 135, 1981-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff, S. L., Mendelsohn, M., Stewart, C. L., Edlund, T. and Jessell, T. M. (1996). Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309-320. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba, A. (1996). Coexpression of HNF-3 beta and Isl-1/2 and mixed distribution of ventral cell types in the early neural tube. Int. J. Dev. Biol. 40, 1081-1088. [PubMed] [Google Scholar]
- Sharma, K., Sheng, H. Z., Lettieri, K., Li, H., Karavanov, A., Potter, S., Westphal, H. and Pfaff, S. L. (1998). LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell 95, 817-828. [DOI] [PubMed] [Google Scholar]
- Sugihara, T. M., Bach, I., Kioussi, C., Rosenfeld, M. G. and Andersen, B. (1998). Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO-4, and CLIM coregulators. Proc. Natl. Acad. Sci. USA 95, 15418-15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y., Dykes, I. M., Liang, X., Eng, S. R., Evans, S. M. and Turner, E. E. (2008). A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat. Neurosci. 11, 1283-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler, J., Harrison, K., Sharma, K., Lettieri, K., Kehrl, J. and Pfaff, S. L. (1999). Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron 23, 675-687. [DOI] [PubMed] [Google Scholar]
- Thaler, J. P., Lee, S. K., Jurata, L. W., Gill, G. N. and Pfaff, S. L. (2002). The LIM homeodomain factor Lhx3 contributes to the specification of motor neuron and Interneuron identity through cell type-specific protein-protein interactions. Cell 110, 237-249. [DOI] [PubMed] [Google Scholar]
- Thaler, J. P., Koo, S. J., Kania, A., Lettieri, K., Andrews, S., Cox, C., Jessell, T. M. and Pfaff, S. L. (2004). A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron 41, 337-350. [DOI] [PubMed] [Google Scholar]
- Thor, S., Andersson, S. G., Tomlinson, A. and Thomas, J. B. (1999). A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature 397, 76-80. [DOI] [PubMed] [Google Scholar]
- Tse, E., Smith, A. J., Hunt, S., Lavenir, I., Forster, A., Warren, A. J., Grutz, G., Foroni, L., Carlton, M. B., Colledge, W. H. et al. (2004). Null mutation of the Lmo4 gene or a combined null mutation of the Lmo1/Lmo3 genes causes perinatal lethality, and Lmo4 controls neural tube development in mice. Mol. Cell. Biol. 24, 2063-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T., Ensini, M., Morton, S. B., Baldassare, M., Edlund, T., Jessell, T. M. and Pfaff, S. L. (1994). Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell 79, 957-970. [DOI] [PubMed] [Google Scholar]
- van Meyel, D. J., O'Keefe, D. D., Jurata, L. W., Thor, S., Gill, G. N. and Thomas, J. B. (1999). Chip and apterous physically interact to form a functional complex during Drosophila development. Mol. Cell 4, 259-265. [DOI] [PubMed] [Google Scholar]
- Visvader, J. E., Mao, X., Fujiwara, Y., Hahm, K. and Orkin, S. H. (1997). The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc. Natl. Acad. Sci. USA 94, 13707-13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman, I. A., Osada, H., Grutz, G. G., Agulnick, A. D., Westphal, H., Forster, A. and Rabbitts, T. H. (1997). The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16, 3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z., Meng, X., Cai, Y., Liang, H., Nagarajan, L. and Brandt, S. J. (2007). Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 21, 942-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Tomita, T., Wines-Samuelson, M., Beglopoulos, V., Tansey, M. G., Kopan, R. and Shen, J. (2006). Notch1 signaling influences v2 interneuron and motor neuron development in the spinal cord. Dev. Neurosci. 28, 102-117. [DOI] [PubMed] [Google Scholar]
- Zeng, C., Justice, N. J., Abdelilah, S., Chan, Y. M., Jan, L. Y. and Jan, Y. N. (1998). The Drosophila LIM-only gene, dLMO, is mutated in Beadex alleles and might represent an evolutionarily conserved function in appendage development. Proc. Natl. Acad. Sci. USA 95, 10637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.