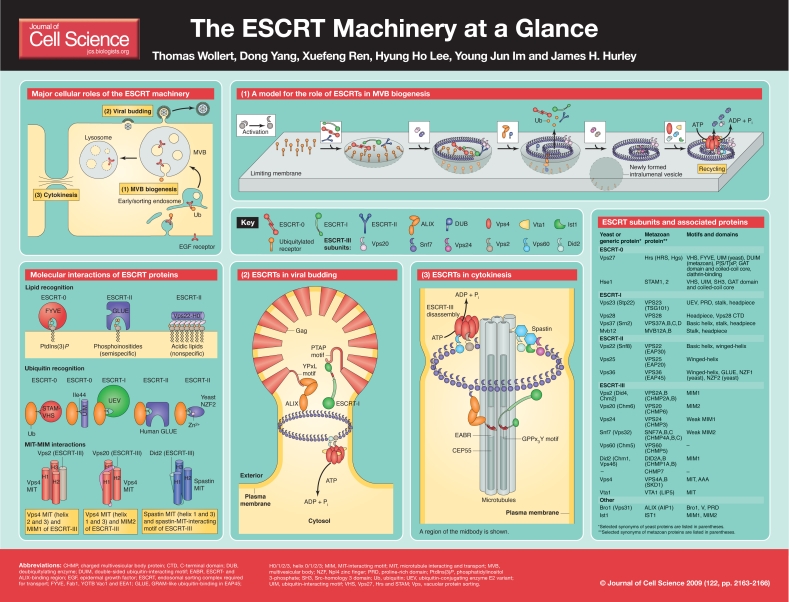
The endosomal sorting complex required for transport (ESCRT) machinery is conserved from Archaea to animals, and catalyzes the scission of membrane necks (Wollert et al., 2009) in the biogenesis of multivesicular bodies (MVBs) (Gruenberg and Stenmark, 2004; Hurley, 2008; Piper and Katzmann, 2007), cytokinesis (Carlton and Martin-Serrano, 2007; Lindas et al., 2008; Morita et al., 2007; Samson et al., 2008) and the budding of enveloped viruses such as HIV-1 (Bieniasz, 2006; Fujii et al., 2007; Morita and Sundquist, 2004). Cleavage occurs from the surface of the membrane that is contiguous with the inside of the membrane neck. The topology of scission mediated by the ESCRT machinery is opposite to that of the dynamin family of membrane-scission proteins, which cleave membrane necks by constricting them from the outside.
The ESCRT machinery consists of the five protein complexes ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III and Vps4-Vta1, and several ESCRT-associated proteins (Hurley, 2008; Saksena et al., 2007; Williams and Urbe, 2007). ESCRT-0, ESCRT-I, ESCRT-II and Vps4-Vta1 are soluble complexes that move between cytosolic and membrane-bound states. By contrast, the subunits of ESCRT-III are soluble monomers that assemble on membranes into tightly bound filaments (Ghazi-Tabatabai et al., 2008), tubes (Hanson et al., 2008; Lata et al., 2008) and spirals (Hanson et al., 2008) that can only be released by the ATP-dependent action of Vps4-Vta1.
Figure 1.
In this article and its accompanying poster, we focus on the fundamentals of the structure of the ESCRT machinery and its molecular interactions with other molecules that explain its function in the diverse biological processes of MVB biogenesis, cytokinesis and virus budding. We hope that this short overview will provide a useful foundation for developmental biologists, clinicians and others that aim to explore the roles of ESCRTs in their research.
ESCRTs in MVB biogenesis and receptor downregulation
The ESCRT proteins were discovered and characterized less than 10 years ago as having a role in the sorting of ubiquitylated cell-surface receptors and other proteins to the vacuole in yeast (Saksena et al., 2007). This sorting process occurs through the formation of MVBs, which are endosomes containing intralumenal vesicles (ILVs) that are destined for degradation in the yeast vacuole or in the mammalian lysosome. The ESCRTs have a central role in the sorting of ubiquitylated receptors, to which they bind directly via their ubiquitin-binding domains. In addition, the ESCRTs also have a direct role in the membrane-remodeling events that lead to ILV formation.
In yeast, ILVs have a mean diameter of 24 nm, and are therefore thought to derive from a roughly circular patch on the limiting membrane with a radius of 24 nm (Nickerson et al., 2006). This value is similar to the dimensions of a single copy each of ESCRT-0, ESCRT-I and ESCRT-II when they are assembled together on the limiting membrane. It is notable that the size of the ESCRTs is on the same order as the dimensions of the ILVs that they produce, and this prompts speculation that as few as one copy each of these ESCRTs might be required to produce a single ILV.
ESCRTs bind to ubiquitylated proteins
Ubiquitylation is the main molecular signal that sends cargo into the ESCRT pathway for lysosomal degradation. The ESCRTs interact with ubiquitylated receptors and other ubiquitylated cargo through various motifs. These include the ubiquitin-interacting motif (UIM) and the double-sided ubiquitin-interacting motif (DUIM) of the yeast and human Hrs subunits of ESCRT-0; the VHS (Vps27, Hrs and STAM) domain and UIM of the STAM subunit of ESCRT-0; the ubiquitin-conjugating enzyme E2 variant (UEV) domain of the Vps23 subunit of ESCRT-I; the GRAM-like ubiquitin binding in EAP45 (GLUE) domain of the VPS36 subunit of human ESCRT-II; and the NZF2 domain of the Vps36 subunit of yeast ESCRT-II (Hurley, 2008; Saksena et al., 2007; Williams and Urbe, 2007). All of these motifs bind to ubiquitin Ile44 and the surrounding residues, which precludes the simultaneous interaction of multiple ESCRTs with a single ubiquitin moiety. The affinities of the individual motifs for monoubiquitin are very low (less than 100 μM) – probably too low to have an important role in targeting ESCRTs to membranes. We favor a model in which multiple ESCRTs co-assemble on membranes and cluster multiple cargo molecules, which may be monoubiquitylated, or polyubiquitylated through the Lys63 residue of ubiquitin.
Membrane lipids recruit ESCRTs to endosomes
ESCRTs are targeted to endosomes through a variety of specific, partially specific and nonspecific interactions with membrane lipids. The FYVE domain of the Hrs subunit of ESCRT-0 is highly specific for phosphatidylinositol 3-phosphate [PtdIns(3)P], whereas the GLUE domain of the Vps36 subunit of ESCRT-II binds to PtdIns(3)P and also to other phosphoinositides. In addition, the first helix of the Vps22 subunit of ESCRT-II binds to acidic phospholipids with no apparent specificity, yet the gain in overall membrane affinity is still important for membrane targeting (Im and Hurley, 2008). Finally, ESCRT-III subunits also bind to acidic lipids with little specificity (Saksena et al., 2009). Membrane binding promotes a conformational change in ESCRT-III subunits that appears to favor their functional polymerization (Saksena et al., 2009). These specific and non-specific interactions are collectively central to the recruitment, assembly and activating conformational changes of the ESCRTs.
The ESCRT-III cycle
Vps4 and the subunits of ESCRT-III are the most conserved elements in the pathway and are directly responsible for membrane cleavage. In the ESCRT-III cycle, soluble ESCRT-III monomeric subunits are recruited from the cytosol to their site of action on the membrane, where they undergo an activating conformational change, recruit various effector proteins and catalyze membrane scission. Subsequent to membrane scission, Vps4 hydrolyzes ATP to drive the recycling of ESCRT-III subunits back to their soluble monomeric form.
In yeast, the four essential ESCRT-III proteins are Vps20, Snf7, Vps24 and Vps2, which assemble in that order (Teis et al., 2008). Two more ESCRT-III proteins, Vps60 and Did2, join later during ESCRT assembly, as does the ESCRT-III-like MIM1 and MIM2-containing protein Ist1 (Agromayor et al., 2009; Bajorek et al., 2009; Dimaano et al., 2008; Rue et al., 2008). Upon membrane binding (Saksena et al., 2009), motifs in the C-termini of ESCRT-III subunits are exposed, allowing them to bind to the microtubule-interacting and transport (MIT) domains of downstream effector proteins (Hurley and Yang, 2008). Together, the short C-termini of ESCRT-III subunits make up a dense region of multiple regulatory signals. Vps4 functions as a 12- to 14-mer, which therefore contains 12-14 MIT domains. One site in the Vps4 MIT domain can bind to the MIM1s (MIT-interaction motif 1) of Vps2 and Did2 (Obita et al., 2007; Stuchell-Brereton et al., 2007), while another site can bind to the MIM2 of Vps20 (Kieffer et al., 2008). The Vps4 MIT domain also binds with very low affinity to the C-termini of most, if not all, of the other ESCRT-III proteins (Kieffer et al., 2008).
Vta1, the non-catalytic subunit of the Vps4-Vta1 complex, contains two MIT domains that bind to late-joining ESCRT-III proteins (Azmi et al., 2008; Shim et al., 2008; Xiao et al., 2008). The full Vps4-Vta1 assembly can thus bind to ESCRT-III with very high cooperativity. The ATPase activity of Vps4, in concert with the multiplicity of binding events between ESCRT-III and the MIT domains of Vps4 and Vta1, is instrumental to ESCRT-III disassembly. The disassembly activity of the Vps4-Vta1 complex allows the subunits of ESCRT-III to be recycled, thereby completing the ESCRT-III cycle and sustaining membrane-scission activity (Wollert et al., 2009).
ESCRTs in HIV-1 budding
The ESCRT proteins are essential for the efficient budding of HIV-1 from cultured cells. The main role of the ESCRTs in viral budding is to cut the neck that connects the virion to the plasma membrane, releasing the virion from the host cell. The HIV-1 Gag p6 protein recruits the host-cell ESCRTs via two late-domain (L-domain) sequence motifs (Fujii et al., 2007; Bieniasz, 2006; Morita and Sundquist, 2004; McDonald and Martin-Serrano, 2009). The most important motif in the Gag p6 protein that mediates ESCRT recruitment is an L-domain with the sequence PTAP, which binds to the UEV domain of the Vps23 subunit of ESCRT-I. Additionally, a YPxL motif near the C-terminus of HIV-1 Gag p6 directly interacts with the V domain of the ESCRT-associated protein ALIX (also known as PDCD6IP and AIP1). The Bro1 domain of ALIX directly activates ESCRT-III by recruiting and activating the Snf7 subunit, which is crucial for ESCRT-III function (McCullough et al., 2008). It is less clear how ESCRT-I activates ESCRT-III; however, it is thought that the main role of ESCRT-I and ALIX is to recruit and activate ESCRT-III. ESCRT-II is apparently not involved in HIV-1 budding, nor is the Vps20 subunit of ESCRT-III, which couples ESCRT-II and ESCRT-III in MVB biogenesis. Ubiquitylation by the ligase NEDD4L is also involved in ESCRT-mediated HIV-1 budding (Chung et al., 2008; Usami et al., 2008). In summary, ESCRT-I and ALIX appear to act as adaptors that connect the viral Gag protein to ESCRT-III, which in turn is directly responsible for membrane cleavage and virion release.
ESCRTs in cytokinesis
During cytokinesis, the ESCRTs play a crucial role in the scission of the membrane neck connecting two daughter cells, both in mammalian cells (Carlton and Martin-Serrano, 2007; Morita et al., 2007; McDonald and Martin-Serrano, 2009) and in a subset of Archaea (Lindas et al., 2008; Samson et al., 2008). In mammalian cytokinesis, the two daughter cells are tethered to one another by a dense microtubule-rich structure known as the midbody. As with HIV-1 budding, ESCRT-I and ALIX are the key adaptors for membrane scission, and ESCRT-II is not involved. GPPx3Y motifs in ESCRT-I and ALIX bind to CEP55, a protein at the midbody (Lee et al., 2008) that is responsible for recruiting the ESCRTs to the site of membrane scission. In turn, ALIX, and probably ESCRT-I, recruit ESCRT-III subunits, which close the neck. However, full closure of the membrane neck is obstructed by the microtubules of the central spindle. ESCRT-III has at least one additional role in the process, which is to recruit the microtubule-severing enzyme spastin (Connell et al., 2008; Yang et al., 2008), which probably cuts the central spindle, together with the ESCRT-III-mediated membrane cleavage.
Perspectives
Recent discoveries have shown that ESCRT-III and Vps4 make up a conserved membrane-scission machine that functions in many different contexts, including MVB biogenesis, HIV-1 budding and cytokinesis. The ESCRT machinery is targeted to the site of action by different means for each of these biological processes: 3-phosphoinositides are central in MVB biogenesis, Gag L-domains in viral budding and CEP55 in cytokinesis. In addition, ubiquitin is involved in most, if not all, of these pathways. Each pathway converges at the ESCRT-III cycle, which probably functions in a similar way in each setting. It will be interesting to see whether the list of biological processes in which the ESCRT machinery has a role expands; for example, the possibility that ESCRTs have a role in sealing autophagosomes is intriguing (Rusten and Stenmark, 2009).
Although beyond the scope of this article, the catalog of ESCRT functions that are involved in health and disease continues to grow rapidly (Saksena and Emr, 2009). In addition to the role in HIV-1 budding described above, genetic defects, alterations in gene expression, mutations or altered interactions involving ESCRTs have been implicated in several pathological conditions, including cancer, frontotemporal dementia, amyotrophic lateral sclerosis, Huntington's disease, Parkinson's disease, hereditary spastic paraplegia and cataracts (Saksena and Emr, 2009). At the molecular level, the structure and interactions of ESCRTs in solution are now understood in great detail. However, upcoming challenges for ESCRT biochemistry and structural biology include gaining a better understanding of the structure and interactions in the membrane-bound setting in which the ESCRTs function. As the fundamental mechanisms of normal ESCRT function become more clear, the long-term challenges in this field will evolve towards understanding how these functions are perturbed in disease.
Supplementary Material
This article is part of a Minifocus on the ESCRT machinery. For further reading, please see related articles: `No strings attached: the ESCRT machinery in viral budding and cytokinesis' by Bethan McDonald and Juan Martin-Serrano (J. Cell Sci. 122, 2167-2177) and `How do ESCRT proteins control autophagy?' by Tor Erik Rusten and Harald Stenmark (J. Cell Sci. 122, 2179-2183).
Research in the Hurley lab is funded by the intramural program of the NIDDK, NIH, and the IATAP program of the NIH. T.W. is an EMBO long-term fellow. Deposited in PMC for release after 12 months.
References
- Agromayor, M., Carlton, J. G., Phelan, J. P., Matthews, D. R., Carlin, L. M., Ameer-Beg, S., Bowers, K. and Martin-Serrano, J. (2009). Essential role of hIST1 in cytokinesis. Mol. Biol. Cell 20, 1374-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi, I. F., Davies, B. A., Xiao, J., Babst, M., Xu, Z. and Katzmann, D. J. (2008). ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell 14, 50-61. [DOI] [PubMed] [Google Scholar]
- Bajorek, M., Morita, E., Skalicky, J. J., Morham, S. G., Babst, M. and Sundquist, W. I. (2009). Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 20, 1360-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz, P. D. (2006). Late budding domains and host proteins in enveloped virus release. Virology 344, 55-63. [DOI] [PubMed] [Google Scholar]
- Carlton, J. G. and Martin-Serrano, J. (2007). Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908-1912. [DOI] [PubMed] [Google Scholar]
- Chung, H. Y., Morita, E., von Schwedler, U., Muller, B., Krausslich, H. G. and Sundquist, W. I. (2008). NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J. Virol. 82, 4884-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell, J. W., Lindon, C., Luzio, J. P. and Reid, E. (2008). Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic 10, 42-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano, C., Jones, C. B., Hanono, A., Curtiss, M. and Babst, M. (2008). Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell 19, 465-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, K., Hurley, J. H. and Freed, E. O. (2007). Beyond Tsg101: the role of Alix in `ESCRTing' HIV-1. Nat. Rev. Microbiol. 5, 912-916. [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai, S., Saksena, S., Short, J. M., Pobbati, A. V., Veprintsev, D. B., Crowther, R. A., Emr, S. D., Egelman, E. H. and Williams, R. L. (2008). Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16, 1345-1356. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. and Stenmark, H. (2004). The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell. Biol. 5, 317-323. [DOI] [PubMed] [Google Scholar]
- Hanson, P. I., Roth, R., Lin, Y. and Heuser, J. E. (2008). Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J. H. (2008). ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol. 20, 4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J. H. and Yang, D. (2008). MIT domainia. Dev. Cell 14, 6-8. [DOI] [PubMed] [Google Scholar]
- Im, Y. J. and Hurley, J. H. (2008). Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev. Cell 14, 902-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer, C., Skalicky, J., Morita, E., De Domenico, I., Ward, D. M., Kaplan, J. and Sundquist, W. I. (2008). Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15, 62-73. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Lata, S., Schoehn, G., Jain, A., Pires, R., Piehler, J., Gottlinger, H. and Weissenhorn, W. (2008). Helical structures of ESCRT-III are disassembled by VPS4. Science 321, 1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. H., Elia, N., Ghirlando, R., Lippincott-Schwartz, J. and Hurley, J. H. (2008). Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science 322, 576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindas, A. C., Karlsson, E. A., Lindgren, M. T., Ettema, T. J. G. and Bernander, R. (2008). A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. USA 105, 18942-18946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough, J., Fisher, R. D., Whitby, F. G., Sundquist, W. I. and Hill, C. P. (2008). ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA 105, 7687-7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, B. and Martin-Serrano, J. (2009). No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 122, 2167-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, E. and Sundquist, W. I. (2004). Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20, 395-425. [DOI] [PubMed] [Google Scholar]
- Morita, E., Sandrin, V., Chung, H. Y., Morham, S. G., Gygi, S., Rodesch, C. K. and Sundquist, W. I. (2007). Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D. P., West, M. and Odorizzi, G. (2006). Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175, 715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita, T., Saksena, S., Ghazi-Tabatabai, S., Gill, D. J., Perisic, O., Emr, S. D. and Williams, R. L. (2007). Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735-739. [DOI] [PubMed] [Google Scholar]
- Piper, R. C. and Katzmann, D. J. (2007). Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23, 519-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue, S. M., Mattei, S., Saksena, S. and Emr, S. D. (2008). Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten, T. E. and Stenmark, H. (2009). How do ESCRT proteins control autophagy? J. Cell Sci. 122, 2179-2183. [DOI] [PubMed] [Google Scholar]
- Saksena, S. and Emr, S. D. (2009). ESCRTs and human disease. Biochem. Soc. Trans. 37, 167-172. [DOI] [PubMed] [Google Scholar]
- Saksena, S., Sun, J., Chu, T. and Emr, S. D. (2007). ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci. 32, 561-573. [DOI] [PubMed] [Google Scholar]
- Saksena, S., Wahlman, J., Teis, D., Johnson, A. E. and Emr, S. D. (2009). Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, R. Y., Obita, T., Freund, S. M., Williams, R. L. and Bell, S. D. (2008). A role for the ESCRT system in cell division in Archaea. Science 322, 1710-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, S., Merrill, S. A. and Hanson, P. I. (2008). Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell 19, 2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton, M., Skalicky, J., Kieffer, C., Karren, M. A., Ghaffarian, S. and Sundquist, W. I. (2007). ESCRT-III recognition by VPS4 ATPases. Nature 449, 740-744. [DOI] [PubMed] [Google Scholar]
- Teis, D., Saksena, S. and Emr, S. D. (2008). Ordered Assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578-589. [DOI] [PubMed] [Google Scholar]
- Usami, Y., Popov, S., Popova, E. and Gottlinger, H. G. (2008). Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J. Virol. 82, 4898-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. L. and Urbe, S. (2007). The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell. Biol. 8, 355-368. [DOI] [PubMed] [Google Scholar]
- Wollert, T., Wunder, C., Lippincott-Schwartz, J. and Hurley, J. H. (2009). Membrane scission by the ESCRT-III complex. Nature 458, 172-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J., Xia, H., Zhou, J., Azmi, I., Davies, B. A., Katzmann, D. J. and Xu, Z. (2008). Structural basis of Vta1 function in the multi-vesicular body sorting pathway. Dev. Cell 14, 37-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., Rismanchi, N., Renvoisé, B., Lippincott-Schwartz, J., Blackstone, C. and Hurley, J. H. (2008). Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15, 1278-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.