Abstract
Oxidation of two anionic phospholipids - cardiolipin (CL) in mitochondria and phosphatidylserine (PS) in extramitochondrial compartments - are important signaling events, particularly during the execution of programmed cell death and clearance of apoptotic cells. Quantitative analysis of CL and PS oxidation products is central to understanding their molecular mechanisms of action. We combined the identification of diverse phospholipid molecular species by ESI-MS with quantitative assessments of lipid hydroperoxides using a fluorescence HPLC-based protocol. We characterized CL and PS oxidation products formed in a model system (cyt c/H2O2), in apoptotic cells (neurons, pulmonary artery endothelial cells) and mouse lung under inflammatory/oxidative stress conditions (hyperoxia, inhalation of single walled carbon nanotubes). Our results demonstrate the usefulness of this approach for quantitative assessments, identification of individual molecular species and structural characterization of anionic phospholipids that are involved in oxidative modification in cells and tissues.
1. Introduction
Oxidative metabolism of small biomolecules is an intrinsic part of aerobic life. Lipids can be utilized as energy-producing fuel but can also undergo a different type of oxidation reaction whereby the addition of oxygen to polyunsaturated fatty acid residues of phospholipids yields oxygenated derivatives [1,2]. For many years, this type of reaction – lipid peroxidation – was mostly considered important in cell and tissue damage as a pathogenic mechanism in degenerative diseases. This deleterious pathway, triggered by massive phospholipid peroxidation, is associated with the disruption of hydrophobic interactions in biological membranes leading to the disorganization of their fundamental functional and structural characteristics – lipid-protein interactions, selective permeability, asymmetry, etc [1–5].
Recently, the role and significance of subtle lipid peroxidation in cell signaling has become more apparent. Oxygenation of polyunsaturated free fatty acids - eicosanoic, docosapentaenoic, docosahexaenoic - induces the production of a variety of potent physiological regulators such as prostaglandins, thromboxanes, and leukotrienes, lipoxenes, resolvins and protectins [6–10]. Appreciation of the signaling functions of oxygenated phospholipids is beginning to evolve. Among several notable examples are studies of their participation in apoptosis, phagocytosis and lymphocyte activation [11–15]. This deficiency in our understanding of the physiological mechanisms of peroxidized phospholipids is due, at least in part, to methodological difficulties in their characterization and quantitative determinations. This is particularly relevant to the detection and analysis of primary products of phospholipid peroxidation – hydroperoxides - that have been limited by their inherent thermal and chemical instability [16] combined with relatively low abundance in cells, tissues and biofluids [17].
Over decades, conventional analytical methods have proven difficult when applied to these compounds. The development of soft-ionization techniques, particularly electrospray ionization (ESI), producing minimal fragmentation and allowing the ionization of non-volatile and thermo- and chemical-labile samples, has offered new opportunities in the direct analysis of (phospho)lipid hydroperoxides by ESI-MS (without prior derivatization) [4,7,10,18,19]. During ESI-MS in the negative ion mode, the abundant generation of carboxylate anions from free acids and free acid hydroperoxides warrants their successful quantitative analysis. Assessments of intact phospholipid hydroperoxides by conventional MS methods are more difficult [16]. One of the key problems is that differences in ionization of individual molecular species of a class may strongly depend on the concentration of lipids due to their aggregation. This is particularly important during analysis of phospholipid oxidation products (hydroperoxides) whose abundance is comparatively very low vs non-oxidized phospholipid species. Therefore direct infusion of lipid extracts may be associated with ionization suppression of lipid hydroperoxides due to aggregation of non-oxidized lipids thus lowering ionization efficiency. Additionally, acyl chain length, degree of acyl chain unsaturation and phospholipid head group structure can also affect the ionization efficiency of phospholipids (reviewed in [20–23]) as well as phospholipid oxidation products. Further, the individual MS signals are dependent on many conditions including overall lipid concentration, solvent composition, and instrument settings. Another complication is that the peroxidation of phospholipids yields multiple products. Hydroperoxides – the primary molecular products of lipid peroxidation – are formed by the addition of hydrogen to peroxyl radical intermediates in polyunsatutarated fatty acids of phospholipids [2,17]; hydroxides are commonly produced by enzymatically catalyzed two electron reduction of hydroperoxides [17]; epoxides originate from epoxidation of hydroperoxides by peroxygenases [24]; carbonyls, as well as short carbon-chain derivatives result from β-scission [25]. Moreover, the lack of heavy isotope-labeled analogs for different oxidized phospholipids limits quantitative MS analysis.
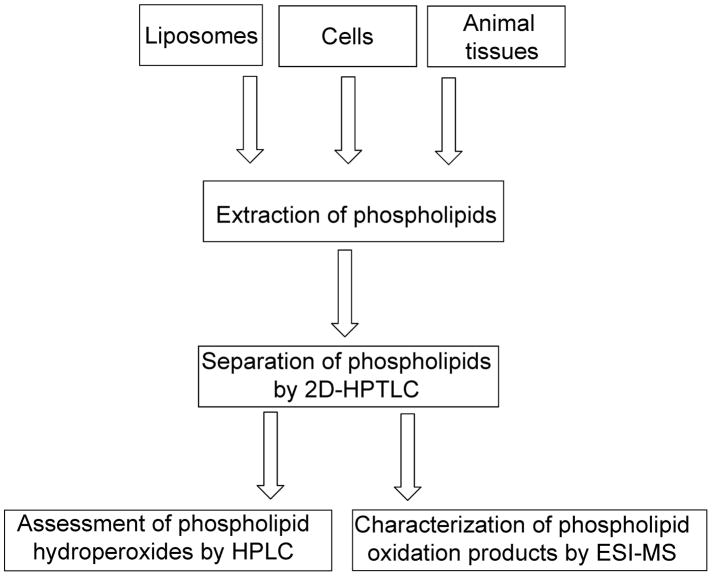
Here, we describe our approach to ESI-MS analysis of hydroperoxy-derivatives of two major anionic phospholipids - cardiolipins (CL) and phosphatidylserines (PS) – found in mitochondrial and extramitochondrial compartments of cells, respectively. We focused our efforts on CL and PS hydroperoxides (CL-OOH and PS-OOH) because they represent the primary peroxidation products whose subsequent oxidative degradation leads to the formation of a large variety of other derivatives. Additionally, we present data on the analysis of hydroxy-CL (CL-OH) and hydroxy-PS (PS-OH) – the products formed by direct reduction of the respective hydroperoxides. To better characterize these phospholipid peroxidation products generated in model system (cyt c/H2O2) and in cells undergoing apoptosis, we employed a combination of identification of diverse phospholipid molecular species by ESI-MS with quantitative assessments of lipid hydroperoxides using a fluorescence HPLC-based protocol. A schematic explanation of our experimental approach is shown in Fig. 1.
Figure 1.
Experimental approach used to identify and characterize oxidized phospholipid molecular species formed in apoptotic cells and tissues of animals.
2. Experimental
2.1. Chemicals and materials
1-Octadecanoyl-2-(4Z, 7Z, 10Z, 13Z, 16Z, 19Z-docosahexaenoyl)-sn-glycero-3-[phospho-L-serine] (sodium salt), 1,2-diheptadecanoyl-sn-glycero-3-[phospho-L-serine] (sodium salt), 1,2-dioleoyl-sn-glycero-3-phosphocholine, 1,1′,2,2′-tetramyristoyl-cardiolipin (sodium salt), and 1,1′,2,2′-tetralinoleoyl-cardiolipin (sodium salt) were from Avanti Polar Lipids Inc. (Alabaster, AL). The concentration of phospholipids was estimated by phosphorus analysis. 9S-hydroperoxy-10E acid, 12Z-octa-decadienoic acid, 13S-hydroperoxy-10E acid, and 12Z-octa-decadienoic acid were obtained from Cayman Chemical Co (Ann Arbor, MI) and were of the highest purity available. Chloroform (HPLC grade), methanol (HPLC grade), acetone (HPLC grade), hexane (HPLC grade), 2-propanol (HPLC grade), ammonium hydroxide, ammonium acetate, acetic acid glacial, CaCl2, EDTA, butylated hydroxytoluene (BHT), SDS, microperoxidase-11 (MP-11), phospholipase A2 and triphenylphosphine (TPP) were from Sigma-Aldrich (St. Louis, MO). HPTLC silica G plates (5×5 cm) were purchased from Whatman, (Schleicher & Schuell, England). N-acetyl-3,7-dihydroxyphenoxazine (Amplex Red) was from Molecular Probes (Eugene, OR).
2.2. Sample preparation
Cell culture and animal tissue
Neurons from fetuses of Sprague-Dawley rats (at embryologic day 16–18) were harvested as described previously [26] and grown in Neurobasal medium containing 2 mM L-glutamine and 100 U/mL penicillin/streptomycin supplemented with 2% B27 at 37°C in a humidified atmosphere (5% CO2 plus 95% air). To induce apoptosis, neuronal cultures were treated by 1 μM STS in culture media for 1h; apoptosis was ascertained by subsequent increases in cyt c release from mitochondria, PS externalization and caspase 3/7 activation [27].
Stretch injury of neurons
Primary cortical neurons were cultured onto thin, clear elastic membranes. The membranes were stretched biaxially once using mechanical loading conditions (100 milliseconds using a peak substrate strain of 60%) that mimic brain deformations during trauma as described previously [28] and observed over a 0–24h time period.
Sheep pulmonary arteries were obtained from a nearby slaughterhouse, and endothelial cells were harvested and purified as described previously [29]. Subpassages of cells were routinely homogeneously positive for DiI-Ac-LDL and factor VIII antigen.
Sheep pulmonary artery endothelial cells (SPAEC) from passage 13 were treated with 100 ng/ml lipopolysaccharide (LPS), at a time (4 h) that preceded significant LPS-induced apoptosis as ascertained by subsequent increases in caspase 3/7.
Hyperoxic Mice
Wild-type C57BL6 male mice (aged 8–10 weeks) were obtained from Jackson Laboratories and housed in a specific pathogen-free environment for 2 days; the animals were placed in a plexiglass chamber with food and water ad libitum and exposed to 100% oxygen for 72 h with an oxygen flow rate of 15 liters per minute. This experimental protocol was in accordance with the standards established by the US Animal Welfare Acts, as set forth in the NIH guidelines and in the Policy and Procedures manual of the University of Pittsburgh Institutional Animal Care and Use Committee experimental guidelines. Oxygen saturation in the chamber was measured periodically with an oxygen analyzer from Vascular Technology, Inc. Mouse lungs were excised following sodium pentobarbital euthanasia of the animals.
C57BL/6 mice were exposed to single walled carbon nanotubes (SWCNT) via inhalation (5 mg/m3, whole body inhalation for 4 consecutive days, 5 h/day) [30].
Lipid extraction, 2D-HPTLC separation and lipid phosphorus determination
Total lipids were extracted from samples by Folch procedure [31]. Lipid extracts were separated and analyzed by 2D-HPTLC [32]. Prior to separation, plates were treated with methanol containing 1 mM EDTA, 100 μM DTPA to bind transition metals from silica. The first solvent system included chloroform:methanol:28% ammonium hydroxide (65:25:5 v/v). To remove the solvent, plates were dried with a forced N2, blower and then they were developed in the second dimension with a solvent system - chloroform:acetone:methanol:glacial acetic acid:water (50:20:10:10:5 v/v). The phospholipids were visualized by exposure to iodine vapors and identified by comparison with authentic phospholipid standards. Lipid phosphorus was determined after mineralization of the residue by HClO4 at 180°C for 30 min by sodium molybdate/ascorbate micro-assay [33]. For Amplex Red and ESI-MS-direct infusion analyses of phospholipid hydroperoxides, the phospholipid spots were visualized by spraying the plates with deionized water. Then spots were scraped from the plates and phospholipids were extracted by choloroform:methanol:water (20:10:2 v/v).
2.3. Fluorescence HPLC assay of phospholipid hydroperoxides
Phospholipid hydroperoxides were quantitatively determined by fluorescence HPLC of products formed in microperoxidase 11-catalyzed reaction with a fluorogenic substrate, Amplex Red, as previously described [27]. Phospholipids were hydrolyzed by porcine pancreatic PLA2 (0.1 U/μl) in 25 mM phosphate buffer, containing 0.8 mM Ca, 0.5 mM EGTA and 0.5 mM SDS (pH 8.0 at RT for 30 min), prior to exposure to reagents for the peroxidase reaction (MP-11/Amplex Red). Amplex Red (50 μM) and MP-11 (1.0 μg/μl) were added to hydrolyzed lipids, and the samples were incubated at 4°C for 40 min. The reaction was terminated by addition of 100 μl of a stop reagent (solution of 10 mM HCl, 4 mM BHT in ethanol). A Shimadzu LC-100AT vp HPLC system equipped with Eclipse XDB-C18 column (5 μm, 150 × 4.6 mm), a fluorescence detector (RF-10Axl) and an auto-sampler (SIL-10AD vp) were used for the analysis of products separated by HPLC. Sample (5 μl) was injected into a column and was eluted by a mobile phase composed of 25 mM KH2PO4 (pH 7.0)/methanol (60:40 v/v) with 1 ml/min flow rate. The resorufin (Amplex red oxidation product) fluorescence was measured at 590 nm after excitation at 560 nm.
2.4. Direct infusion ESI-MS analysis
ESI-MS analysis of phospholipids was performed by direct infusion into linear ion-trap mass spectrometer (LXQ™ with the Xcalibur operating system (Thermo Electron, San Jose, CA)). After 2D-HPTLC separation, phospholipids were evaporated under N2, re-suspended in chloroform:methanol 1:1 v/v (20 pmol/μl), and directly utilized for the acquisition of negative-ion ESI mass spectra at a flow rate of 5 μl/min. The ESI probe was operated at a voltage differential of 5.0 kV. Capillary temperature was maintained at 70 or 150°C. MS/MS analysis of individual PL species was employed in full range zoom (200–1800 m/z) in negative ion mode. The MS/MS spectra were acquired using isolation width 1.0 m/z, 5 microscans with a maximum injection time 200 ms. For analysis of CL structure, the scan time setting of the ion trap for full MS (in the ranges of 700–1000 m/z and 1200–1600 m/z) was set at 50 microscans with a maximum injection time of 1000 msec. MSn analysis was performed using an isolation width of 1 m/z, 5 microscans with a maximum injection time of 1000 ms. Two ion activation techniques were used for MS analysis: collision-induced dissociation (CID, Q=0.25, low mass cut off at 28% of the precursor m/z) and pulsed-Q dissociation technique (PQD), with Q=0.7, and no low mass cut off for the analysis of low molecular weight fragment ions. Additionally, for the analysis of oxidized PL molecular species (hydroperoxy- and hydroxy-), we treated the reaction products with a reductant, triphenylphosphine (TPP), to convert CL–OOH and PS-OOH into CL–OH and PS-OH, respectively. Isotopic corrections were performed by entering the chemical composition of each species into the Qual browser of the Xcalibur (operating system), using the simulation of the isotopic distribution to make adjustments for the major peaks. To minimize isotopic interferences between isolated masses M+2, the MS/MS spectra were acquired using an isolation width of 1.0 m/z. Based on MSn fragmentation data, chemical structures of lipid molecular species were analyzed and confirmed by using ChemDraw.
Statistics
The results are presented as mean ± S.D. values from at least three experiments, and statistical analyses were performed by either Student’s t-test or one-way ANOVA. To estimate the significance of differences in accumulation of phospholipid hydroperoxides by Amplex Red procedure in liposomes a Student’s t-test was used. In experiments with cells and animals, the significance of differences was estimated by one-way ANOVA. The statistical significance of differences was set at p< 0.05.
3. Results and discussion
Quantitative assessments of lipid hydroperoxides
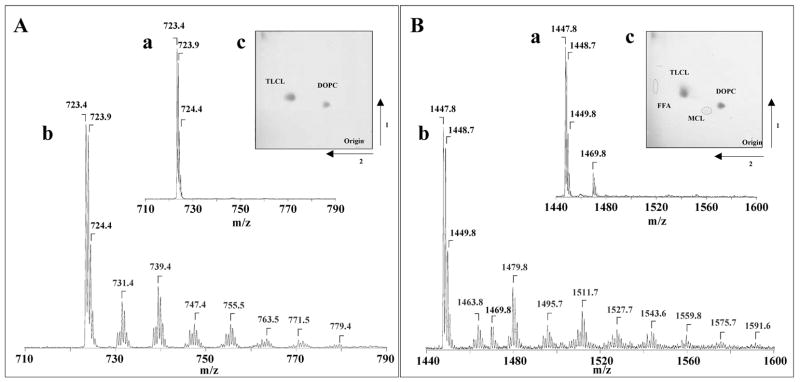
2D-HPTLC has been extensively used to separate the phospholipids into different molecular classes. The incubation of liposomes, containing TLCL and DOPC with cyt c/H2O2 triggered the oxidation of TLCL. 2D-HPTLC revealed that non-oxidized TLCL and its major oxidation products co-migrated together (Fig. 2A(c), 1B(c), Table 1). Under the conditions used, the total amount of TLCL-OOH accumulated was estimated as 174.6 + 10.5 pmol/nmol CL and 2.6 + 0.8 pmol/nmol CL for oxidized CL and control, respectively (Table 1). For comparison, data illustrating total amounts of CL-OOH and PS-OOH - determined by fluorescence HPLC-based protocol – in several types of cells triggered to apoptosis in vitro and in vivo are also shown in Table 1.
Figure 2.
Typical negative-ion ESI mass spectra for doubly [M-2H]2− and singly [M-H]− charged ions of TLCL molecular species before (A(a); B(a)) and after its oxidation induced by cyt c/H2O2 (A(b); B(b)), respectively. Insert shows typical 2D-HPTLC of total lipids extracted from liposomes, containing TLCL and DOPC, before (A, inset) and after 30 min incubation at 37°C with cyt c/H2O2 (B, inset). CL – cardiolipin, PC- phosphatidylcholine, MCL- monolyso-CL, FFA- free fatty acids. TLCL/DOPC liposomes (250 μM/250 μM) were incubated in 50 mM Na-phosphate buffer (pH 7.4) containing 100 μM DTPA in the presence of cyt c (5 μM) and H2O2 (100 μM). At the end of incubation, lipids were extracted by Folch procedure and MS analysis was performed. CL-OOH formation was confirmed by the reduction of CL-OOH into CL-OH using TPP (1 mg/ml, for 20 min at 4°C); thus formed CL-OH was analyzed by ESI-MS.
Table 1.
Detection of CL- hydroperoxides and PS-hydroperoxides in liposomes and different cells and tissues from animals exposed to pro-oxidant or pro-apoptotic stimuli.
| Sample ID | Conditions | CL-OOH pmol/nmol CL | PS-OOH pmol/nmol PS |
|---|---|---|---|
| Liposomes PC(C18:1/C18:1)/CL(C18:2)4, n=4 |
Control | 2.6 ± 0.8 | --- |
| Cyt c/H2O2, 30 min | 174.6 ± 10.5* | --- | |
| Liposomes PC(C18:1/C18:1)/PS(C16:0/C22:6), n=3 |
Control | --- | 2.2 ± 0.8 |
| Cyt c/H2O2, 60 min | --- | 37.2 ± 3.2* | |
| Rat brain cortical neurons, n=6 | Control | 6.6 ± 2.4 | 3.6 ± 1.4 |
| STS, 1 μM, 60 min | 74.3 ± 6.8* | 25.8 ± 1.2* | |
| Rat brain cortical neurons, n=3 | Stretch | 107.0 ± 23.0* | 19.9 ± 4.2* |
| SPAEC, n=3 | Control | 1.6 ± 0.4 | 2.9 ± 0.4 |
| LPS, 100 ng/ml, 4 h | 21.5 ± 3.1* | 12.9 ± 2.3* | |
| Mouse lung homogenate, n=3 | Control | 4.5 ± 0.3 | 2.3 ± 0.7 |
| Hyperoxia, 72 h | 33.8 ± 8.0* | 7.6 ± 1.7* | |
| SWCNT, 1 day | 45.0 ± 4.1* | 7.3 ± 2.9* | |
| SWCNT, 7 day | 87.1 ± 9.7* | 3.7 ± 1.1 |
SPAEC, sheep pulmonary endothelial cells; CL-OOH, cardiolipin hydroperoxdes; PS-OOH, phosphatidylserine hydroperoxides; Cyt c, cytochrome c; STS, staurosporine; LPS, lipopolysacharide; SWCNT, single walled carbon nanotubes.
p< 0.05 vs corresponding control.
Identification of CL hydroxy- and hydroperoxy- molecular species in model systems
Identification of oxidized CL and PS molecular species was performed by their mass-charge ratio (MS1) and fragmentation properties (MSn) using ESI-MS analysis. In negative ion mode, CL produces two anionic charges that form both singly charged [M-H]− and doubly charged [M-2H]−2 ions with corresponding signals of non-oxidized TLCL with m/z 1447 and 723, respectively (Fig 2A, 2B). In addition, the sodium adduct of the TLCL singly-charged ion with m/z 1469 was also observed (Fig. 2B(b)).
ESI-MS assessment of TLCL oxidation products revealed the accumulation of different hydroxy-, hydroperoxy-, di-hydroxy- and hydroxy/hydroperoxy-derivatives of TLCL as evidenced by the appearance of doubly charged signals with m/z of 731, 739, 747, 755, 763, 770, and 779, and corresponding singly charged signals with m/z 1463, 1479, 1495, 1511, 1527, 1543, 1559, respectively (Fig. 2A, 2B).
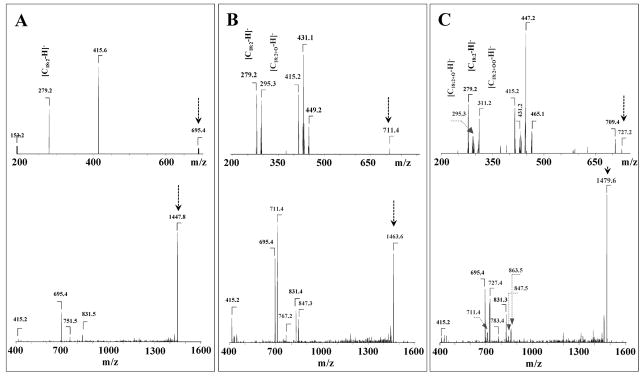
Typical ions formed during the fragmentation process of CL (a, b, a+56, or b+136) were identified by MS/MS analysis as described by Hsu and co-authors [34]. The MSn CID fragmentation of TLCL was performed on each of ions (a, b, a+56 and b+136) to assign fatty acids and their positions (Fig. 3, Table 2). Additionally, using the pulsed-Q- dissociation technique, we also obtained MS spectra of TLCL ion fragments including a, b, a+56; a+136; b+56; b+136, and fatty acid carboxylate anions. For example, MS2 fragmentation (m/z Full scan → 447) of a parent ion with m/z 1447 resulted in a major ion with m/z 695 (a=b) with characteristic ion fragments (a+56, a+136), at m/z 751 and 831 (Fig. 3A). MS3 fragmentation of ions with m/z 659, 751 and 831 yielded carboxylate ion with m/z 279, leading to identification of the symmetric structure (C18:2/C18:2/C18:2/C18:2)-TLCL.
Figure 3.
Typical MS2 spectra of singly charged TLCL molecular species of non-oxidized TLCL with m/z 1447 (A, lower panel) and oxidized TLCL with m/z 1463 and 1479, corresponding to mono-hydroxy- and hydroperoxy-TLCL (B, C, lower panel), respectively. MS3 spectra of TLCL fragments of non-oxidized ion fragment with m/z 695 (A, upper panel) and oxidized ion fragments with m/z 711 and 727 (B, C, upper panel), originated from singly charged TLCL molecular species with m/z 1447, 1463 and 1479, respectively. The major product ion with m/z 279 found after fragmentation of the molecular ion with m/z 695 corresponds to C18:2. Product ions with m/z 279 and 295 detected after fragmentation of the molecular ion with m/z 711 represent C18:2 and C18:2-OH, respectively. Product ions with m/z 279, 295 and 311 observed in the spectra after fragmentation of the molecular ion with m/z 727 correspond to C18:2, C18:2-OH (trace), C18:2-OOH and C18:2-OH+OH (minor), respectively.
Table 2.
Major oxidized cardiolipin and phosphatidylserine molecular species from several sources induced by different oxidized stimuli.
| Molecular species | [M-H]− m/z | Identified Acyl Chains | Molecular species | [M-H]− m/z | Identified Acyl Chains |
|---|---|---|---|---|---|
| Cardiolipin (TLCL) | |||||
| Diacyl species control | Oxidized Diacyl species by Cyt c/H2O2 | ||||
| 72:8 | 1447.8 | (C18:2)1/(C18:2)1/(C18:2)1/(C18:2)1 | 72:8 | 1463.8 | (C18:2)1/(C18:2+O)1/(C18:2)1/(C18:2)1 |
| 72:8 | 1479.8 | (C18:2)1/(C18:2+OO)1/(C18:2)1/(C18:2)1 (C18:2+O)1/(C18:2+O)1/(C18:2)1/(C18:2)1 (C18:2)1/(C18:2+O+O)1/(C18:2)1/(C18:2)1 |
|||
| 72:8 | 1495.8 | (C18:2+O)1/(C18:2+OO)1/(C18:2)1/(C18:2) | |||
| 72:8 | 1511.8 | (C18:2+OO)1/(C18:2+OO)1/(C18:2)1/(C18:2) | |||
| 72:8 | 1527.8 | (C18:2+O)1/(C18:2+OO)1/(C18:2+OO)1/(C18:2) | |||
| 72:8 | 1543.8 | (C18:2+O)1/(C18:2+OO)1/(C18:2+OO)1/(C18:2+O) | |||
| 72:8 | 1559.8 | (C18:2+O)1/(C18:2+OO)1/(C18:2+OO)1/(C18:2+OO) | |||
| Phosphatidylserine (C18:0/C22:6) | |||||
| Diacyl species control | Oxidized Diacyl species by Cyt c/H2O2 | ||||
| 40:6 | 834.6 | (C18:0)/(C22:6) | 40:6 | 850.6 866.7 882.5 898.5 |
(C18:0)/(C22:6+O) (C18:0)/(C22:6+OO) (C18:0)/(C22:6 + 3O) (C18:0)/(C22:6+4O) |
| Cardiolipin from neuronal cells | |||||
| Diacyl species control | Oxidized Diacyl species by STS | ||||
| 74:9 | 1474.0 | (C16:0)1/(C18:1)1/(C18:2)1/(C22:6)1 | 74:9 | 1506.0 | (C16:0)1/(C18:1)1/(C18:2)1/(C22:6+OO)1 |
| 74:8 | 1476.0 | (C16:0)1/(C18:1)1/(C20:4)1/(C20:3)1;.(C16:0)1/(C18:1)2/(C22:6)1 | 74:8 | 1492.0 | (C16:0)1/(C18:1)1/(C20:4+O)1/(C20:3)1; (C16:0)1/(C18:1)2/(C22:6+O)1; |
| Phosphatidylserine from neuronal cells | |||||
| Diacyl species control | Oxidized Diacyl species by STS, or Stretch | ||||
| 40:6 | 834.6 | (C18:0)/(C22:6) | 40:6 | 850.6 866.7 882.5 |
(C18:0)/(C22:6+O) (C18:0)/(C22:6+OO) (C18:0)/(C22:6 + 3O) |
| Cardiolipin from SPAEC | |||||
| Diacyl species | Oxidized Diacyl species by LPS | ||||
| 70:5 | 1426.5 | (C16:0)1/(C18:2)1/(C18:2)1/(C18:1)1 | 70:5 | 1458.5 | (C16:0)1/(C18:2)1/(C18:2+OO)1/(C18:1)1 |
| 72:5 | 1454.0 | (C18:0)1/(C18:1)1/(C18:2)1/(C18:2)1 | 72:5 | 1486.0 | (C18:0)1/(C18:1)1/(C18:2+OO)1/(C18:2)1 |
| 74:7 | 1478.0 | (C18:0)1/(C18:1)1/(C18:2)1/(C20:4)1 | 74:7 | 1510.0 | (C18:0)1/(C18:1)1/(C18:2+OO)1/(C20:4)1 |
| 76:4 | 1512.0 | (C18:2)1/(C18:2)1/(C20:0)1/(C20:0)1 | 76:4 | 1544.0 | (C18:2)1/(C18:2+OO)1/(C20:0)1/(C20:0)1 |
| Phosphatidylserine from SPAEC | |||||
| Diacyl species control | Oxidized Diacyl species by LPS | ||||
| 38:4 | 810.6 | (C18:0)/(C20:4) | 38:4 | 826.6 | (C18:0)/(C20:4+O) |
| 38:4 | 842.6 | (C18:0)/(C20:4+OO) | |||
| 40:6 | 834.6 | (C18:0)/(C22:6) | 40:6 | 850.6 | (C18:0)/(C22:6+O) |
| 40:6 | 866.6 | (C18:0)/(C22:6+OO) | |||
| Cardiolipin from mouse lung | |||||
| Diacyl species control | Oxidized Diacyl species by hyperoxia | ||||
| 70:6 | 1424.0 | (C16:1)1/(C18:2)1/(C18:2)1/(C18:1)1 | 70:6 | 1556.9 | (C16:1)1/(C18:2)1/(C18:2+OO)1/(C18:1)1 |
| 72:7 | 1450.0 | (C18:2)1/(C18:2)1/(C18:2)1/(C18:1)1 | 72:7 | 1482.0 | (C18:2)1/(C18:2)1/(C18:2+OO)1/(C18:1)1; |
| 74:10 | 1472.0 | (C16:0)1/(C18:2)1/(C20:4)1/(C20:4)1; | 74:10 | 1504.0 | (C16:0)1/(C18:2+OO)1/(C20:4)1/(C20:4)1; |
| 76:12 | 1498.0 | (C16:1)1/(C18:2)1/(C20:4)1/(C22:5)1 | 76:12 | 1540.0 | (C16:1)1/(C18:2+OO)1/(C20:4)1/(C22:5)1 |
| 78:11 | 1526.0 | (C18:0)1/(C18:2)1/(C20:4)1/(C22:5)1 | 78:11 | 1558.0 | (C18:0)1/(C18:2+OO)1/(C20:4)1/(C22:5)1 |
| Phosphatidylserine from mouse lung | |||||
| Diacyl species control | Oxidized Diacyl species by Hyperoxia, or SWCNT | ||||
| 40:6 | 834.5 | (C18:0)/(C22:6) | 40:6 | 866.7 | (C18:0)/(C22:6+OO) |
+ O, molecular species containing hydroxy-group; +OO, molecular species containing hydroperoxy-group; +O+O, molecular species containing two hydroxy-groups.
SPAEC, sheep pulmonary endothelial cells; Cyt c, cytochrome c; STS, staurosporine; LPS, lipopolysacharide; SWCNT, single walled carbon nanotubes.
MS2 fragmentation of the first abundant TLCL oxidized peak with m/z 1463 resulted in two major ions with m/z 695 (a) and 711 (b) (Fig. 3B). The major ion at m/z 695 originated from a non -oxidized fragment of TLCL and gave two additional ions (a+56 and a+136) with m/z 751 and 831, respectively. The second major ion with m/z 711 represents an oxidized fragment of TLCL with two additional ions (b+56 and b+136) at m/z 767 and 847, respectively (Fig. 3B). MS3 fragmentation (m/z Full scan→ 1469→711) of the ion at m/z 711 yielded carboxylate anions of a non-oxidized product with m/z 279 and of an oxidation product with m/z 295 that corresponds to mono-hydroxy-linoleic acid (Fig. 3B, upper panel). Detailed analysis (MS4 fragmentation) of the ion with m/z 295 (m/z Full scan→ 1469→711→295) is presented in Fig. 4A. Ions with m/z 415, 431 and 449 correspond to [b-(C18:2+O-H)]−, [b-(C18:2-H)]− and [b-(C18:2-H)+H2O]−.
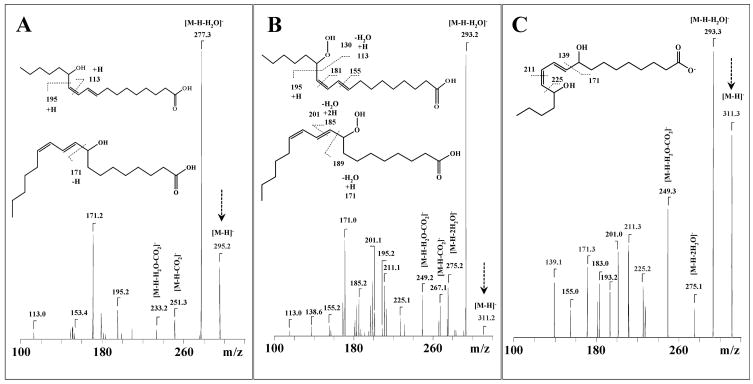
Figure 4.
Identification of mono-hydroxy- and hydroperoxy- TLCL products produced during incubation with cyt c/H2O2. MS4 spectra of TLCL fragments for molecular ion with m/z 295 (m/z Full scan→ 1469→711→295) (A), and m/z 311 (m/z Full scan→1479→727→311) (B) correspond to C18:2-OH, and C18:2-OOH or C18:2-OH+OH (minor), respectively. MS3 spectra of oxidized TLCL fragments after their reduction by TPP for molecular ion with m/z 311 (m/z Full scan→739→311) (C) represent C18:2-OH+OH Carboxylate ion with m/z 311 was detected as a minor fraction of di-hydroxy TLCL oxidized product (C18:2-OH-OH) in a much larger signal from hydroperoxy- molecular ion (singly charged 1479 and doubly charge 739); the dihydroxy-TLCL signal became the major product after treatment by TPP.
MS2 fragmentation of the second abundant TLCL oxidized peak with m/z 1479, resulted in two major ions with m/z 695, 727 and one minor signal with m/z 711 (Fig. 3C). The ion with m/z 727 represents oxidized fragment of TLCL with two additional ions (b+56 and b+136) at m/z 783 and 863, respectively (Fig. 3B). The ion with m/z 695 represents a non-oxidized TLCL fragment, whereas ions with m/z 727 and 711 produced oxidized TLCL fragments corresponding to hydroperoxy- or di-hydroxy- and mono-hydroxy- molecular species of TLCL. MS3 fragmentation (m/z Full scan→1479→727) of an ion at m/z 727 yielded a non-oxidized carboxylate anion at m/z 279 and two oxidized anions at m/z 311 and 295, corresponding to hydroperoxy- or di-hydroxy- and mono-hydroxy-linoleic acid (Fig. 3B, upper panel). Detailed MS4 fragmentation analysis (m/z Full scan→1479→727→311) of the ion with m/z 311 is shown on Fig. 4B.
MSn analysis of TLCL oxidized cluster with m/z 1479 revealed the presence of different TLCL molecular species, containing oxygenated fatty acids C18:2, C18:2-OOH, C18:2-OH+OH and C18:2-OH, as follows: (C18:2)1/(C18:2)1/(C18:2)1/(C18:2+OO)1 and (C18:2)1/(C18:2)1/(C18:2+O)1/(C18:2+O)1 (Table 2). Results of MS4 fragmentation of oxidized carboxylate ions with m/z 295 and 311, corresponding to mono-hydroxy-, hydroperoxy- or di-hydroxy- carboxylate ions, respectively, are presented on Fig. 4A, 4B, 4C. Oxidized carboxylate ions showed trivial signals due to the loss of water, CO2, or a combination of water plus CO2. For example, MS4 CID fragmentation (m/z Full scan→1463→711→295) of carboxylate anion with m/z 295 [M-H]− generated product ions with m/z 277, 251 and 233 that corresponded to [M-H-H2O]−, [M-H-CO2]−, and [M-H-H2O+CO2]−, respectively (Fig. 4A). Product ions with m/z 153 and m/z 171 were formed after cleavage of 9S-OH-C18:2 on both sides of the hydroxyl group [35,36]. Product ions at m/z 195 and m/z 113 are characteristic of 13 regio-isomers 13S-OH-C18:2 (Fig. 4A) [37]. MS4 CID fragmentation (m/z Full scan→1479→727→311) of carboxylate anion with m/z 311 [M-H]− generated product ions with m/z 293, 275, 267 and 249 that corresponded to [M-H-H2O]−, [M-H-2H2O]−, [M-H-CO2]−, and [M-H-H2O+CO2]−, respectively (Fig. 4B). Vinylic cleavage of the C12-C13 bond yields the fragment with m/z 195, whereas cleavage of the double-bond allylic of the hydroperoxy-group yields the fragment with m/z 113 [37]. A similar fragmentation pattern has been observed during analysis of free fatty acid hydroperoxides in negative and in positive ion modes [35,36,38]. In the case of 9S-regioisomers, a characteristic product ion with m/z 171 (m/z 189 - H2O) and m/z 155 from the fission of the corresponding C9–C10 bond and subsequent elimination of H2O2 was detectable [38]. Additionally, fragments of a carboxylate ion with m/z 311 corresponding to di-hydroxy-C18:2, were observed in the spectrum (Fig. 4B); these fragments became abundant after reduction of hydroperoxy- group of linoleic acid in TLCL by TPP (Fig. 4C).
Reduction of hydroperoxy-TLCL to hydroxy-TLCL in a model system
In a separate series of experiments, we performed detailed analysis of TLCL oxidation products (TLCL-OOH and TLCL-OH) formed by cyt c/H2O2 after their reduction with TPP (Fig. 4A, B). Among different TLCL oxidation products, a smaller peak corresponding to mono-hydroxy-linoleate-containing TLCL with m/z 731 significantly increased and appeared as a dominant peak after treatment with TPP (Fig. 5A(a), 5B(a)). MS2 analysis of oxidized TLCL molecular ion at m/z 731 before and after TPP treatment revealed carboxylate ions of C18:2 (m/z 279) and C18:2–OH (m/z 295) (Fig. 5A(b), 5B(b)). Trace amounts of molecular ions corresponding to C18:2–OH were also observed in the MS2 spectrum obtained from the TLCL oxidation product with m/z 739 (Fig. 5A(c), Table 2). After TPP treatment, the major carboxylate ion with m/z 311 was converted into carboxylate ion with m/z 295 (Fig 5A(c), 5B(c)). Thus, hydroperoxy-TLCL molecular species were converted into respective hydroxy-TLCL molecular species (Fig. 5A(a), 5B(a)).
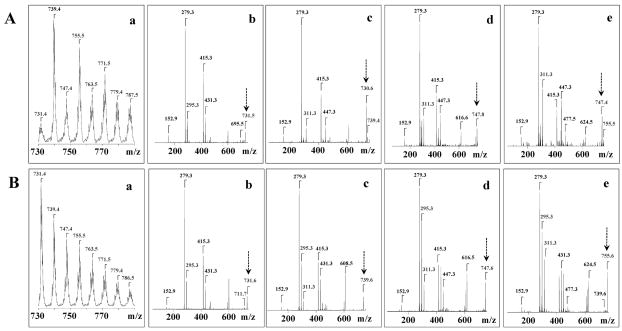
Figure 5.
ESI-MS spectra and MSn analysis of oxidized TLCL before and after reduction of hydroperoxy-TLCLs oxidation products to hydroxy-TLCLs by TPP. Panel A represents MS spectra of TLCL oxidation products before treatment with TPP. Panel B represents MS spectra of TLCL oxidation products after treatment with TPP. a – MS1 spectra of doubly charged ions of TLCL; b, c, d, e – MS2 spectra after CID fragmentation of oxidized molecular ions of TLCL with m/z 731, 739, 747 and 755, respectively.
Fragmentation of the molecular ion with m/z 739 that originated from TPP-treated oxidized TLCL revealed product ions with m/z 279, 295, and 311 corresponding to C18:2, C18:2–OH, and C18:2–OH+OH, respectively (Fig. 4C, Fig. 5B(c)). This suggests that molecular species with m/z 739 consist of both mono-hydroxy-TLCL and di-hydroxy- TLCL. The fragment ions with m/z 295 were detected in the MS2 spectra obtained from m/z 739, 747, and 755 of the TLCL oxidation products treated with TPP (Fig. 5B(c), 5B(d), 5B(e)). After reduction of hydroperoxy-derivatives of TLCL by TPP, the carboxylate ion with m/z 311 was converted into an ion of hydroxy-derivative with m/z 295. In the MS3 spectrum, the carboxylate ion with m/z 311 corresponded to 9, 14-dihydroxy-C18:2 acid and 9, 10-dihydroxy-C18:2 acids with characteristic fragmentation ions with (m/z 139, 171, 225, 249, 275, 293, 311) and (m/z 139, 171, 201, 249, 275, 293, 311), respectively, (Fig. 4C). Cleavage of the C13–C14, yielded a fragment with m/z 225, whereas cleavage of the C12–C13 double-bond produced a fragment with m/z 211. Cleavage of the C9–C10 bond generated fragments with m/z 171 and 139. This interpretation is in line with data published by Lang et al., [39] characterizing fragmentation of 9,14- and 9,10-dihydroxy derivatives of linoleic acid using LC-MS [39].
Identification of PS hydroxy- and hydroperoxy- molecular species in model system
In the next series of experiments, we performed oxidation of PS (C18:0/C22:6) by cyt c/H2O2 and then conducted structural analysis of the oxidation products formed. The total amount of accumulated PS-OOH estimated by fluorescence HPLC was as 42.8 + 3.7 pmol/nmol PS after oxidation vs 0.8 + 0.6 pmol/nmol PS in control (Table 1). Non-oxidized PS, in the negative ionization mode revealed in the MS1 spectrum the major molecular de-protonated ion [M-H]− at m/z 834, whereas oxidized PS elicited several molecular species that were represented by ions [M-H]− with m/z 850; 866; 882, that corresponded to mono-hydroxy-, hydroperoxy- and dihydroxy-, hydroxy-hydroperoxy- and tri-hydroperoxy-PS, respectively (Fig. 6A – 6H).
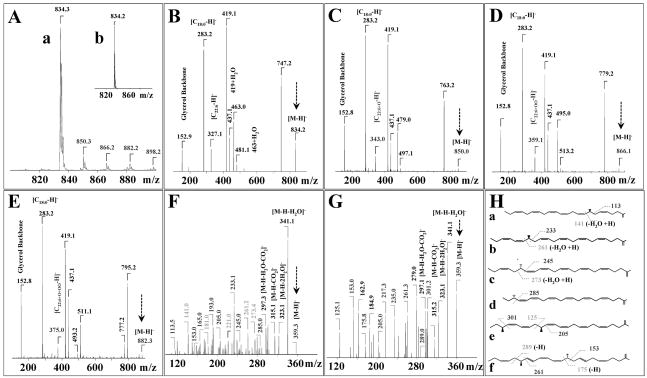
Figure 6.
Identification of mono-hydroxy- and hydroperoxy- PS (C18:0/C22:6) products formed during incubation with cyt c/H2O2. (A) Full MS spectra of oxidized (a) and non-oxidized PS (b, inset) molecular species. MS2 spectra of non-oxidized PS (B) and oxidized PS (C, D, E) molecular clusters, corresponding to m/z 834, 850, 866 and 882, respectively. MS3 spectra (m/z Full scan→866→359) of PS carboxylate ions with m/z 359 before (F) and after (G) treatment with TPP. Possible fatty acid structures are shown on panel H: a - 7-hydroperoxy-C22:6, b - 16-hydroperoxy-C22:6, c - 17-hydroperoxy-C22:6, d - 20-hydroperoxy-C22:6, e -14, 20-dihydroxy-C22:6, f - 10, 17-dihydroxy-C22:6.
MS2 PQD or CID fragmentations of non-oxidized PS at m/z 834 yielded a strong peak with m/z 747 caused by a loss of the serine group. Further MS2 fragmentation of this ion resulted in appearance of two lyso-products ions with m/z 463 and 419 and two free fatty acid molecular fragments with m/z 283 and 327 corresponding to carboxylate anions of C18:0 and C22:6, respectively (Fig. 6B). MS2 analysis of oxidized PS molecular species showed that they contained carboxylate ions of mono-hydroxy-C22:6, hydroperoxy- C22:6 and di-hydroxy- C22:6 and a combination of hydroxy-hydroperoxy- C22:6 and tri-hydroxy-C22:6 at m/z 343; 359; and 375, respectively (Fig. 6C, 6D, 6E). The structure of C22:6 oxidized species was further confirmed by the CID technique commonly used for fatty acid hydroperoxides analysis [35,37,38]. For example MS2 PQD fragmentation of de-protonated [M-H]− ion peak with m/z 866 showed the appearance of carboxylate ion at m/z 359. Subsequent MS3 CID fragmentation of this molecular ion, corresponding to C22:6-OOH and C22:6-OH+OH, resulted in the first typical set of product ions at m/z 341; 323, 315 and 297, corresponding to loss of water (one or two molecules), CO2, and a combination of water plus CO2, respectively (Fig. 6F). More detailed analysis of MS3 fragmentation of carboxylate ion with m/z 359 revealed multiple hydroperoxy- products: 7-OOH-C22:6; 10-OOH-C22:6; 11-OOH-C22:6; 13-OOH-C22:6; 14-OOH-C22:6; 16-OOH-C22:6; 17-OOH-C22:6; 20-OOH-C22:6. For example, cleavage of bonds (C6–C7), (C9–C10), (C10–C11), (C12–C13), (C13–C14), (C16–C17) and (C19–C20) on the carboxylic side of the hydroperoxy-group resulted in typical fragments with m/z 113; 153; 165; 193; 205; 245; and 285, respectively [7]. When cleavage of these bonds occurred on the methyl side of the hydroperoxy-group, the ion fragments formed had m/z 141; 181; 193; 221; 233; 261; 273, respectively (Fig. 6F). The schematic fragmentation of hydroperoxy-isomers of C22:6 is shown on Fig. 6H.
TPP treatment of PS oxidation products caused the disappearance of ion fragments corresponding to hydroperoxy- carboxylate ions from the MS3 spectrum (m/z Full scan→866→359) (Fig. 6G). In the MS3 spectrum (Fig. 6G, 6H), ion fragments, corresponding exclusively to di-hydroxy- derivatives of C22:6 (m/z 359): 7, 17-dihydroxy-C22:6; 10, 17-dihydroxy-C22:6 and 14, 20-dihydroxy-C22:6 were observed [7]. As an example, cleavage of 14, 20-dihydroxy-C22:6 at C19–C20 on the carboxylic side of the hydroxy-group resulted in the appearance of an ion with m/z 301 (Fig. 6G, 6H). Further, cleavage of C13–C14 bond from the carboxylic side of the acyl chain produced two ion fragments with m/z 205 and 155, whereas cleavage of C14–C15 bond from the methyl side of hydroxy-group gave two ion fragments with m/z 125 and 235, respectively (Fig. 6G, 6H).
Formation and identification of hydroxy- and hydroperoxy- CL, PS species during apoptosis
The characterization of hydroperoxides and hydroxy-derivatives from individual CL and PS species in model systems is instrumental to identifying of their primary oxidation products in cells and tissues during apoptosis. We applied ESI-MS analysis to investigate mass-spectrometric profiles of anionic phospholipids (CL and PS) in several types of cells triggered to undergo apoptosis by different stimuli as illustrated by the following examples.
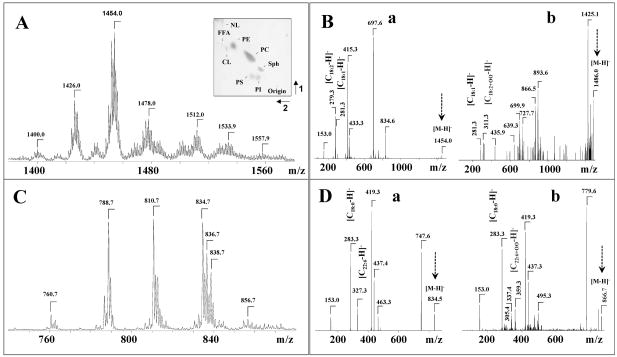
LPS treatment of sheep pulmonary artery cells (SPAEC) induced prominent accumulation of phospholipid hydroperoxides in CL and PS (Table 1). The total amount of CL hydroperoxides was increased more than 10-fold after LPS treatment up to 21.5 from 1.6 pmol CL-OOH/nmol CL in the control (Table 1). Several molecular species of CL with m/z 1426, 1454, 1478 and 1512 underwent peroxidation to yield respective hydroperoxy-CL with m/z 1458, 1486, 1510 and 1544 (Fig. 7, Table 2). For example, oxidized molecular species with m/z 1486 containing hydroperoxy-linoleate as one of acyls-(C18:0)1/(C18:1)1/(C18:2+OO)1/(C18:2)1 - originated from the major non-oxidized CL containing (C18:0)1/(C18:1)1/(C18:2)1/(C18:2)1 fatty acids (m/z 1454) (Fig 7A, 7B; Table 2). The total level of PS hydroperoxides was increased 4-fold after LPS treatment to 12.9 from 2.9 pmol PS-OOH/nmol PS in control (Table 1). Hydroperoxy-molecular species of PS were detected mainly for ion peaks with m/z 842 (C18:0/C20:4+OO), and 866 (C18:0/C22:6+OO), which originated from ions with m/z 810 (C18:0/C20:4), and 834 (C18:0/C22:6), respectively (Fig. 7C, 7D, Table 2). Moreover, mono-hydroxy- molecular species with m/z 826 (C18:0/C20:4+O), and 850 (C18:0/C22:6+O) were also observed in the MS spectra after LPS exposure (Table 2).
Figure 7.
ESI-MS spectra of molecular species of CL (A, B) and PS (C, D) isolated from SPAEC before and after exposure of cells to LPS. Typical ESI-MS2 spectra of CL (B(b)) and PS (D(b)) of oxidized molecular species with m/z 1486 (CL) and 866 (PS) originated from dominant non-oxidized molecular species with m/z 1454 (CL) (B(a)) and 834 (PS) (D(a)), respectively. Insert: Typical 2D-HPTLC of lipids extracted from SPAEC.
Mitochondrial cyt c release is a key event in the mitochondria-dependent cell death pathway during hyperoxia-induced lung injury [40–42]. Selective oxidation of CL and PS was found in hyperoxic mouse lung during acute lung injury (72 h, 99.9% of oxygen) (Table 1). MS analysis of CL oxidation products identified CL molecular species containing hydroperoxy-acyls - C18:2-OO (Table 2). PS oxidation products mostly were represented by their molecular species containing C22:6-OO (Table 2).
Recently, using a number of different cell culture models of oxidant- and non-oxidant-induced apoptosis, we demonstrated that selective early oxidation of CL is the hallmark of the mitochondrial stage of apoptotic program [12,43]. During random non-enzymatic lipid peroxidation, phospholipids containing mostly polyunsaturated fatty acid residues such as docosahexaenoic, docosapentaenoic, arachidonic, undergo oxidative attack at much higher rates than those containing linoleic acid residues. This general rule was not fulfilled during CL oxidation in SPAEC treated with LPS, as well as in the lungs of mice exposed to SWCNT or hyperoxia. In these cases, oxygenation occurred predominantly in CL species containing linoleic residues ((C16:0)1/(C18:2)2/(C18:1)1; (C18:0)1/(C18:1)2/(C18:2)1; (C18:0)1/(C18:1)1/(C18:2)1/(C20:4)1; (C18:2)2/(C20:0)1/(C20:0)1; (C16:1)1/(C18:2)1/(C20:4)1/(C22:5)1; (C18:0)1/(C18:2)1/(C20:4)1/(C22:5)1) in spite of the fact that more unsaturated species of CL were also present in these cells/tissues, albeit at lower concentrations [44]. It is tempting to speculate that this selective peroxidation of linoleate-containing species of CL was catalyzed by cyt c during the progression of apoptosis. Binding of cyt c with abundant and oxidizable linoleate-containing CLs defines this preferential peroxidation due to proximity of oxidizable fatty acid residues to reactive intermediates of the peroxidase complex.
In neuronal cells, both CL and PS are enriched with highly oxidizable polyunsaturated molecular species [27,45–47]. Analysis of the total content of hydroperoxides in anionic phospholipids (CL and PS) revealed their robust oxidation in stretched neurons or neuronal cells exposed to STS (Table 1). ESI-MS analysis of CL and PS after exposure of neuronal cells to STS revealed a variety of oxygenated species that contained hydroxy- and hydroperoxy-groups (Table 2). For example, molecular ion at m/z, 1506 corresponded to hydroperoxy-CL molecular species (C16:0)1/(C18:1)1/(C18:2)1/(C22:6+OO)1, originating from an ion with m/z 1474 and corresponding to (C16:0)1/(C18:1)1/(C18:2)1/(C22:6)1 molecular species. PS species containing oxidized C22:6 were also observed in STS-treated neurons (Table 2). MSn analysis of ions with m/z 850; 866 and 882 established that they represented hydroxy-, hydroperoxy-, and hydroxy-hydroperoxy-derivatives of PS and could be fragmented to carboxylate ions with signals at m/z 343; 359 and 375, respectively. These primary PS oxidation products originated from the molecular cluster of PS with m/z 834 (C18:0/C22:6) (Table 2). In the brain and neuronal cells, the abundances of highly unsaturated CL species containing C22:6, C22:5 and C20:4 are comparable or higher than those of C18:2-containing species of CL. Not surprisingly, these highly unsaturated species of CL underwent significant peroxidation. It is likely that cyt c also acted as a major catalyst of these oxidation reactions in which polyunsaturated C22:6 species of CL participated in the formation of cyt c/CL peroxidase complexes.
Important characterization of CL and PS oxidation products can be performed by ESI-MS analysis using lipids extracted from tissues of animals exposed to pro-oxidant, pro-inflammatory, or pro-apoptotic stimuli. For example, CL and PS hydroperoxides were accumulated in the lung of C57BL/6 mice exposed via inhalation to single walled carbon nanotubes (whole body inhalation at 5mg/cm3 for 4 consecutive days, 5 h/day). CL and PS hydroperoxides were detected in the lungs at days 1 and 7 after inhalation (Table 1, 2).
Our previous work identified CL and its oxidation products as important participants and signaling molecules in the intrinsic apoptotic cell death program [12,48]. Early in apoptosis, CL becomes readily available for interactions with cyt c. This interaction results in the formation of cyt c/CL complexes which utilize CL as the preferred oxidation substrate. This results in massive peroxidation of CL in mitochondria [12,27,43,49]. Oxidized molecular species of CL (CL-OOH) triggers the release of cyt c and other pro-apoptotic factors from mitochondria. While a small fraction of the released cyt c is utilized in apoptosome formation, peroxidase catalytic competence of cyt c can be realized in extra-mitochondrial reactions of cyt c with another anionic phospholipid, PS [12]. The peroxidation of PS takes place in plasma membranes before its externalization [50] and functions to generate an essential “eat-me” signal for macrophages to engulf and clear apoptotic cells [51]. Thus, PS oxidation occurs only after oxidation of CL and the release of cyt c into the cytosol but before PS externalization [12]. Given that inhalation of SWCNT induces a markedly increased number of alveolar macrophages reaching a maximum on day 7 post-exposure [30] we suggested that effective clearance of cells with oxidized and externalized PS is mostly responsible for the low steady-state levels of peroxidized PS. At the same time, oxidation of CL in early apoptotic cells precedes their clearance by professional phagocytes. It is possible that quantitative analysis of CL oxidation products may be a specific and robust biomarker of oxidative damage to tissues.
Overall, our results demonstrate that combined employment of ESI-MS and fluorescence HPLC for analysis of phospholipid hydroperoxides can be useful for qualitative assessments, identification of individual molecular species and structural characterization of phospholipids that are involved in oxidative modification in cells and tissues. We found that in CL, isolated from LPS treated SPAEC and lung of mice exposed to hyperoxia, the oxidation of linoleic acid residues was predominant whereas in STS treated or stretched neuronal cells mostly docosahexaenoic acid residues underwent oxidative modification. Further, doubly oxygenated linoleic and docosahexaenoic residues were predominant in MS spectra. Similarly, oxidation of tetra-linoleoyl CL by cyt c in the presence of hydrogen peroxide in a model system yielded mainly doubly oxygenated linoleic acid residues. Significant further work and synthesis of appropriate standards are required for more detailed, quantitative and complete molecular characterization of different products during phospholipid peroxidation in cells and tissues.
Acknowledgments
Supported by grants from NIH HL70755, NIOSH OH008282, HD057587, NS061817, DAMD 17-01-2-637, Pennsylvania Department of Health SAP 4100027294, AHA-0535365N, Human Frontier Science Program, NORA 927Z1LU, NORA NTRC927ZGFY, Project NANOMMUNE No 214281 FP7-NMP-2007-SMALL-1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLAIMERS: The findings and conclusions in this report are those of the authors and do not necessarily represent the view of the National Institute for Occupational Safety and Health.
References
- 1.Kagan VE. Lipid peroxidation in biomembranes. CRC Press: Boca Raton, FL; 1988. [Google Scholar]
- 2.Niki E, Yoshida Y, Saito Y, Noguchi N. Biochem Biophys Res Commun. 2005;338:668. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 3.Fruhwirth GO, Loidl AHA. Biochim Biophys Acta. 2007;1772:718. doi: 10.1016/j.bbadis.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Domingues MR, Reis ADP. Chem Phys Lipids. 2008;156:1. doi: 10.1016/j.chemphyslip.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Megli FM, Russo L. Biochim Biophys Acta. 2008;1778:143. doi: 10.1016/j.bbamem.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 6.Bannenberg G, Arita M, Serhan CN. ScientificWorldJournal. 2007;7:1440. doi: 10.1100/tsw.2007.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong S, Lu Y, Yang R, Gotlinger KH, Petasis NA, Serhan CN. J Am Soc Mass Spectrom. 2007;18:128. doi: 10.1016/j.jasms.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwab JM, Chiang N, Arita M, Serhan CN. Nature. 2007;447:869. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serhan CN. Br J Pharmacol. 2008;153:S200. doi: 10.1038/sj.bjp.0707489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masoodi M, Mir AA, Petasis NA, Serhan CN, Nicolaou A. Rapid Commun Mass Spectrom. 2008;22:75. doi: 10.1002/rcm.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyurina YY, Serinkan FB, Tyurin VA, Kini V, Yalowich JC, Schroit AJ, Fadeel B, Kagan VE. J Biol Chem. 2004;279:6056. doi: 10.1074/jbc.M309929200. [DOI] [PubMed] [Google Scholar]
- 12.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Nature Chem Biol. 2005;4:223. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg ME, Sun M, Zhang R, Febbraio M, Silverstein R, Hazen SL. J Exp Med. 2006;203:2613. doi: 10.1084/jem.20060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maskrey BH, Bermudez-Fajardo A, Morgan AH, Stewart-Jones E, Dioszeghy V, Taylor GW, Baker PR, Coles B, Coffey MJ, Kuhn H, O’Donnell VB. J Biol Chem. 2007;282:20151. doi: 10.1074/jbc.M611776200. [DOI] [PubMed] [Google Scholar]
- 15.Pope S, Land JM, Heales SJR. Biochim Biophys Acta. 2008;1777:794. doi: 10.1016/j.bbabio.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Yin H, Porter NA. Antioxidants Redox Signal. 2005;7:170. doi: 10.1089/ars.2005.7.170. [DOI] [PubMed] [Google Scholar]
- 17.Girotti AW. Free Radic Biol Med. 2008;44:956. doi: 10.1016/j.freeradbiomed.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison KA, Davies SS, Marathe GK, McIntyre T, Prescott S, Reddy KM, Falck JR, Murphy RC. J Mass Spectrom. 2000;35:224. doi: 10.1002/(SICI)1096-9888(200002)35:2<224::AID-JMS933>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 19.Pulfer M, Murphy RC. Mass Spectrometry Reviews. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 20.Hermansson M, Uphoff A, Kkel R, Somer P. Anal Chem. 2005;77:2166. doi: 10.1021/ac048489s. [DOI] [PubMed] [Google Scholar]
- 21.Peterson BL, Cummings BS. Biomed Chromatogr. 2006;20:227. doi: 10.1002/bmc.563. [DOI] [PubMed] [Google Scholar]
- 22.Hou W, Zhou H, Elisma F, Bennett AAL, Figeys D. 2008;7:395. doi: 10.1093/bfgp/eln042. [DOI] [PubMed] [Google Scholar]
- 23.Roberts LD, McCombie G, Titman CM, Griffin JL. Journal of Chromatography B. 2008;871:174–181. doi: 10.1016/j.jchromb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Blée E, Wilcox AL, Marnett LJ, Schuber F. J Biol Chem. 1993;268:1708. [PubMed] [Google Scholar]
- 25.Catalá A. Chem Phys Lipids. 2009;157:1. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Du L, Bayir H, Lai Y, Zhang X, Kochanek PM, Watkins SC, Graham SH, Clark RS. J Biol Chem. 2004;279:38563–38570. doi: 10.1074/jbc.M405461200. [DOI] [PubMed] [Google Scholar]
- 27.Tyurin VA, Tyurina YY, Feng W, Mnuskin A, Jiang J, Tang M, Zhang X, Zhao Q, Kochanek PM, Clark RSB, Bayir H, Kagan VE. J Neurochem. 2008;107:1614. doi: 10.1111/j.1471-4159.2008.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusardi TA, Wolf JA, Putt ME, Smith DH, Meaney DF. J Neurotrauma. 2004;21:61. doi: 10.1089/089771504772695959. [DOI] [PubMed] [Google Scholar]
- 29.Hoyt DG, Mannix RJ, Rusnak JM, Pitt BR, Lazo JS. Am J Physiol (Lung Cell Mol Physiol) 1995;13:L171. doi: 10.1152/ajplung.1995.269.2.L171. [DOI] [PubMed] [Google Scholar]
- 30.Shvedova AA, Kisin E, Murray AR, Johnson VJ, Gorelik O, Arepalli S, Hubbs AF, Mercer RR, Keohavong P, Sussman N, Jin J, Yin J, Stone S, Chen BT, Deye G, Maynard A, Castranova V, Baron PA, Kagan VE. Am J Physiol Lung Cell Mol Physiol. 2008;295:L552. doi: 10.1152/ajplung.90287.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folch J, Lees M, Sloan-Stanley GH. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 32.Rouser G, Fkeischer S, Yamamoto A. Lipids. 1970;5:494. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- 33.Böttcher CSF, Van Gent CM, Fries C. Anal Chim Acta. 1961;24:203. [Google Scholar]
- 34.Hsu FF, Turk J, Rhoades ER, Russell DG, Shi YX, Groisman EA. J Am Soc Mass Spectr. 2004;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 35.MacMillan DK, Murphy RC. J Am Soc Mass Spectrom. 1995;6:1190. doi: 10.1016/1044-0305(95)00505-6. [DOI] [PubMed] [Google Scholar]
- 36.Kerwin JL, Wiens AM, Ericsson LH. J Mass Spectrometry. 1996;31:184. doi: 10.1002/(SICI)1096-9888(199602)31:2<184::AID-JMS283>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Haefliger OP, Sulzer JW. Chromatographia. 2007;65 [Google Scholar]
- 38.Schneider C, Schreier P, Herderich M. Lipids. 1997;32:331. doi: 10.1007/s11745-997-0041-0. [DOI] [PubMed] [Google Scholar]
- 39.Lang I, Gobel C, Porzel A, Heilmann I, Feussner I. Biochem J. 2008;410:347. doi: 10.1042/BJ20071277. [DOI] [PubMed] [Google Scholar]
- 40.Freeman BA, Crapo JD. J Biol Chem. 1981;256:10986–10992. [PubMed] [Google Scholar]
- 41.Mantell LL, Horowitz S, Davis JM, Kazzaz JA. Ann N Y Acad Sci. 1999;887:171– 180. doi: 10.1111/j.1749-6632.1999.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 42.Pagano A, Donati Y, Metrailler I, Argiroffo CB. Am J Physiol (Lung Cell Mol Physiol) 2004;30:L275. doi: 10.1152/ajplung.00181.2003. [DOI] [PubMed] [Google Scholar]
- 43.Tyurina YY, Tyurin VA, Epperly MW, Greenberger JS, Kagan VE. Free Radic Biol Med. 2008;44:299. doi: 10.1016/j.freeradbiomed.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 44.Tyurina YY, Tyurin VA, Kapralova VI, Amoscato AA, Epperly MW, Greenberger JS, Kagan VE. Meth Mol Biol. 2009 doi: 10.1007/978-1-60761-325-1_9. (in press) [DOI] [PubMed] [Google Scholar]
- 45.Bayir H, Tyurin VA, Tyurina YY, Viner R, Ritov V, Amoscato AA, Zhao Q, Zhang XJ, Janesko-Feldman KL, Alexander H, Basova LV, Clark RS, Kochanek PM, Kagan VE. Ann Neurol. 2007;62:154. doi: 10.1002/ana.21168. [DOI] [PubMed] [Google Scholar]
- 46.Cheng H, Mancuso DJ, Jiang X, Guan S, Yang J, Yang K, Sun G, Gross RW, Han X. Biochemistry. 2008;47:5869. doi: 10.1021/bi7023282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiebish MA, Han X, Cheng H, Lunceford A, Clarke CF, Moon H, Chuang JH, Seyfried TN. J Neurochem. 2008;106:299. doi: 10.1111/j.1471-4159.2008.05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tyurin VA, Tyurina YY, Kochanek PM, Hamilton R, DeKosky ST, Greenberger JS, Bayir H, Kagan VE. Methods Enzymol. 2008;442:375. doi: 10.1016/S0076-6879(08)01419-5. [DOI] [PubMed] [Google Scholar]
- 49.Belikova NA, Jiang J, Tyurina YY, Zhao Q, Epperly MW, Greenberger J, Kagan VE. Int J Radiat Oncol Biol Phys. 2007;69:176. doi: 10.1016/j.ijrobp.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 50.Tyurina YY, Kawai K, Tyurin VA, Liu SX, Kagan VE, Fabisiak JP. Antioxid Redox Signal. 2004;6:209. doi: 10.1089/152308604322899288. [DOI] [PubMed] [Google Scholar]
- 51.Savill J, Gregory C, Haslett C. Science. 2003;302:1516. doi: 10.1126/science.1092533. [DOI] [PubMed] [Google Scholar]