Abstract
Although extracellular application of lysophosphatidic acid (LPA) has been extensively documented to produce a variety of cellular responses through a family of specific G protein-coupled receptors, the in vivo organismal role of LPA signaling remains largely unknown. The first identified LPA receptor gene, lpA1/vzg-1/edg-2, was previously shown to have remarkably enriched embryonic expression in the cerebral cortex and dorsal olfactory bulb and postnatal expression in myelinating glia including Schwann cells. Here, we show that targeted deletion of lpA1 results in approximately 50% neonatal lethality, impaired suckling in neonatal pups, and loss of LPA responsivity in embryonic cerebral cortical neuroblasts with survivors showing reduced size, craniofacial dysmorphism, and increased apoptosis in sciatic nerve Schwann cells. The suckling defect was responsible for the death among lpA1(−/−) neonates and the stunted growth of survivors. Impaired suckling behavior was attributable to defective olfaction, which is likely related to developmental abnormalities in olfactory bulb and/or cerebral cortex. Our results provide evidence that endogenous lysophospholipid signaling requires an lp receptor gene and indicate that LPA signaling through the LPA1 receptor is required for normal development of an inborn, neonatal behavior.
Lysophosphatidic acid (LPA) and sphingosine-1-phosphate are components of serum responsible for proliferative and Rho-mediated morphological effects on cells in culture (1–4). These compounds also induce numerous other cellular effects, including neurotransmitter release, tumor cell invasion, chemotaxis, changes in ion conductance, changes in gap- and tight-junction communication, and inhibition of both cell differentiation and apoptosis (reviewed in refs. 5 and 6). We previously identified the lysophospholipid (“lp”) receptor gene (lpA1/vzg-1/Edg2) that showed enriched expression in proliferating regions of the embryonic cerebral cortex and developing olfactory bulb (7, 8). Subsequent studies led to the identification of seven additional related lp/Edg receptor genes by ourselves and other groups (reviewed in refs. 9 and 10). The primary evidence that these genes encode specific LPA or sphingosine-1-phosphate receptors comes from heterologous expression studies where specific receptor binding and cellular responsivity to LPA or sphingosine-1-phosphate is conferred (e.g., refs. 11–13).
Despite identification of lp receptor genes, there remains no evidence that any lp gene is required for normal developmental or physiological processes. Clarification of this issue is of particular importance in view of the postulated second messenger roles of some lysophospholipid species (6). However, the expression pattern of each gene suggests potential in vivo functions. A role for lpA1 in cerebral cortical neurogenesis was suggested by its specific expression pattern in the embryonic ventricular zone, a location where cell proliferation and neurogenesis occurs (7). Cell lines derived from this region respond to LPA with morphological changes including neurite retraction/cell rounding (7). In addition, primary cultured embryonic cortical neuroblasts respond to LPA with ionic conductance and morphological changes (14, 15). These results implicated lpA1 and LPA signaling in the normal biology of cortical neuroblasts. A second function for lpA1 was suggested by its expression in oligodendrocytes of the postnatal brain and in Schwann cells that are found in peripheral nerves such as the sciatic nerve (8, 16); these glia are the myelinating cells of the nervous system. In vitro studies demonstrated that LPA acted as a survival factor for Schwann cells in culture, probably through the LPA1 receptor (16). To examine the actual biological functions of lpA1 and LPA signaling in intact animals, we generated mice with a homozygous null lpA1 mutation and analyzed them for LPA signaling defects and developmental abnormalities.
Materials and Methods
Targeting lpA1.
Mouse 129SvJ genomic clone isolation, characterization, and subcloning were described previously (17). A 5.7-kb EcoRI/NotI genomic subclone containing exon 3 was used as a source for fragments to insert into the pFlox vector (18). Cloning took place in three steps: (i) the 3.5-kb EcoRI/XbaI 3′ “long arm” from clone vzg5.5 was ligated into the SalI/XbaI sites of pFlox, (ii) the 1.0-kb BglII/BamHI from clone BX1.2 containing exon 3 was ligated into the BamHI site, and then (iii) the 1.1-kb SalI/SalI fragment from clone BN1.1 was ligated into the XhoI site. After checking this plasmid by PCR, Southern blotting, and sequencing, a 10-kb NotI/NotI linear fragment was isolated, and 20 μg were electroporated into R1 embryonic stem cells. Both homologous recombination and Cre-mediated deletion were detected by PCR initially and confirmed by Southern blot analysis. Of 228 G418-resistant colonies screened, six were positive for homologous recombination (efficiency 1:38 or 2.6%). Two of these six were electroporated with a Cre-expression plasmid. Of 72 gancyclovir-resistant clones analyzed, 10 had deleted both exon 3 and neomycin-resistant/thymidine kinase genes (type I deletion). One type I clone (28–5#11) was confirmed to be euploid and was injected into blastocysts, resulting in chimeric mice backcrossed with C57BL/6J females. Intercrosses were begun immediately with these mice, as well as with mice generated from one additional backcross.
Southern Blot and PCR Genotyping of Knockout Mice.
To genotype mice, Southern blots were made using BglII-digested genomic DNA (gDNA) isolated from embryonic or P21 tails. The 650-bp PX650 probe was obtained from the XX1.5 plasmid by digestion with PvuII and XbaI (17). The exon 3 probe was generated by PCR from a cDNA plasmid using primers 513 M and 513T (17). For PCR genotyping of mice, 0.4 μl (100–200 ng) of gDNA was added as template in hot-start 20 μl of PCR, then cycled 35 times (95°C, 30 s; 56°C, 30 s; and 72°C, 2 min) with a final incubation of 72°C for 5 min before cooling to 4°C. The PCR mix consisted of 60 mM Tris (pH 8.5)/15 mM (NH4)2SO4/2.0 mM MgCl2/0.5 mM each oligonucleotide/0.25 mM each dNTP/0.4 U Taq. Primers in the PCR mix were as follows: 513QL, 5′-GCCAATCCAGCGAAGAAGTC-3′; vzg.il1, 5′-GGTATTCTTAATTCTAGAGGATCAGC-3′; vzg.is2, 5′-TATAGGAGTCTTGTGTTGCCTGTCC-3′.
Northern Blots and Reverse Transcriptase–Polymerase Chain Reaction (RT-PCR).
Northern blotting and RT-PCR conditions have been described previously (16, 19). The number of cycles for all of the PCR shown were adjusted so product was not maximized, as compared with quantities obtained with excess product as template. All cDNA template amounts were standardized to yield identical quantities of β-actin product, which was amplified with primers that do not amplify any pseudogenes from gDNA (20). All reactions used primers located in different exons, eliminating the possibility of gDNA contamination. Primers used were the following: lpA1, 513C/513T (17); lpA2, edg6f (5′-GACCACACTCAGCCTAGTCA-3′) and edg6e3c′ (5′-ACATCCAGAGATAACACAGTAA-3′); lpB1, edg1p (5′-CCGTCAGTCGCCGACAACAA-3′) and edg1b (5′-GTAGAGGATGGCGATGGAAA-3′).
Analysis of Cerebral Cortical Cell Cultures.
Cortical cultures were made essentially as described (14, 15). Briefly, the cerebral cortices from embryonic (E) day 12–13 mice were dissected, and meninges were carefully removed. Tissues were transferred to culture medium and gently triturated with a fire-polished Pasteur pipette into small clusters (<100 cells per cluster). Cells were seeded onto glass coverslips (50–200 clusters on 12-mm coverslips or 5–20 clusters on the center of 40-mm coverslips) precoated with 2 μg/cm2 of Cell-Tak (Becton Dickinson) and cultured at 37°C under 5% CO2 for 18 h. For time-lapse video-enhanced differential interference contrast microscopy, the coverslips were mounted onto a heat-controlled perfusion chamber (BioOptiks) set at 37°C and observed with an inverted microscope (Axiovert 135, Carl Zeiss). Either vehicle (0.1% fatty acid-free BSA) or LPA was added for 15 min. The differential interference contrast images were detected with a cooled charge-coupled device color camera (DEI-47, Carl Zeiss). Cluster area was determined using Scion Image software, with changes calculated as percentage of initial area (t = 0 min). To detect proliferation, cultures were treated with or without LPA for 18 h and pulsed with 40 μM BrdUrd for the last 30 min, followed by staining with α-BrdUrd antibody (Boehringer Mannheim). Only cells in clusters were counted.
Analysis of Sciatic Nerves.
For apoptotic DNA in situ end-labeling plus (ISEL+; ref. 21), sciatic nerves (eight nerves from four different animals of each genotype) were isolated from young adult mice and rapidly frozen in Tissue Tek (Miles). Cryostat sections were cut at 10 μm and then fixed, processed, and labeled as described (21). Microscope fields (×100 objective) were counted along the entire length of each nerve section (>16,000 cells total for each genotype). For electron microscopy, sciatic nerves from adult mice were removed, and ≈2- to 3-mm pieces were immersed in 3% glutaraldehyde in 0.1 M cacodylate buffer at 4°C for 24 h. These were embedded in Scipoxy 812 resin (Energy Beam Sciences, Agawan, MA), and 80-nm sections were cut and stained with saturated ethanol/uranyl acetate and bismuth subnitrate. Grids were examined and photographed using a Zeiss electron microscope.
Results and Discussion
Targeted Deletion of lpA1.
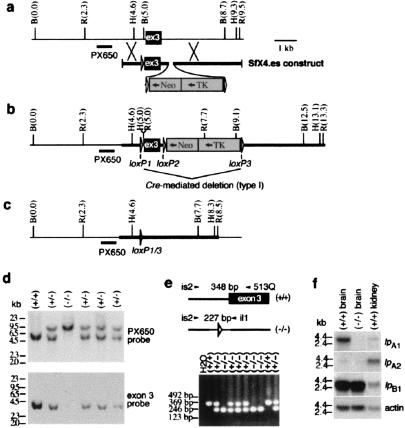
Mouse lpA1 consists of at least 5 exons, with 68% of the coding region (including transmembrane domains I–VI) located in exon 3 (17). We therefore targeted exon 3 for deletion, using the Cre-lox system, which allows embryonic stem cells to be generated without a residual selectable marker gene and its associated constitutive promoter (18). In the event that exons 2 and 4 were spliced together, all coding sequence contributed from exon 4 would be out of frame. Mutant embryonic stem cell clones were obtained by homologous recombination of a replacement vector containing loxP sites flanking both exon 3 and the neomycin-resistant/thymidine kinase genes (Fig. 1 a and b). Expression of Cre recombinase in one of these clones allowed deletion of exon 3 and neomycin-resistant/thymidine kinase (Fig. 1c). After injection into blastocysts, chimeric mice were mated to C57BL/6J mice and progeny intercrossed. We found that some lpA1(−/−) mice could survive and reproduce, as determined by verification of homozygosity through Southern blot and PCR analyses of gDNA (Fig. 1 d and e). Northern blot analysis of lpA1(−/−) adult brain RNA confirmed the lack of exon 3 expression (Fig. 1f). Because the up-regulation of related LPA receptor genes might compensate for the absence of lpA1, we determined whether transcript levels of lpA2/edg-4 (10, 17, 19, 22, 23), lpA3/edg-7 (refs. 10, 23, and 24; data not shown), or lpB1/edg-1 (25, 26) were altered in lpA1(−/−) adult brain. No changes were observed in expression levels of any of these genes (Fig. 1f).
Figure 1.
Targeted disruption of lpA1 by homologous recombination. (a) Map of the wild-type lpA1 locus including exon 3 (ex3, black box), the targeting construct, and location of probe (PX650) used for Southern blot. B, BglII; R, EcoRI; H, HindIII. (b) Map of the initial homologously recombined locus. (c) Map of the homologously recombined locus after transient expression of Cre recombinase in an embryonic stem cell clone has deleted all DNA between loxP1 and loxP3 sites. (d) Southern blot analysis of BglII-digested genomic DNA (10 μg per lane) from mice generated by crossing heterozygotes. After initial hybridization with PX650, the same blot was stripped and rehybridized with an exon 3 probe. (e) PCR assay containing three primers (vzg.is2, 513Q, and vzg.il1) detecting inheritance of wild-type lpA1 (348 bp) and deletion mutation (227 bp). (f) Northern blot (20 μg per lane) of RNA from adult brain probed with the indicated gene (wild-type kidney shown as positive control for lpA2 and lpB1; only an exon 3 probe was used for lpA1).
Gross Abnormalities in lpA1(−/−) Mice.
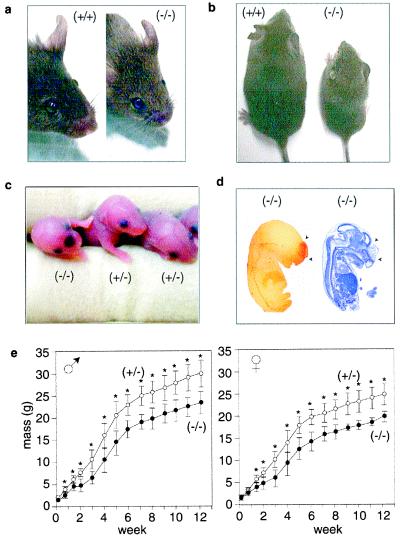
Juvenile and adult lpA1(−/−) mice could always be distinguished from their nonhomozygous siblings by their shorter snouts, more widely spaced eyes, and smaller size (Fig. 2 a and b). A quantitative assessment of craniofacial deformity in adults using a measure independent of overall head size (eye-to-nose tip length/interocular distance) supported qualitative impressions (lpA1(−/−): 1.04 ± 0.16 vs. lpA1(+/−): 1.47 ± 0.05; P < 0.05; unpaired t test). X-ray analysis of skeletal structures in adult lpA1(−/−) mice revealed only relative size differences in the frontal area rather than missing bones, cleft palate, or other abnormalities (data not shown). The underlying cause of the dysmorphism is likely related to loss of lpA1 that is normally expressed within developing facial bone tissue (C. McGiffert and J.C., unpublished observation). An additional factor may be frontal hematomas observed in a small fraction (4 of 160) of embryonic and neonatal lpA1(−/−) mice (Fig. 2 c and d) because neonatal pups with such hematomas showed more pronounced craniofacial dysmorphism later in life. Hematomas might be caused by defects in LPA-induced endothelial cell monolayer barrier formation (27) or a localized role in angiogenesis similar to that hypothesized for LPB receptors (28). The second obvious gross phenotype of adult lpA1(−/−) mice, smaller size, was found to be significant from several days after birth and into adulthood for both males and females, with up to a 30% reduction relative to control siblings (Fig. 2e). This primarily reflected size differences in all structures because of decreased postnatal growth rate (Fig. 2b). However, lower relative amounts of fat also contributed to the total mass differences (data not shown), possibly because of defective LPA signaling in adipose cell development (29).
Figure 2.
Gross abnormalities in lpA1(−/−) mice. (a) Craniofacial deformity in adult lpA1(−/−) mice. Note the blunted snout and more widely spaced eyes. (b) Overall size decrease in lpA1(−/−) mice. Two 9-week-old male siblings are shown. (c) Frontal cranial hemorrhage in a newborn lpA1(−/−) pup but not in littermates. (d) Sagittal sections of an E13 lpA1(−/−) embryo with frontal cranial hemorrhage (the earliest time point this phenotype was observed). Sections shown were stained to identify erythrocytes (left) using peroxidase (pseudocolored red) or Nissl substance (right) using cresyl violet. (e) Total body mass measurements for lpA1(−/−) mice (●) and control lpA1(+/−) littermates (○) from lpA1(+/−) X lpA1(+/−) crosses. Data points are means ± SD; *, P < 0.05 (repeated measures were analyzed with ANOVA). Mass measurements of lpA1(+/+) littermates overlapped those shown for lpA1(+/−) and are not presented for clarity.
Increased Apoptosis in lpA1(−/−) Sciatic Nerve.
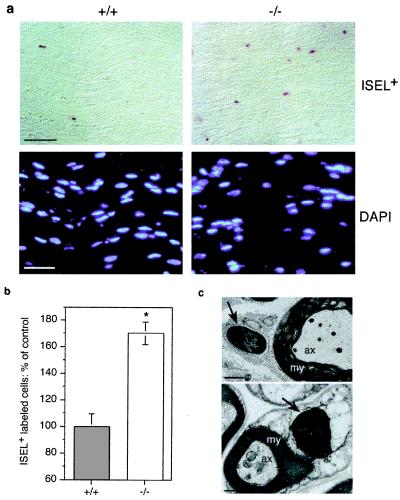
To examine whether there might be a defect in Schwann cell survival in vivo, we examined young adult sciatic nerve sections for cells undergoing apoptosis using ISEL+ that labels fragmented DNA (21). These analyses demonstrated that there was an approximately 80% increase in the percentage of ISEL+-labeled cells in lpA1(−/−) mice compared with wild-type mice (Fig. 3 a and b). The presence of apoptotic cells was confirmed by observing condensed nuclei using electron microscopy (Fig. 3c). In addition to increased apoptosis of Schwann cells in vivo, fewer cells were also apparent in the sections (Fig. 3a). In support of an LPA signaling defect as the primary cause of the increased apoptosis rates, we determined that cultured Schwann cells had severely diminished LPA responses in a morphological assay (J.A.W., N.F., and J.C., unpublished observation). Despite these defects, we did not observe grossly abnormal movement in lpA1(−/−) mice, which might be expected from severe myelination defects. This may reflect a relatively minor extent of Schwann cell/myelin loss because the actual increase in Schwann cell apoptosis (18% in lpA1(−/−) mice as opposed to 10% in wild-type mice) still leaves the majority of Schwann cells intact.
Figure 3.
Increased apoptosis and decreased Schwann cell numbers in lpA1(−/−) sciatic nerves. (a) Sections of young adult lpA1(−/−) sciatic nerves were stained for fragmented DNA (using ISEL+) or nuclei (using DAPI). (b) Quantified apoptosis increases in lpA1(−/−) sciatic nerves. Bars are means ± SEM from four animals of each genotype; *, P < 0.005 (unpaired t test). (c) Electron microscopic confirmation of Schwann cell apoptosis in lpA1(−/−) sciatic nerves. Note black nuclei indicating chromatin condensation (arrows). ax, axon; my, myelin sheath. [Bars = 40 μm (a), 1 μm (c, top), 0.6 μm (c, bottom).]
Neonatal Death in lpA1(−/−) Mice.
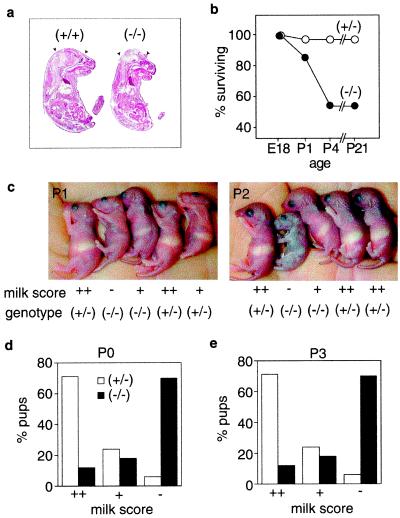
Genotyping litters from both lpA1(+/−) × lpA1(+/−) and lpA1(+/−) × lpA1(−/−) crosses demonstrated that about half of both male and female lpA1(−/−) mice died sometime before 3 weeks of age (Table 1). This effect was not caused by a homozygous maternal phenotype because the semilethality was similar for both lpA1(+/−) and lpA1(−/−) mothers (Table 1 and data not shown). To determine whether pups were dying embryonically, we genotyped litters before birth, E12–18. Here, the expected ratios were present, indicating pups were being lost sometime between late embryonic development and 3 weeks of age (Table 1). During this analysis, we observed a small fraction (3 of 61) of lpA1(−/−) embryos with exencephaly (Fig. 4a). Exencephaly is a neonatal lethal phenotype that is also found in a small fraction of other null-mutant mice (30). Although we have no explanation for this effect, it does suggest that LPA signaling may have a previously unrecognized role in neural tube closure. Whereas one might speculate that the lpA1(−/−) lethality is related to frontal hematomas, we observed that several newborn pups with frontal hematomas survived, with hematomas dissipating after several days.
Table 1.
Inheritance of the lpA1 deletion allele
| Parental genotypes | Offspring
genotypes
|
|||
|---|---|---|---|---|
| +/+ | +/− | −/− | Expected −/− | |
| At weaning (approximately P21) | ||||
| +/+ × +/− | 206 (49%) | 216 (51%) | 0 (0%) | 0 |
| +/− × +/− | 89 (28%) | 194 (60%) | 39 (12%) | 94 |
| +/− × −/− | 0 (0%) | 166 (62%) | 99 (38%) | 166 |
| Sex ratios (female:male) | 140:155 | 280:296 | 63:75 | |
| Embryonically (E12–18) | ||||
| +/− × +/− | 41 (25%) | 80 (49%) | 41 (25%) | 41 |
| +/− × −/− | 0 (0%) | 27 (51%) | 26 (49%) | 27 |
Numbers of individual genotyped progeny from the indicated crosses are shown (P, postnatal day; E, embryonic day). Indicated in parentheses is the percentage (rounded to the nearest percent) of each genotype from a particular cross. The results demonstrate that sometime between birth (P0, which occurs at E19) and 3 weeks of age, approximately half of the lpA1(−/−) mice are lost.
Figure 4.
Neonatal deaths and impaired suckling in lpA1(−/−) mice. (a) Sagittal sections of an E18 exencephalic lpA1(−/−) embryo and control littermate stained with hematoxylin/eosin. Note disorganized brain structure and lack of skull (arrowheads). (b) Survival plot for lpA1(−/−) mice and control lpA1(+/−) littermates from late embryonic ages through weaning (n = 38 for lpA1(−/−) pups; n = 37 for lpA1(+/−) pups). (c) Observed lack of milk in stomachs of dead and/or dying lpA1(−/−) pups. Relative milk quantities in bellies were scored either as full/near full (++), intermediate (+), or empty/near empty (−). Note dead lpA1(−/−) pup at P2. (d) Pup milk scores at P0 (8–20 h after birth) or (e) P3 correlated with genotype (n = 34–38 for lpA1(−/−) pups; n = 37 for lpA1(+/−) pups).
Suckling Defects in lpA1(−/−) Neonatal Pups.
To determine the primary cause of lpA1(−/−) deaths, 10 litters from lpA1(+/−) × lpA1(−/−) crosses were observed from just after birth through weaning. A survival plot is presented in Fig. 4b. Of the 38 lpA1(−/−) pups genotyped [approximately the expected number based on 37 lpA1(+/−) littermates], six were dead immediately after birth. Two of these were partially cannibalized by the time of observation (possibly related to exencephaly), whereas the other four did not commence normal breathing (but were otherwise grossly normal). Twelve additional lpA1(−/−) pups died within the first 4 postnatal days, and all of them lacked milk in their stomachs (e.g., Fig. 4c). Observation of these pups before their death indicated that they were not suckling. The craniofacial dysmorphism was not apparent in the dead lpA1(−/−) pups (nor any neonatal pups) and thus was not causally related to the deaths. To determine whether the impaired suckling phenotype was present in all lpA1(−/−) pups rather than a subset, milk quantity was correlated with genotype at two postnatal ages (P0 and P3). The majority of lpA1(−/−) pups had little or no milk in their stomachs, unlike control lpA1(+/−) littermates, which normally had full stomachs (Fig. 4 c and d). Thus, the suckling defect was present in nearly all lpA1(−/−) pups. This result can explain the decreased size of surviving lpA1(−/−) pups relative to control littermates and is consistent with results from olfactory bulbectomized pups or mouse olfactory mutants (e.g., Golf and cyclic nucleotide-gated channel mutants) that together show suckling defects, decreased size, and partial lethality (31–33).
Neonatal suckling in rats and mice is a complex behavior that depends on several brain structures as well as nerves and muscles required to extract milk from the nipple (31, 34, 35). Olfactants/pheromones present on the nipple are the primary sensory cue used in orientation and localization, although tactile sensation is also required. Once a nipple is located, a rooting reflex program is activated, which involves rhythmic mouth suckling movements and swallowing. Structures involved in the overall behavior include the olfactory epithelia, olfactory bulb, various cranial ganglia, the nerves/muscles involved in rhythmic mouth contractions and swallowing, and perhaps the vomeronasal organ and olfactory cortex (entorhinal/pyriform cortex and amygdala). To determine more precisely what behavioral defect impaired suckling in lpA1(−/−) pups, we observed their nipple searching and tactile reflexes. All lpA1(−/−) pups exhibited a normal rooting reflex in response to manual stimulation of their mouth region, indicating normal tactile sensation and motor control. Although we found that lpA1(−/−) pups searched for nipples normally, they often did not find them, similar to the phenotype observed with Golf and cyclic nucleotide-gated channel mutant mice (32, 33) (data not shown). These results indicated that the impaired suckling was induced by a lack of olfactant detection and/or processing.
Olfactant detection and processing involves olfactory epithelia, bulb, and possibly cortex. To determine whether general histological abnormalities in these structures might explain the impaired behavior, we perfusion-fixed neonatal lpA1(−/−) pups that had little milk in their stomachs and stained coronal head sections with cresyl violet. These studies did not reveal obvious abnormalities in olfactory/vomeronasal epithelia, olfactory bulb, or cortex, except occasionally, slightly smaller cortical width (data not shown). Expression of lpA1 in the embryonic brain is most prominent in the dorsal cerebral cortex (7) but also extends into the anterior aspect of the dorsal telencephalon that in part gives rise to the olfactory bulb (8). Although no clear differences between lpA1(+/+) and lpA1(−/−) brain histological sections assayed for cells in S-phase or apoptosis were observed, several embryonic lpA1(−/−) brains/olfactory bulbs analyzed showed reduced wall thickness (data not shown).
Lack of Normal LPA but Increased Basic Fibroblast Growth Factor (bFGF) Responsivities in lpA1(−/−) Neuroblasts.
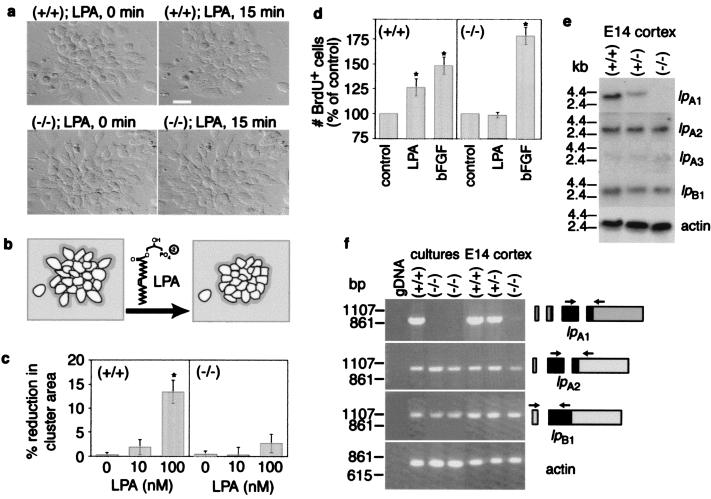
To investigate the possible mechanism of a growth defect in embryonic neuroblasts, we examined whether LPA signaling was normal. Additionally, there has been some debate regarding the identity of the lpA genes as encoding endogenous LPA receptors, especially in view of a report implicating a molecularly dissimilar LPA receptor gene (PSP24) (36). Primary cultures from both lpA1(−/−) and lpA1(+/+) embryonic (E12–13) cerebral cortex were examined for LPA morphological and proliferative responses. Proliferating cells in these cultures existed predominantly in small clusters (Fig. 5a) and were immunoreactive for the intermediate filament protein, nestin (data not shown), consistent with being neuroblasts (37). Wild-type cultures responded to 100 nM LPA with cluster compaction (quantifiable as a reduction in cluster area) (15), reflecting both cell migration toward, and cell rounding within, clusters (Fig. 5 a and b). By comparison, lpA1(−/−) clusters exposed identically to LPA were severely attenuated in this response, with quantified area changes not significantly different from vehicle-treated controls (Fig. 5 a and c). A second LPA response, increased cell proliferation, was analyzed by treatment with vehicle solution or 100 nM LPA for 18 h followed by labeling with BrdUrd for the last 30 min. Although cells from lpA1(+/+) embryos responded to LPA with a 25% increase in BrdUrd incorporation relative to vehicle treatment, lpA1(−/−) cells did not respond (Fig. 5d). Although both lpA1(+/+) and lpA1(−/−) cultures exhibited proliferative responses to bFGF stimulation as expected (38), lpA1(−/−) cultures showed increased responsivity (Fig. 5d). These data indicate that LPA1 mediates endogenous LPA effects in cerebral cortical neuroblasts and suggest that loss of normal LPA-dependent proliferation in lpA1(−/−) cortex may in part be compensated for by increased proliferative responses to bFGF.
Figure 5.
Lack of normal LPA responsiveness in lpA1(−/−) cortical neuroblast cultures and expression of additional LPA receptor gene transcripts. (a) Time-lapse differential interference contrast images of cortical neuroblasts treated for 15 min with 100 nM LPA. (Bar = 20 μm) (b) Schematic diagram of observed cluster compaction in response to LPA and measured areas (inside dark outline of cluster). (c) Quantified relative changes in neuroblast cluster culture areas. Data show mean ± SEM; n >15 embryos of each genotype. *, P < 0.05 (ANOVA with Fisher's post hoc test). (d) Effects of 18-h LPA (100 nM) or bFGF (20 ng/ml) treatment on neuroblast proliferation as measured by BrdUrd incorporation. Data show mean ± SEM; n >12 embryos of each genotype. *, P < 0.05 (ANOVA with Fisher's post hoc test). (e) Northern blot detection of lpA1, lpA2, lpA3, and lpB1 in E14 cortex. (f) RT-PCR detection of lpA1, lpA2, and lpB1 in cortical cultures and E14 brains. Relative locations of primers in each gene are indicated to the right (boxes indicate exons; black areas indicate coding region). lpA3 was not readily detectable in the cultures by RT-PCR. Linear range β-actin RT-PCR amplification products are shown as a control for template cDNA quantities.
The existence of two homologous high-affinity receptors (LPA2 and LPA3) and a putative low-affinity (LPB1/Edg-1) LPA receptor (39) suggests that each may mediate distinct LPA-dependent functions. Thus, we examined expression of germane lp receptor genes in the mutant embryonic cerebral cortex and cortical cell cultures. Interestingly, despite the loss of LPA-dependent cell proliferation in these cultures, we detected relatively abundant transcript levels of lpA2 and lpB1 and low levels of lpA3 (Fig. 5 e and f). The expression of these genes suggests that other LPA responses might still be found in some lpA1(−/−) cells, and in fact, we have observed LPA-induced, distinct morphological changes in other cell subtypes from lpA1(−/−) brain (N.F. and J.C., unpublished observation). Such differing effects mediated by LPA receptor subtypes are supported by comparative expression studies of LP receptors in neuronal cell lines (23). There was no obvious change in expression level of any of the examined receptors in lpA1(−/−) cells (Fig. 5 e and f). These data indicate that specific, nonredundant LPA effects are mediated by the LPA1 receptor.
We have demonstrated using genetics that lpA1 is required for normal neonatal suckling behavior. The defect in suckling is responsible for both the decreased size of the mice and the majority of lethality associated with the homozygous lpA1 null allele. Enriched expression of lpA1 in the dorsal telencephalic proliferative zone and loss of LPA responsivity in cells from this region suggest that altered development of the cerebral cortex and/or olfactory bulb may be responsible for the suckling defect. Increases in Schwann cell apoptosis within the intact sciatic nerve of lpA1 null animals lend further support for LPA signaling in the survival of myelinating cells. Overall, our results provide the first evidence that endogenous LPA signaling is receptor mediated, that a single lp receptor gene can have nonredundant biological roles, and that receptor-mediated LPA signaling is required for normal developmental processes.
Acknowledgments
We thank Drs. Colin Fletcher and Joe Gleeson for critically reading the manuscript, the lab of Dr. Jamey Marth for providing the pFlox vector and helpful discussions on the targeting protocol, Forrest Liu at the University of California, San Diego, core facility for embryonic stem cell work and blastocyst injections, Carol Akita for the maintenance of mice, and Casey Cox for copyediting the manuscript. This work was funded by the National Institute of Mental Health (to J.C., J.J.A.C., and J.A.W.), the National Science Foundation (to D.K.), the Lucille P. Markey Charitable Trust (to J.J.A.C.), the Uehara Foundation (to N.F.), and an institutional award from the Howard Hughes Medical Institute (to J.C.).
Abbreviations
- LPA
lysophosphatidic acid
- gDNA
genomic DNA
- ISEL+
in situ end-labeling plus
- RT-PCR
reverse transcriptase–polymerase chain reaction
- bFGF
basic fibroblast growth factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.van Corven E J, Groenink A, Jalink K, Eichholtz T, Moolenaar W H. Cell. 1989;59:45–54. doi: 10.1016/0092-8674(89)90868-4. [DOI] [PubMed] [Google Scholar]
- 2.Ridley A J, Hall A. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 3.Eichholtz T, Jalink K, Fahrenfort I, Moolenaar W H. Biochem J. 1993;291:677–680. doi: 10.1042/bj2910677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatomi Y, Igarashi Y, Yang L, Hisano N, Qi R, Asazuma N, Satoh K, Ozaki Y, Kume S. J Biochem (Tokyo) 1997;121:969–973. doi: 10.1093/oxfordjournals.jbchem.a021681. [DOI] [PubMed] [Google Scholar]
- 5.Moolenaar W H, Kranenburg O, Postma F R, Zondag G C. Curr Opin Cell Biol. 1997;9:168–173. doi: 10.1016/s0955-0674(97)80059-2. [DOI] [PubMed] [Google Scholar]
- 6.Spiegel S. J Leukocyte Biol. 1999;65:341–344. doi: 10.1002/jlb.65.3.341. [DOI] [PubMed] [Google Scholar]
- 7.Hecht J H, Weiner J A, Post S R, Chun J. J Cell Biol. 1996;135:1071–1083. doi: 10.1083/jcb.135.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiner J A, Hecht J H, Chun J. J Comp Neurol. 1998;398:587–598. [PubMed] [Google Scholar]
- 9.Chun J, Contos J J A, Munroe D. Cell Biochem Biophys. 1999;30:213–242. doi: 10.1007/BF02738068. [DOI] [PubMed] [Google Scholar]
- 10.Contos, J. J. A., Ishii, I. & Chun, J. (2000) Mol. Pharmacol., in press. [DOI] [PubMed]
- 11.Fukushima N, Kimura Y, Chun J. Proc Natl Acad Sci USA. 1998;95:6151–6156. doi: 10.1073/pnas.95.11.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson J R, Wu J J, Goddard J G, Tigyi G, Kawanishi K, Tomei L D, Kiefer M C. J Biol Chem. 1998;273:1506–1510. doi: 10.1074/jbc.273.3.1506. [DOI] [PubMed] [Google Scholar]
- 13.Zhang G, Contos J J, Weiner J A, Fukushima N, Chun J. Gene. 1999;227:89–99. doi: 10.1016/s0378-1119(98)00589-7. [DOI] [PubMed] [Google Scholar]
- 14.Dubin A E, Bahnson T, Weiner J A, Fukushima N, Chun J. J Neurosci. 1999;19:1371–1381. doi: 10.1523/JNEUROSCI.19-04-01371.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukushima, N., Weiner, J. A. & Chun, J. (2000) Devel. Biol., in press. [DOI] [PubMed]
- 16.Weiner J A, Chun J. Proc Natl Acad Sci USA. 1999;96:5233–5238. doi: 10.1073/pnas.96.9.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contos J J A, Chun J. Genomics. 1998;51:364–378. doi: 10.1006/geno.1998.5400. [DOI] [PubMed] [Google Scholar]
- 18.Marth J D. J Clin Invest. 1996;97:1999–2002. doi: 10.1172/JCI118634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Contos J J A, Chun J. Genomics. 2000;64:155–169. doi: 10.1006/geno.2000.6122. [DOI] [PubMed] [Google Scholar]
- 20.Raff T, van der Giet M, Endemann D, Wiederholt T, Paul M. BioTechniques. 1997;23:456–460. doi: 10.2144/97233st02. [DOI] [PubMed] [Google Scholar]
- 21.Blaschke A J, Staley K, Chun J. Development. 1996;122:1165–1174. doi: 10.1242/dev.122.4.1165. [DOI] [PubMed] [Google Scholar]
- 22.An S, Bleu T, Hallmark O G, Goetzl E J. J Biol Chem. 1998;273:7906–7910. doi: 10.1074/jbc.273.14.7906. [DOI] [PubMed] [Google Scholar]
- 23.Ishii I, Contos J J A, Fukushima N, Chun J. Mol Pharmacol. 2000;58:895–902. doi: 10.1124/mol.58.5.895. [DOI] [PubMed] [Google Scholar]
- 24.Bandoh K, Aoki J, Hosono H, Kobayashi S, Kobayashi T, Murakami-Murofushi K, Tsujimoto M, Arai H, Inoue K. J Biol Chem. 1999;274:27776–27785. doi: 10.1074/jbc.274.39.27776. [DOI] [PubMed] [Google Scholar]
- 25.Zondag G C, Postma F R, Etten I V, Verlaan I, Moolenaar W H. Biochem J. 1998;330:605–609. doi: 10.1042/bj3300605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M J, Van Brocklyn J R, Thangada S, Liu C H, Hand A R, Menzeleev R, Spiegel S, Hla T. Science. 1998;279:1552–1555. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- 27.English D, Kovala A T, Welch Z, Harvey K A, Siddiqui R A, Brindley D N, Garcia J C. J Hematother Stem Cell Res. 1999;8:627–634. doi: 10.1089/152581699319795. [DOI] [PubMed] [Google Scholar]
- 28.Lee M J, Thangada S, Claffey K P, Ancellin N, Liu C H, Kluk M, Volpi M, Sha'afi R I, Hla T. Cell. 1999;99:301–312. doi: 10.1016/s0092-8674(00)81661-x. [DOI] [PubMed] [Google Scholar]
- 29.Valet P, Pagès C, Jeanneton O, Daviaud D, Barbe P, Record M, Saulnier-Blache J S, Lafontan M. J Clin Invest. 1998;101:1431–1438. doi: 10.1172/JCI806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris M J, Juriloff D M. Teratology. 1997;56:177–187. doi: 10.1002/(SICI)1096-9926(199709)56:3<177::AID-TERA1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 31.Risser J M, Slotnick B M. Physiol Behav. 1987;40:545–549. doi: 10.1016/0031-9384(87)90042-4. [DOI] [PubMed] [Google Scholar]
- 32.Brunet L J, Gold G H, Ngai J. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- 33.Belluscio L, Gold G H, Nemes A, Axel R. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 34.Blass E M, Teicher M H. Science. 1980;210:15–22. doi: 10.1126/science.6997992. [DOI] [PubMed] [Google Scholar]
- 35.Eisthen H L. Brain Behav Evol. 1997;50:222–233. doi: 10.1159/000113336. [DOI] [PubMed] [Google Scholar]
- 36.Guo Z, Liliom K, Fischer D J, Bathurst J C, Tomei L D, Kiefer M C, Tigyi G. Proc Natl Acad Sci USA. 1996;93:14367–14372. doi: 10.1073/pnas.93.25.14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cattaneo E, McKay R. Nature (London) 1990;347:762–765. doi: 10.1038/347762a0. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh A, Greenberg M E. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 39.Lee M J, Thangada S, Liu C H, Thompson B D, Hla T. J Biol Chem. 1998;273:22105–22112. doi: 10.1074/jbc.273.34.22105. [DOI] [PubMed] [Google Scholar]