Abstract
Certain plant viruses encode suppressors of posttranscriptional gene silencing (PTGS), an adaptive antiviral defense response that limits virus replication and spread. The tobacco etch potyvirus protein, helper component-proteinase (HC-Pro), suppresses PTGS of silenced transgenes. The effect of HC-Pro on different steps of the silencing pathway was analyzed by using both transient Agrobacterium tumefaciens-based delivery and transgenic systems. HC-Pro inactivated PTGS in plants containing a preexisting silenced β-glucuronidase (GUS) transgene. PTGS in this system was associated with both small RNA molecules (21–26 nt) corresponding to the 3′ proximal region of the transcribed GUS sequence and cytosine methylation of specific sites near the 3′ end of the GUS transgene. Introduction of HC-Pro into these plants resulted in loss of PTGS, loss of small RNAs, and partial loss of methylation. These results suggest that HC-Pro targets a PTGS maintenance (as opposed to an initiation or signaling) component at a point that affects accumulation of small RNAs and methylation of genomic DNA.
Keywords: HC-Pro, PTGS Suppression, Short RNA, Methylation
Posttranscriptional gene silencing (PTGS) or RNA interference occurs in a wide variety of organisms, including plants, animals, and fungi (1, 2). The PTGS process involves recognition of a target RNA and initiation of a sequence-specific RNA degradation pathway in the cytoplasm. Targets for PTGS may be recognized because of the presence of extensive double-stranded RNA (dsRNA) structure or because of an aberrant feature of the RNA (1). Small RNAs of 21–23 nt, corresponding to both sense and antisense strands of the target, are consistently associated with PTGS (2–6). It was proposed that these short RNAs provide specificity for target RNA degradation through association with an RNaseIII-like enzyme (2, 6). In plants, PTGS of transgenes is typically associated with methylation of nuclear DNA corresponding to the transcribed region of the target RNA, although transcription levels of the transgene are generally unaffected (1). In addition, systemic signaling to trigger PTGS at a distance can occur in plants, presumably through transport of a nucleic acid signal (7–9). Several genes encoding factors required for PTGS or RNA interference have been isolated, and these include proteins with similarities to an RNA-dependent RNA polymerase (5, 10, 11), a RecQ-like DNA helicase (10), an RNaseD-like protein (12), and a protein encoded by the piwi/sting/argonaute/zwille/eIF2C gene family (13).
Although PTGS in plants has been studied most extensively by using transgenes, viruses are both initiators and targets of PTGS. Infection by a range of viruses results in PTGS-like responses, even in the absence of homologous nuclear sequences (14–17). Virus-induced PTGS presumably reflects an adaptive defense mechanism whereby a sequence-specific response limits the extent of virus infection. Among the early indications that the PTGS response could have antiviral effects was the discovery that several plant viruses encode a PTGS suppressor (18–21). The potyviruses encode helper component-proteinase (HC-Pro), a multifunctional protein required for maintenance of genome replication, long-distance movement through plants, and polyprotein processing (22–24). Expression of HC-Pro in plants by either a transgene or a virus vector is sufficient to inhibit PTGS of a transgene (19–21). It was proposed that the PTGS-suppressing activity of HC-Pro accounts for the requirement of HC-Pro in replication and long-distance movement (19).
In this article, the effects of HC-Pro on specific steps in the PTGS process were determined. The results show that HC-Pro partially reverses silencing in cells with a preexisting silenced transgene, suggesting that HC-Pro inhibits a function required for maintenance of the silenced state. Data also reveal that HC-Pro acts to inhibit a step upstream from production of short RNAs in the silencing pathway and to reduce silencing-associated transgene methylation.
Materials and Methods
Transgenic Plants and Plasmids.
Nicotiana tabacum plants expressing a nontranslatable β-glucuronidase (GUS) gene under the control of the 35S promoter and terminator have been characterized previously (Fig. 1A; ref. 19). The homozygous transgenic lines 407 and 422 are posttranscriptionally GUS-silenced, whereas the 446 line contains the same transgene but is nonsilenced. The transgenic line U-6B contains a polyprotein including the P1 proteinase, HC-Pro, and the N terminus of the P3 protein and has been described (25). Several lines were derived from crosses between the 407 GUS-silenced line and the U-6B P1/HC-Pro-expressing line (19). A brief description of the plants used in this study is provided in Table 1.
Figure 1.
Diagram of constructs and Agrobacterium injection strategy. (A) Key portion of the construct used to produce transgenic plants containing a posttranscriptionally silenced GUS sequence. The GUS coding sequence was interrupted by a stop codon/frameshift mutation after codon four (19). The construct, therefore, contains a nontranslatable (nt) GUS sequence and directs no GUS activity in transgenic plants. (B) Key portions of the plasmids used for transient expression in Agrobacterium infiltration assays. The GUS + HC-Pro plasmid contains two independent expression cassettes. (C) Agrobacterium infiltration strategy. Two zones in each leaf were infiltrated with combinations of Agrobacterium cultures containing empty pSLJ755I5 vector, single-cassette GUS plasmid, or dual-cassette GUS + HC-Pro plasmid. Leaves were detached at 4 days after infiltration and subjected to GUS activity assay or immunoblot analysis.
Table 1.
Summary of transgenic plants
| Plant | PTGS of the GUS Transgene | Description |
|---|---|---|
| Parental Plants | ||
| U-6B | NA* | Homozygous HC-Pro locus |
| 407† | + | Homozygous GUS-silencing locus |
| 422† | + | Homozygous GUS-silencing locus |
| 446† | − | Homozygous GUS-nonsilencing locus |
| F3 Progeny of U-6B × 407 | ||
| Line 13‡ | NA* | No transgenes |
| Line 7§ | + | Homozygous GUS-silencing locus, HC-Pro-null |
| Line 17¶ | + | Homozygous GUS-silencing locus, HC-Pro-null |
| Line 17HC¶ | − | Homozygous GUS-silencing locus, homozygous or hemizygous HC-Pro locus |
Not applicable.
Plants were transformed with the same nontranslatable GUS transgene. Plants were characterized as posttranscriptionally GUS-silenced (lines 407 and 422) or nonsilenced (line 446; ref. 19).
Progeny of a self-pollinated F2 plant that lacked both the GUS and HC-Pro transgenes.
Progeny of a self-pollinated F2 plant in which the GUS-silencing locus was homozygous and the HC-Pro locus was absent.
Progeny of a self-pollinated F2 plant in which the GUS-silencing locus was homozygous and the HC-Pro locus was hemizygous. These plants either lacked the HC-Pro locus (line 17) or contained the HC-Pro locus in the homozygous or hemizygous state (line 17HC).
The plasmids pRTL2-GUS and pRTL2–0027 expressing the GUS and tobacco etch virus P1/HC-Pro coding sequences, respectively, have been described (25, 26). These plasmids contain the 35S promoter and terminator sequences. The plasmid pSLJ755I5-GUS was constructed by inserting the expression cassette from PstI-digested pRTL2-GUS into the binary plasmid pSLJ755I5 (27). The dual expression plasmid, pSLJ755I5-GUS + HC-Pro, was constructed by insertion of the HindIII-digested expression cassette from pRTL2–0027 into pSLJ755I5-GUS. The resulting plasmid, pSLJ755I5-GUS + HC-Pro, contained the GUS and tobacco etch virus P1/HC-Pro coding sequences under the control of independent 35S promoters (Fig. 1B). The three binary plasmids, pSLJ755I5, pSLJ755I5-GUS, and pSLJ755I5-GUS + HC-Pro, were referred to as vector, GUS, and GUS + HC-Pro plasmids, respectively. Each of these plasmids was introduced by triparental mating into the virulent Agrobacterium strain GV2260 and the avirulent strain C58C1D.
Agrobacterium Injection.
Individual Agrobacterium colonies were grown for 20 h in 5-ml cultures (Luria broth, 100 μg/ml rifampicin, 12.5 μg/ml tetracycline) at 30°C. This was used to inoculate a 50-ml culture (Luria broth, 20 μM acetosyringone/10 mM Mes, pH 5.7/12.5 μg/ml tetracycline), which was grown for 16–20 h at 30°C. The bacteria were pelleted by centrifugation, resuspended in infiltration medium (10 mM MgCl2/10 mM Mes, pH 5.7/150 μM acetosyringone) to 0.5 OD at 600 nm, and incubated at room temperature for a minimum of 3 h (28). By using a 3-ml syringe, the Agrobacterium solution was injected into leaves through an incision.
For silencing-release assays, two zones on apposing half-leaves of either nontransgenic (line 13) or GUS-silenced (line 7) tobacco plants were injected with combinations of Agrobacterium containing empty vector, GUS, or GUS + HC-Pro plasmids. Leaves were harvested at 4 days after injection and GUS activity was visualized by infiltration with the colorimetric substrate, 5-bromo-4-chloro-3-indolyl β-D-glucuronide. After overnight incubation at room temperature, the leaves were cleared in 75% (vol/vol) ethanol at 70°C. Leaves were photographed, and images were processed electronically by using Adobe photoshop.
For immunoblot analysis, leaves were infiltrated with a single Agrobacterium culture, and injection foci were collected 3 days after injection. Four foci were pooled, ground in liquid nitrogen, and resuspended in five volumes (wt/vol) buffer (40 mM sodium phosphate, pH 7.0/10 mM EDTA/0.1% Triton X-100/0.1% N-lauryl sarcosine/10 mM β-mercaptoethanol) with proteinase inhibitors (0.5 mM PMSF/1 μg/ml aprotinin/1 μg/ml leupeptin). Protein samples (100 μg) were subjected to immunoblot analysis by using anti-GUS and anti-HC-Pro serum.
Nucleic Acid Isolation.
Total RNA was extracted from mature tobacco leaves (2.0 g) as described (19). High-molecular weight RNA was precipitated in the presence of 2 M lithium chloride. Low-molecular weight RNA was precipitated from the 2 M lithium chloride supernatant in the presence of 75% (vol/vol) ethanol, resuspended in sterile water, and reprecipitated in the presence of 70% (vol/vol) ethanol and 0.1 M sodium acetate. The precipitate was washed with 70% (vol/vol) ethanol and resuspended in 40 μl of diethyl pyrocarbonate-treated deionized water. Alternatively, small RNAs were isolated by anion exchange chromatography (RNA/DNA Midi Kit; Qiagen, Chatsworth, CA) after removal of large RNA by precipitation with 2 M lithium chloride and DNA by precipitation in 6% (vol/vol) polyethylene glycol 8000 and 0.8 M sodium chloride.
Genomic DNA was isolated as described (19) and digested with restriction endonucleases in 16-h reactions. The DNA was extracted from the reaction mixture by using phenol/chloroform/isoamyl alcohol, precipitated in the presence of 70% (vol/vol) ethanol and resuspended in 20 μl of sterile deionized water.
Gel Blot Analysis.
High molecular weight RNA (10 μg) and genomic DNA (20 μg) samples were subjected to blot hybridization analysis, as described (19). Radiolabeled probes for specific GUS sequences were made by random priming reactions in the presence of [32P]dATP (29), and radioincorporation was measured. RNA blots were stripped and reprobed by using a tobacco ribosomal RNA probe labeled with [32P]dATP, and DNA blots were reprobed by using a tobacco eIF4E probe labeled with [32P]dATP (30).
Small RNAs (40–50 μg) were separated by electrophoresis in 15% polyacrylamide gels containing 7 M urea in 45 mM Tris-borate, pH 8.0/1 mM EDTA, electroblotted in 90 mM Tris2-borate, pH 8.0/2 mM EDTA, pH 8 to Hybond-N membranes (Amersham Pharmacia) for 1 h at 40 V, and UV crosslinked (1200 μJ, Stratalinker; Stratagene). The membrane was prehybridized in 50% (vol/vol) formamide, 10× Denhardt's solution, 0.5 mg/ml sheared salmon sperm DNA, 1% SDS, 3× SSC, and 50 mM phosphate buffer at 35°C for at least 1 h. Fresh hybridization solution containing a GUS probe radiolabeled with [32P]dATP then was added, and the blot was incubated at 35°C for 16 h. Blots were washed subsequently at 50°C in a solution of 2× SSC and 0.2% SDS. Ethidium bromide staining before blot transfer was used to confirm equal loading of RNA samples.
Results
HC-Pro Suppresses the Maintenance Phase of PTGS.
Previous analyses of silencing suppression by HC-Pro depended on delivery of HC-Pro into plants through either a stable transgene or an RNA virus vector (18–21). However, determination of whether HC-Pro suppresses an initiation/recognition step, a maintenance step, or a signaling step in PTGS with either type of delivery system is complicated by several factors. By using the transgenic system, both the HC-Pro and silencing loci are present in most or all cells during development. Suppression of initiation, maintenance, or signaling of PTGS would result in the same phenotype, namely, lack of silencing in the mature plant. By using a virus vector, interpretation of silencing-suppression phenotypes may be influenced or clouded by the presence of a suppressor encoded by the vector itself.
To enable analysis of PTGS suppression in a tissue-autonomous manner and in the absence of a virus vector, a transient system was developed by using Agrobacterium-mediated delivery of HC-Pro and a silencing reporter into a plant containing a PTGS locus. Transgenic N. tabacum plants (line 7) containing a posttranscriptionally silenced, defective GUS gene were produced and have been characterized (19). The modified 35S-GUS transgene contains nonsense and frameshift mutations near the 5′ end and does not encode an active protein (Fig. 1A). Single-gene or dual-gene expression plasmids encoding GUS alone (GUS) or GUS plus HC-Pro (GUS + HC-Pro), respectively, were produced and introduced into Agrobacterium. The HC-Pro construct actually encoded a larger region of the tobacco etch virus polyprotein, including the P1 proteinase adjacent to the N terminus of HC-Pro and part of the P3 protein adjacent to the C terminus (Fig. 1B). The N and C termini of HC-Pro are normally formed by autoproteolytic cleavage of the viral polyprotein by P1 proteinase and HC-Pro, respectively (31, 32). Therefore, the cassette was predicted to yield mature, accurately processed HC-Pro after expression in plant cells (see below).
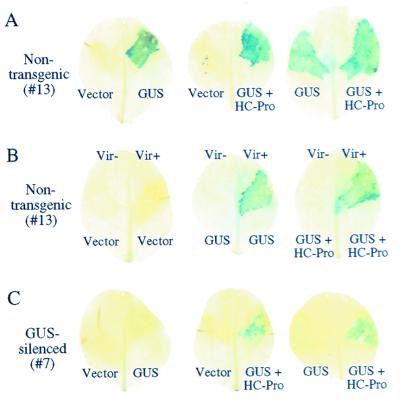
To confirm that the 35S-GUS expression cassettes in the single and dual vectors were functional, cultures of Agrobacterium containing the expression plasmids or empty vector were injected into leaves of nontransgenic N. tabacum plants (line 13). In all experiments, leaves were injected with combinations of experimental and control cultures on apposing half-leaves (Fig. 1C) and infiltrated with a GUS colorimetric substrate at 4 days after injection. Tissues receiving each plasmid with a 35S-GUS expression cassette, but not empty vector, generated GUS activity (Fig. 2A). To rule out that the GUS activity was caused by expression in bacteria, the experiment was done with both T-DNA transfer-competent (Vir+) and T-DNA transfer-defective (Vir−) strains of Agrobacterium. GUS activity was detected only in tissues injected with Vir+ bacteria containing 35S-GUS cassettes (Fig. 2B). Based on microscopic examination, virtually all cells within the infiltration zone contained GUS activity, suggesting that the Agrobacterium-mediated delivery of T-DNA in this system was extremely efficient.
Figure 2.
Suppression of PTGS by transient Agrobacterium-mediated delivery of HC-Pro. GUS encoded by single-GUS or dual-GUS + HC-Pro cassettes was detected by histochemical assay in leaf tissue at 4 days after infiltration. (A) Control series of Agrobacterium injection assays with nontransgenic plants (line 13). (B) Dependence of GUS activity on delivery of T-DNA by Agrobacterium. Plasmids were introduced into Vir+ and Vir− strains of Agrobacterium, followed by injection into nontransgenic plants. (C) Agrobacterium injection assays with the same series shown in A, but with GUS-silenced plants (line 7). Note that GUS activity occurs only in leaves infiltrated with Agrobacterium containing the dual GUS + HC-Pro plasmid.
The GUS and GUS + HC-Pro expression cassettes were then introduced into leaves of silencing line 7. No GUS activity was detected in half-leaves receiving the vector alone or the single-gene 35S-GUS cassette (Fig. 2C), indicating that the GUS-silencing phenotype of line 7 was maintained after introduction of a functional GUS gene. However, GUS activity was detected in half-leaves injected with Agrobacterium containing the dual GUS + HC-Pro plasmid (Fig. 2C), although the intensity of the histochemical signal generally was less than in leaves of nontransgenic line 13 plants.
To analyze further GUS and HC-Pro in the silencing and nonsilencing lines after Agrobacterium-mediated gene transfer, tissue from the injection zone was excised and subjected to immunoblot analysis by using anti-HC-Pro and anti-GUS sera. In nontransgenic tissue, GUS accumulated after injection of Agrobacterium containing either the single or dual expression cassette (Fig. 3, lanes 3–6), whereas HC-Pro accumulated only in tissue receiving the dual cassette (Fig. 3, lanes 5 and 6). In tissue from silencing line 7, no GUS protein was detected after introduction of the single 35S-GUS cassette (Fig. 3, lanes 9 and 10). In contrast, GUS protein was detected in tissue receiving the GUS + HC-Pro construct, although, like the histochemical assay, the level of accumulation of GUS in tissue from line 7 was significantly lower than in tissue from the nonsilencing line 13 (Fig. 3, lanes 11 and 12). These data indicate that PTGS can be inactivated by HC-Pro, at least partially, at a postrecognition/postinitiation step in a cell- or tissue-autonomous fashion that does not require systemic signaling. These results suggest that HC-Pro suppresses the maintenance phase in the PTGS process.
Figure 3.
Immunoblot analysis of GUS and HC-Pro after Agrobacterium-mediated delivery into GUS-silenced and nontransgenic plant lines. Normalized, total detergent-soluble protein extracts were prepared from tissue injected with Agrobacterium carrying vector alone or plasmids containing the single-GUS or dual-GUS + HC-Pro expression cassettes. Lanes 1–6, nontransgenic line 13. Lanes 7–12, GUS-silenced line 7. Samples consisted of pools of tissue from four injection zones. Two samples are shown for each treatment. Immunoblot results with anti-HC-Pro (Upper) and anti-GUS sera (Lower) are shown.
HC-Pro Suppresses a Step Before Accumulation of Short RNAs.
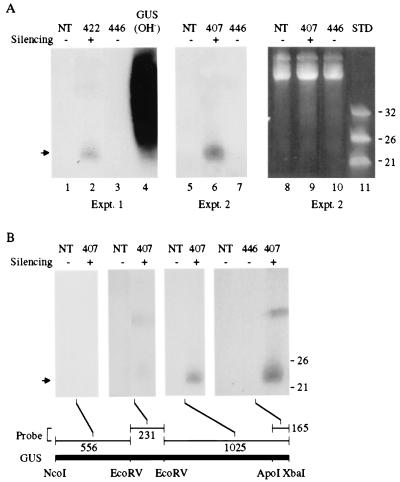
To determine whether PTGS of the nontranslatable GUS gene in transgenic lines 422 and 407 [the parent from which the GUS-silencing locus was derived in line 7 (Figs. 2 and 3)] was associated with small RNAs, low-molecular weight RNA was extracted and analyzed by blot hybridization with a radiolabeled GUS probe. A discrete band of material that migrated between the 21- and 26-nt single-stranded DNA standards was detected in GUS PTGS lines 422 and 407 in independent experiments (Fig. 4A, lanes 2 and 6). No such species was detected in nontransgenic plants (Fig. 4A, lanes 1 and 5) or in plants from line 446 (Fig. 4A, lanes 3 and 7), which contains the same GUS transgene as in lines 407 and 422 but does not display PTGS (19). The GUS-related nucleic acid species was sensitive to RNaseA but insensitive to DNaseI (data not shown), indicating that the material was RNA. By using probes normalized for total radioactivity and RNA extracts from line 407, the small RNAs hybridized preferentially to sequences corresponding to the 3′ proximal region of the GUS coding sequence, with the most intense hybridization occurring with a probe for the 3′ terminal 165 nucleotides (Fig. 4B). Little or no hybridization with small RNAs from line 407 was detected by using probes representing the 5′ proximal 787 nucleotides of the GUS sequence. The GUS sequence-related RNAs likely correspond to the PTGS-specific small RNAs identified by Hamilton and Baulcombe (3).
Figure 4.
Detection of short RNAs in GUS-silenced transgenic plants. Low molecular weight RNA was extracted from leaves of either nontransgenic (NT), GUS-silenced (422 and 407), or GUS-nonsilenced (446) plants. Equal amounts of each RNA sample were subjected to electrophoresis in denaturing 15% polyacrylamide gels, stained with ethidium bromide, blotted to a nylon membrane, and hybridized using various 32P-labeled GUS DNA fragments as probes. The PTGS status of each plant is indicated above the lanes. (A) Two experiments analyzing small RNAs with a full-length, 32P-labeled GUS probe. In vitro transcribed GUS RNA was hydrolyzed (OH−) and used as a hybridization control. The arrow indicates the position of short RNA. (Right) Ethidium bromide staining of the gel used in experiment 2. DNA oligonucleotides (21, 26, and 32 nt) were used as standards (STD). (B) Analysis of small RNAs with normalized (2 × 106 cpm) 32P-labeled probes corresponding to different regions of the GUS coding sequence. The positions of the 21 and 26 nt DNA standards are shown at the right.
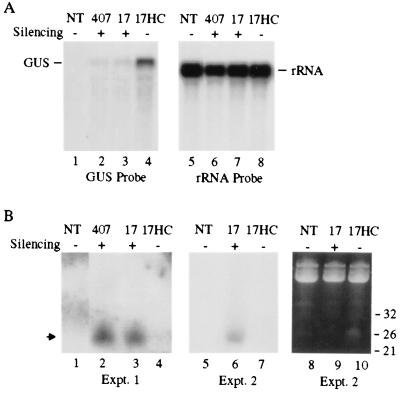
To test the effect of HC-Pro on accumulation of short RNAs associated with PTGS of the GUS sequence, F3 progeny plants from a cross between the 407 line and a transgenic plant expressing the P1/HC-Pro region of the tobacco etch virus genome were analyzed. Plants from the F2 and F3 generations were characterized with respect to transgene configuration and GUS-silencing phenotype (Table 1). Progeny plants containing either no transgenes [referred to as nontransgenic (NT) or line 13] or a homozygous GUS-, HC-Pro-null transgene configuration (line 7) were nonsilenced or GUS-silenced, respectively (Fig. 2). In addition, an F2 plant that was homozygous at the GUS transgene locus and hemizygous at the HC-Pro transgene locus (line 17a) was identified. The F3 progeny from line 17a were all homozygous at the GUS-silencing locus but either lacked the HC-Pro transgene (line 17, silenced) or were homozygous/hemizygous at the HC-Pro locus (line 17HC, silencing suppressed). Both high and low molecular weight RNAs were isolated from leaf tissue of NT, 407 parental, line 17, and line 17HC plants and subjected to blot hybridization by using a GUS-sequence probe. As shown previously (19), the level of GUS mRNA in silencing-suppressed line 17HC plants was considerably higher than the levels in GUS-silenced 407 and line 17 plants (Fig. 5A). In contrast, the level of small RNAs in line 17HC plants was considerably lower than the levels in 407 or line 17 plants. This result was observed consistently in multiple experiments, two of which are shown in Fig. 5B. These data indicate that silencing suppression by HC-Pro blocks accumulation of silencing-specific small RNAs.
Figure 5.
HC-Pro suppresses accumulation of short RNAs. (A) Blot analysis of GUS mRNA in either nontransgenic (NT), GUS-silenced (407 and 17), or silencing-suppressed (17HC) plants. High-molecular weight RNA was isolated, normalized (10 μg/lane), subjected to electrophoresis, blotted to a nylon membrane, and hybridized by using 32P-labeled full-length GUS DNA as a probe. The blot was stripped and reprobed with a 32P-labeled DNA probe specific for rRNA. The positions of both GUS and rRNA mRNAs are indicated. (B) Blot analysis of short RNA. Low-molecular weight RNA was isolated and analyzed as described in the legend for Fig. 4. The results from two independent experiments (Expt.) are shown. The arrow indicates the position of silencing-specific short RNA. For presentation purposes, the data shown in A and B are composite images from noncontiguous lanes from a single blot.
HC-Pro Partially Affects PTGS-Associated Methylation of Transgene DNA.
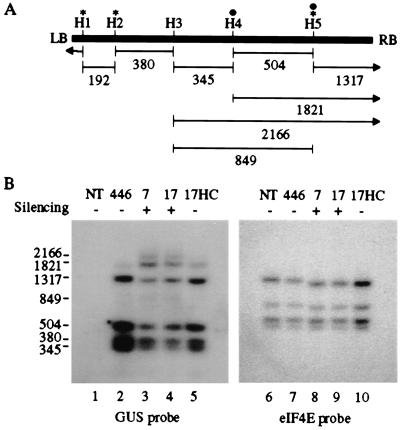
Previous experiments revealed that delivery of HC-Pro into plants containing a posttranscriptionally silenced reporter gene does not reverse cytosine methylation associated with the transcribed sequence of the transgene (33). These observations were based on introduction of HC-Pro with an RNA virus vector, when PTGS of the transgene was already operative, and cells at the time of inoculation contained previously methylated DNA. To determine whether the presence of HC-Pro affects methylation of a GUS-silencing locus during development and through multiple generations, the cytosine-methylation status of the GUS transgene in 446, line 7, line 17, and line 17HC plants was analyzed by using methylation-sensitive restriction enzymes followed by DNA blot hybridization. Most methylation-sensitive restriction enzymes tested, however, did not reveal consistent methylation patterns in the GUS-silenced line 7 and line 17 plants (data not shown). Only HaeIII revealed consistent cytosine methylation of the GUS sequence in line 7 and line 17 plants, as revealed by the appearance of partial-digestion products (Fig. 6B, lanes 3 and 4). Three DNA fragments (2,166, 1,821, and 849 nt) were identified as partial digestion products containing 3′ proximal-GUS coding sequences based on analysis of size and on further hybridization analysis with specific probes (Fig. 6 and data not shown). These products arose through inhibition of digestion at HaeIII sites H4 and/or H5 (Fig. 6A). No partial digestion products corresponding to those predicted if HaeIII sites H1, H2, or H3 were methylated were identified. As shown by stripping the blot and reanalyzing with an eIF4E-specific probe (Fig. 6B, lanes 6–10), all HaeIII sites in a control gene were digested to completion, indicating that the GUS-transgene results were not simply caused by insufficient HaeIII reaction conditions. Based on densitometric scans of blots from three independent experiments and normalization of results based on the nucleotide length of each fragment, cytosine residues at approximately 10% and 12% of H4 sites, and 54% and 40% of H5 sites, were methylated in GUS-silenced lines 7 and 17, respectively.
Figure 6.
HC-Pro partially suppresses methylation of target DNA. (A) Schematic representation of DNA corresponding to the GUS coding sequence. Positions of HaeIII restriction sites (H1-H5) and sizes (in nucleotides) of the expected digestion products are illustrated. Sites marked by an asterisk contain cytosines in a symmetrical (CpNpG) configuration. Filled circles indicate HaeIII sites that were cytosine methylated in GUS-silenced plants. The right (RB) and left (LB) borders of the GUS transgene are indicated. (B) Blot analysis of genomic DNA in nontransgenic (NT), GUS-silenced transgenic (7 and 17), GUS-nonsilenced transgenic (446), and GUS-silencing suppressed (no. 17HC) plants. Blots were hybridized with a 32P-labeled probe specific for the GUS gene. The blot was stripped and rehybridized with a 32P-labeled DNA probe specific for the eIF4E coding sequence.
In contrast to results with GUS-silenced line 7 and line 17 plants, evidence for methylation was obtained only for site H5 in the GUS transgene in line 17HC plants (Fig. 6B, lane 5). Results consistent with cytosine methylation at 20% of H5 sites were obtained based on averages from three experiments. Similarly, evidence for cytosine methylation in the nonsilenced 446 GUS transgene was obtained for a low percentage of H5 sites (8%; Fig. 6B, lane 2). These results indicate that silencing suppression by HC-Pro results in a decrease in the extent of silencing-associated methylation of DNA.
Discussion
The point of HC-Pro-mediated suppression in the PTGS pathway was analyzed by using a series of transient and transgenic assays. These experiments were done in the absence of a replicating virus vector. The results, therefore, are free from the potential complications associated with extraneous suppressors that may be encoded by a virus vector.
Three conclusions were drawn from these results. First, HC-Pro suppresses one or more maintenance steps in the PTGS pathway. Transient delivery of HC-Pro by Agrobacterium injection into tissue of a plant with a silenced GUS transgene was sufficient to inhibit silencing in a cell- or tissue-autonomous manner, indicating that suppression occurs beyond the point of initiation of silencing and without the need for systemic signaling. In contrast, the cucumber mosaic virus 2b protein was suggested to suppress a signaling step, but not a maintenance step, in the silencing pathway (20). The transient expression data also imply that HC-Pro targets or suppresses a factor that is required on a continual basis or that is relatively labile. Silencing suppression by HC-Pro in the transient assay, however, was not complete. Incomplete silencing suppression could be caused by a number of factors, including the inability of HC-Pro to suppress silencing in all cells, a quantitative effect reflecting the activity of residual silencing factors, or the sampling of tissue at a time point at which silencing suppression was incomplete.
Second, HC-Pro inhibits a step required for accumulation of small RNAs in the PTGS pathway. Transgenic plants with a silenced GUS gene accumulated small RNAs of approximately 21–26 nts in length, which almost certainly correspond to small RNAs associated with PTGS or RNA interference identified by others (2–6). By using an in vitro RNA interference system from Drosophila cells, Zamore et al. (6) demonstrated that the small RNAs likely derive from dsRNA inducer molecules rather than from degradation of the target mRNA. Small RNAs are proposed to be produced by cleavage of a dsRNA precursor by an RNaseIII-like enzyme, to remain associated with the nuclease, and to confer sequence specificity to the nuclease (2, 6). Introduction of HC-Pro through a genetic cross substantially reduced levels of small RNA. The most straightforward interpretation of this result is that HC-Pro suppresses a step upstream of, or at the point of, production of small RNAs. Such a step could be recognition of an inducer dsRNA molecule, modification of an inducer molecule, interaction of the putative RNaseIII-like nuclease with the dsRNA, or processing of the dsRNA to form the small RNAs. Alternatively, small RNA production may depend on a feedback-amplification loop from a point downstream of initial production of limited quantities of small RNAs. In this case, HC-Pro could conceivably affect any point in the loop.
Third, HC-Pro reduces the level of cytosine methylation of a transgene sequence that is a PTGS target. When HC-Pro was expressed from a transgenic plant with a GUS PTGS locus, the relative level of cytosine methylation at two HaeIII sites (one symmetrical and one nonsymmetrical) near the 3′ end of the GUS-transcribed sequence was lower than in plants containing only the GUS PTGS locus. Methylation at one site (H5), however, was not eliminated entirely in the presence of HC-Pro. On the other hand, a low level of methylation also was detected at the H5 in the GUS transgene locus from plant 446, which did not display PTGS. The basis for PTGS-associated methylation is not yet clear, although it likely involves an RNA-mediated feedback mechanism from the cytoplasm (33). Sites of methylation at the GUS PTGS locus in line 7 and line 17 plants correlated roughly with the GUS sequence represented among the small RNAs, because both methylation sites and small RNAs were associated with 3′ proximal sequences of the transgene or GUS RNA. One possible interpretation of these results is that methylation of the PTGS transgene locus is guided by small RNAs that diffuse from the cytoplasm and interact with chromosomal DNA (1). Inhibition of small RNA accumulation by HC-Pro, therefore, would lead to reduced methylation of the transcribed region of the PTGS transgene.
The specific factor or factors in the PTGS pathway that are affected by HC-Pro remain to be determined. It is possible that HC-Pro interacts with a PTGS structural or regulatory factor. Structural factors that are proposed to function upstream of small RNA accumulation in the pathway include the host RNA-dependent RNA polymerase (5, 34, 35) and a dsRNA-binding ribonuclease (2, 6). Regulatory factors may influence the production, activation, or accumulation of functional forms of these factors. The identities and functions of PTGS pathway components and the effects of HC-Pro on their activities are key problems to address.
Acknowledgments
We thank Brian Staskawicz and Doug Dahlbeck for supplying Agrobacterium strains GV2260 and C58C1D and for helpful comments on setting up the injection system. We thank Jonathan Jones for providing pSLJ755I5. We are grateful to Julia Gothard and Sue Vogtman for excellent help in the greenhouses. This work was supported by United States Department of Agriculture Grant 98-35303-6485 and National Institutes of Health Grants AI 43288 and AI 27832.
Abbreviations
- HC-Pro
helper component-proteinase
- PTGS
posttranscriptional gene silencing
- GUS
β-glucuronidase
- dsRNA
double-stranded RNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230334397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230334397
References
- 1.Sijen T, Kooter J M. BioEssays. 2000;22:520–531. doi: 10.1002/(SICI)1521-1878(200006)22:6<520::AID-BIES5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 2.Bass B L. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton A J, Baulcombe D C. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 4.Hammond S M, Bernstein E, Beach D, Hannon G J. Nature (London) 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 5.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe D C. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 6.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 7.Sonoda S, Nishiguchi M. Plant J. 2000;21:1–8. doi: 10.1046/j.1365-313x.2000.00645.x. [DOI] [PubMed] [Google Scholar]
- 8.Voinnet O, Vain P, Angell S, Baulcombe D C. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 9.Palauqui J C, Vaucheret H. Proc Natl Acad Sci USA. 1998;95:9675–9680. doi: 10.1073/pnas.95.16.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cogoni C, Macino G. Science. 1999;286:2342–2344. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- 11.Smardon A, Spoerke J M, Stacey S C, Klein M E, Mackin N, Maine E M. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 12.Ketting R F, Haverkamp T H, van Luenen H G, Plasterk R H. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 13.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 14.Ratcliff F G, Harrison B D, Baulcombe D C. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 15.Ratcliff F G, MacFarlane S A, Baulcombe D C. Plant Cell. 1999;11:1207–1216. doi: 10.1105/tpc.11.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Covey S N, Al-Kaff N S, Lángara A, Turner D S. Nature (London) 1997;385:781–783. [Google Scholar]
- 17.Al-Kaff N S, Covey S N, Kreike M M, Page A M, Pinder R, Dale P J. Science. 1998;279:2113–2115. doi: 10.1126/science.279.5359.2113. [DOI] [PubMed] [Google Scholar]
- 18.Voinnet O, Pinto V M, Baulcombe D C. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasschau K D, Carrington J C. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 20.Brigneti G, Voinnet O, Wan-Xiang L, Ding S W, Baulcombe D C. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Anandalakshmi R, Pruss G J, Ge X, Marathe R, Smith T H, Vance V B. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cronin S, Verchot J, Haldeman-Cahill R, Schaad M C, Carrington J C. Plant Cell. 1995;7:549–559. doi: 10.1105/tpc.7.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasschau K D, Cronin S, Carrington J C. Virology. 1997;228:251–262. doi: 10.1006/viro.1996.8368. [DOI] [PubMed] [Google Scholar]
- 24.Klein P G, Klein R R, Rodríguez-Cerezo E, Hunt A G, Shaw J G. Virology. 1994;204:759–769. doi: 10.1006/viro.1994.1591. [DOI] [PubMed] [Google Scholar]
- 25.Carrington J C, Freed D D, Oh C-S. EMBO J. 1990;9:1347–1353. doi: 10.1002/j.1460-2075.1990.tb08249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Restrepo M A, Freed D D, Carrington J C. Plant Cell. 1990;2:987–998. doi: 10.1105/tpc.2.10.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones J D, Shlumukov L, Carland F, English J, Scofield S R, Bishop G J, Harrison K. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- 28.Tai T H, Dahlbeck D, Clark E T, Gajiwala P, Pasion R, Whalen M C, Stall R E, Staskawicz B J. Proc Natl Acad Sci USA. 1999;96:14153–14158. doi: 10.1073/pnas.96.24.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feinberg A P, Vogelstein B. Anal Biochem. 1983;257:8569–8572. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 30.Schaad, M. C., Anderberg, R. J. & Carrington, J. C. (2000) Virology273, in press. [DOI] [PubMed]
- 31.Verchot J, Koonin E V, Carrington J C. Virology. 1991;185:527–535. doi: 10.1016/0042-6822(91)90522-d. [DOI] [PubMed] [Google Scholar]
- 32.Carrington J C, Cary S M, Parks T D, Dougherty W G. EMBO J. 1989;8:365–370. doi: 10.1002/j.1460-2075.1989.tb03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones A L, Hamilton A J, Voinnet O, Thomas C L, Maule A J, Baulcombe D C. Plant Cell. 1999;11:2291–2302. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cogoni C, Macino G. Nature (London) 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 35.Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel J B, Jouette D, Lacombe A M, Nikic S, Picault N, et al. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]