Abstract
The symbiotic interaction between Medicago truncatula and Sinorhizobium meliloti results in the formation of nitrogen-fixing nodules on the roots of the host plant. The early stages of nodule formation are induced by bacteria via lipochitooligosaccharide signals known as Nod factors (NFs). These NFs are structurally specific for bacterium–host pairs and are sufficient to cause a range of early responses involved in the host developmental program. Early events in the signal transduction of NFs are not well defined. We have previously reported that Medicago sativa root hairs exposed to NF display sharp oscillations of cytoplasmic calcium ion concentration (calcium spiking). To assess the possible role of calcium spiking in the nodulation response, we analyzed M. truncatula mutants in five complementation groups. Each of the plant mutants is completely Nod− and is blocked at early stages of the symbiosis. We defined two genes, DMI1 and DMI2, required in common for early steps of infection and nodulation and for calcium spiking. Another mutant, altered in the DMI3 gene, has a similar mutant phenotype to dmi1 and dmi2 mutants but displays normal calcium spiking. The calcium behavior thus implies that the DMI3 gene acts either downstream of calcium spiking or downstream of a common branch point for the calcium response and the later nodulation responses. Two additional mutants, altered in the NSP and HCL genes, which show root hair branching in response to NF, are normal for calcium spiking. This system provides an opportunity to use genetics to study ligand-stimulated calcium spiking as a signal transduction event.
Calcium signaling takes many forms and is used by diverse biological systems. Despite its chemical simplicity as a single ionized atom, the calcium ion is an effective signal because its background in the cytoplasm is typically low, and because there exist calcium-binding effector proteins with quite specific downstream consequences. Thus elevated calcium can selectively activate distinct cellular functions. Calcium spiking is a distinctive behavior characterized by sharp upswings of cytoplasmic calcium ion concentration, followed by decreases back to a relatively unchanged background level, and is interpreted as release from and reuptake into a calcium ion reservoir or release and uptake from external sources (1). Calcium spiking accompanies signal transduction in animal cells (1) and has recently been discovered in some plant cells (2). The combination of cell imaging, pharmacological, and biochemical approaches points to several conclusions about the upstream events leading to calcium release and the calcium oscillation machinery, but these studies also reveal much variability in the regulation of calcium spiking from one system to another (reviewed by refs. 1, 3, and 4). What events may lie downstream of calcium spiking is a question of high interest but with few specific mechanistic answers so far. In leukemic tissue culture cell lines, manipulation of the frequency of forced calcium spikes affects both gene expression profiles and expression levels (5–7), pointing to a major connection between calcium spiking and transcription machinery. A model for how protein activity might mediate calcium oscillation frequency is suggested by the in vitro observation that the frequency of calcium transients altered persistence of the kinase activity of CaMKinaseII (8). In the majority of cell systems studied, however, little if anything is yet known about the components that translate calcium spiking into specific downstream events.
A genetic approach would allow an unbiased dissection of the steps upstream and downstream of calcium spiking, making it possible to identify components of the calcium spiking response and to reveal unsuspected roles for calcium spiking in cellular processes. An ideal system for genetic studies of calcium spiking would involve a triggered response associated with a facultative behavior. This would permit recovery of a broad range of mutant individuals, which should bracket steps upstream and downstream of calcium spiking. The legume–Rhizobium symbiosis offers such an opportunity to study the possible role of calcium spiking in a developmental process. Nodule formation in legumes is remarkable in that the development of a fully formed plant organ is activated by a bacterium (9). Many of the components of the nodulation process can be triggered by chemical signals synthesized by the bacterium, termed Nod factors (NFs) (10–12). These bacterial signals are lipochitooligosaccharides synthesized through the action of bacterial nod genes; they feature an N-acetyl d-glucosamine backbone modified by residues that confer specificity of action on particular host plants (13, 14). The host plant responds to bacterial NFs by diverse molecular and cellular events, including deformed growth of root hairs, reinitiation of mitoses in root cortical cells, and expression of several genes called early nodulins, or ENODs, in the epidermis, cortex, and pericycle of the root (for a recent review, see ref. 15). Genetic analyses have defined plant loci required for the multiple responses of legumes to Rhizobium cells and NFs (R.C., T. Timmers, C.G., F. Maillet, C.G., R.V.P., D.C., G. Truchet and J.D., unpublished results; refs. 16–20).
The signaling events that lead from NFs to cellular responses and changes in host gene expression are not well understood, but clues may be found in a series of rapid responses to NF (21). Membrane depolarization and ion fluxes (22–26) occur in alfalfa root hairs within 1–5 minutes after NF application (21). Three to six minutes later, periodic oscillation of the cytoplasmic calcium levels, or calcium spiking, begins (27). Calcium spiking is the only NF-induced rapid response that has been characterized in multiple legume genera and species (28). This behavior was absent in a nonnodulating line of alfalfa, MN-NN1008, which lacks any morphological response to bacteria or NF (27). In this study, we used Nod− Medicago truncatula mutants blocked at different stages before infection to examine whether calcium spiking correlates with specific nodulation responses.
The occurrence of calcium spiking as an early event in the nodulation response provides several important experimental opportunities: (i) calcium spiking can be used as a means to dissect the nodulation process and to place into order mutants that are phenotypically similar, and (ii) the genetic tractability of the system allows study of the role of calcium spiking in a complex multistage process. Calcium spiking may be causative in the signal transduction chain or may be a correlative event. A comprehensive genetic study can assess these possibilities and can identify steps that are upstream and downstream of the calcium spiking event.
Materials and Methods
Plant Growth and Preparation.
Wild-type M. truncatula cv. Jemalong seeds were surface sterilized in 95% ethanol for 45 min, followed by treatment with 5.25% sodium hypochlorite for 45 min. Mutant seeds were scarified for 5 min with concentrated sulfuric acid and rinsed twice in sterile water, followed by 3-min sterilization in 5.25% sodium hypochlorite and 5 rinses in sterile water. The seeds were germinated overnight and then transferred to plates containing Buffered Nodulation Medium (BNM; ref. 22) with 12% agar (Sigma) and 0.1 μM l-α-(2-aminoethoxyvinyl) glycine (Sigma). Two to three plants were transferred to a 2-ml BNM bath on a 48 × 60-mm coverslip.
Calcium Oscillation Assay.
Calcium imaging was carried out on an inverted Nikon TE200 microscope, equipped with a FITC filter (41 High Q series, excitation 480 ± 20 nm, dichroic 505 long pass, emission 535 ± 25 nm; Chroma Technology, Brattleboro, VT), a neutral density filter (0.6 optical density), and a ×20 CFI plan apochromat objective. Root hair cells were injected with Oregon green–dextran dye (Molecular Probes) by iontophoresis, as described in ref. 27. Cells that showed active streaming were chosen for subsequent imaging. Multiple cells on a single plant were imaged simultaneously. Fluorescence imaging was performed using a Princeton Instruments (Trenton, NJ) charge-coupled device (CCD)-1360SO cooled 1-MHz CCD (Sony front-illuminated interline array) camera. Images were acquired every 5 seconds, with a 200- to 250-ms exposure time and 2 × 2 binning, using metafluor software from Universal Imaging (West Chester, PA). After 10–30 min of imaging to collect the baseline for calcium in unstimulated cells, purified NodRmIV(C16:2,Ac, S) NF was added to a final concentration of 1 nM. The cells were imaged for at least 30 min after addition of NF. Raw fluorescence data were corrected for background according to ref. 27.
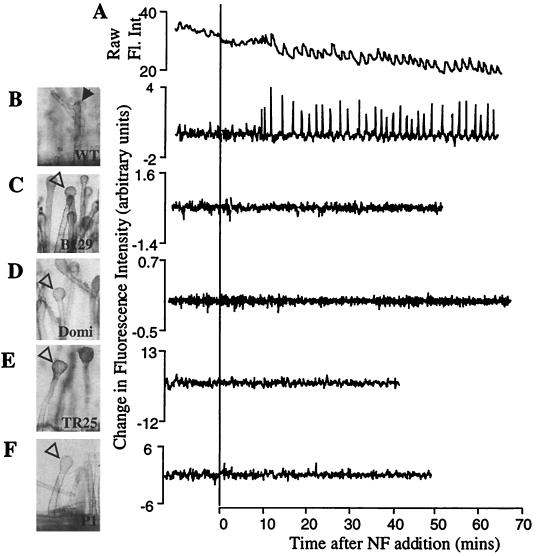
The amplitude of calcium spikes has been defined in Medicago sativa (alfalfa) by using the ratiometric dye Fura-2-dextran and was found to extend from a baseline of 40 nM to a peak of greater than 600 nM (27). In M. truncatula, we have found that the combination of Fura-2-dextran and ultraviolet light was toxic to root hair cells (E. Engstrom, S. Shaw, and S.R.L., unpublished data). The injection of two dextran-coupled long-wavelength dyes can be used for the study of single cells. However, the cell-to-cell variability of the dye ratio as a result of iontophoretic injections and the differential rates of photobleaching of the two dyes over the extended period of these experiments disallowed a quantitative comparison of calcium spike amplitude between cells. Therefore, we do not assess the amplitude of calcium spikes in this study. Ratiometric dyes also control for changes in fluorescence because of changes in local dye concentration. In Oregon green–dextran-injected root hairs, the baseline fluorescence is uneven (Fig. 1A). We therefore transformed the raw fluorescence data by using the function Y = X(n+1) − Xn, where Y is the point-to-point change in fluorescence, and X(n+1) and Xn are the intensity measurements at time points (n + 1) and (n). This function stabilizes the baseline fluctuations and emphasizes rapid changes in the fluorescence data that typify calcium spiking (Fig. 1 A and B).
Figure 1.
The root hair phenotypes and calcium spiking response of wild-type, dmi1 mutants, and dmi2 mutants. Root hair morphology was scored 16 h after application of 1 nM NF. Closed arrows point to root hair branches. Open arrowheads indicate swollen root hair tips. Traces represent raw fluorescence intensity, corrected for background (A), and change in fluorescence from one time point to the next (Xn+1 − Xn) (B–F) of single root hairs injected with Oregon green–dextran. (A) Wild-type M. truncatula, raw data; (B) wild-type M. truncatula; the same cell as A. However, the data were transformed by using the function Y = Xn+1 − Xn; (C) dmi1 mutant B129; (D) dmi1 mutant Domi; (E) dmi2 mutant TR25; (F) dmi2 mutant P1.
A cell was counted as positive for calcium spiking when five or more consecutive spikes with amplitudes of more than twice the standard deviation of the cell's baseline fluorescence were detected. Calcium spikes were counted as consecutive when they occurred no more than 2 minutes apart. The lag to onset of calcium spiking was calculated by measuring the time from addition of NF to the first observed calcium spike. The period between calcium spikes was calculated by measuring the time between two spike peaks for 10–15 spikes per cell.
Results
Wild-Type M. truncatula Shows Calcium Spiking in Response to NF.
To examine the calcium response in early nodulation mutants of the model legume M. truncatula, we first investigated whether wild-type M. truncatula root hair cells displayed calcium spiking. M. truncatula plants, like their alfalfa relatives (27), displayed calcium spiking when treated with purified Sinorhizobium meliloti NF (Fig. 1 A and B). Because of the nonratiometric nature of the dye used in this study, we cannot calculate the amplitudes of the calcium spikes (see Materials and Methods), and therefore in this study only the lag before the onset of spiking and the period between calcium spikes were quantified. Calcium spiking in wild type is initiated after a lag of an average 11.1 ± 3.4 min and has an average periodicity of 102 ± 24 seconds. As was seen in alfalfa (27), calcium spiking in M. truncatula continues for as long as the cells are imaged (at least 2 h; data not shown).
Calcium Spiking in dmi1, dmi2, and dmi3 Mutants.
On the basis of their symbiotic phenotypes, the three complementation groups dmi1, dmi2, and dmi3 of M. truncatula have been proposed to encode proteins that act early in the NF perception and signal transduction pathway (16). All three mutants are unable to form nodules, lack cortical cell division, and do not form infection structures in response to S. meliloti. These mutants also show a dramatically reduced root hair response to NF. Root hairs of wild-type plants respond to NF by inducing root hair deformation and branching (Fig. 1B). In contrast, dmi1, dmi2, and dmi3 mutants show root hair tip swelling (Figs. 1 C–F and 3A) but are blocked for branching (16). All three complementation groups show a complete block in the induction of the symbiotically induced genes ENOD11 and ENOD40 and a dramatically reduced induction of RIP1 (16). Furthermore, all three classes of mutants show a block in the association with mycorrhizal (Myc) fungi (16) (see Table 2). This block in the Myc phenotype is relatively tight in all Nod− alleles of dmi1 and dmi3. However, some dmi2 alleles are leaky for mycorrhizal invasion and/or nodulation (V. Gianinazzi-Pearson and D. Morandi, personal communication).
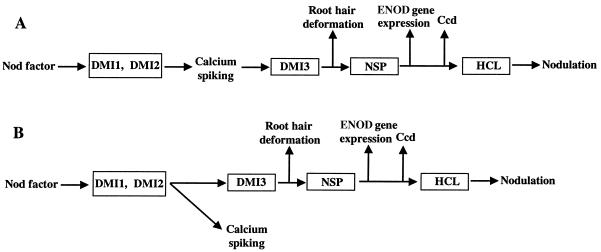
Figure 3.
Alternative models for placement of calcium spiking with respect to NF signal transduction. (A) Data in this paper show that DMI1 and DMI2 encode genes required in common upstream of calcium spiking and of multiple nodulation behaviors. Calcium spiking may lie directly upstream of one or more of the known nodulation responses, such as transcription or morphogenesis. In this case, DMI3 may lie downstream of calcium spiking and could potentially function as a calcium-sensitive effector protein that translates calcium signals to other cellular components. (B) Calcium spiking may occur as a branch off of the pathway that ultimately leads to root hair branching, early nodulin (ENOD) gene expression, and cortical cell division (ccd). The DMI1 and DMI2 loci lie upstream of the branch point, and DMI3 must lie downstream, as should NSP. HCL might act downstream of earlier NF signal transduction events or in a separate pathway altogether. The events or components required for NF-induced root hair swelling in the dmi1, dmi2, and dmi3 mutants must lie upstream of calcium spiking. If calcium spiking is not in the main signal transduction pathway but is stimulated by NF and by early steps in signal transduction, it may nonetheless have a function related to nodulation.
Table 2.
Symbiotic phenotypes and calcium response in M. truncatula mutants
| Locus | DMI1 | DMI2 | DMI3 | NSP | HCL | Wild type |
|---|---|---|---|---|---|---|
| Mutants | (Domi, B129) | (TR25, P1) | (TRV25) | (B85) | (B56) | N.A. |
| Calcium spiking | − | − | + | + | + | + |
| Root hair deformation | Swelling | Swelling | Swelling | Branching | Branching | Branching |
| Gene expression* | − | − | − | − | + | + |
| Cortical cell division | − | − | − | − | + | + |
| Curling in response to live bacteria | − | − | − | − | − | + |
| Myc association | − | −† | − | + | + | + |
We analyzed dmi1, dmi2, and dmi3 mutants for the induction of calcium spiking in response to NF. For mutants exhibiting altered calcium spiking, two allelic mutants were studied to be sure that the same mutation was responsible for both the altered calcium spiking and the nodulation defect. The dmi1 allelic mutants Domi (dmi1–1) and B129 (dmi1–2) were both unable to induce calcium spiking in response to NF: zero of 55 root hairs on 17 B129 plants and zero of 32 root hairs on 9 Domi plants showed calcium spiking (Fig. 1 C and D; Table 1). We assayed two dmi2 allelic mutants: TR25 (dmi2–1) that is Nod− and Myc− and P1 (dmi2–2) that is Nod− but appears to be slightly leaky in soil-grown plants and shows an intermediate level of mycorrhizal invasion. TR25 was completely blocked for calcium spiking: zero of 38 root hair cells on 9 plants showed calcium spiking (Fig. 1E; Table 1). P1 showed an absence of calcium spiking in 45 of 60 cells tested (Fig. 1F; Table 1). However, 5 of the 60 cells showed calcium spiking, and 10 cells showed an intermediate behavior that we could not define as either calcium spiking or unresponsive (representative traces for the different P1 phenotypes are available at www.pnas.org). We conclude that dmi1 and dmi2 mutants are blocked for calcium spiking, but that the P1 allele appears to be leaky for calcium spiking, nodulation, and mycorrhizal invasion.
Table 1.
NF-induced calcium responses of the Nod− mutants
| Locus | Mutant | Allele | Number of cells spiking/ total cells (plants) | Lag, mins | Period between spikes, secs |
|---|---|---|---|---|---|
| DMI1 | Domi | dmi1-1 | 0/32 (9) | ||
| B129 | dmi1-2 | 0/55 (17) | |||
| DMI2 | TR25 | dmi2-1 | 0/38 (9) | ||
| P1 | dmi2-2 | 5/60 (13) | |||
| DMI3 | TRV25 | dmi3-1 | 38/38 (8)* | 11.4 ± 3.8 | 102 ± 27 |
| NSP | B85 | nsp-1 | 13/24 (7)* | 14.8 ± 4.7 | 90 ± 14 |
| HCL | B56 | hcl-1 | 30/41 (10)* | 10.0 ± 3.1 | 102 ± 18 |
| Wild type | 38/46 (14)* | 11.1 ± 3.4 | 102 ± 24 |
All plants displayed calcium spiking.
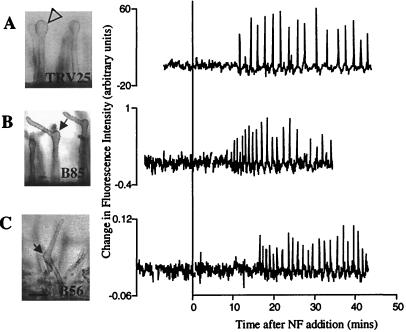
We analyzed a dmi3 mutant, TRV25 (dmi3–1), for its calcium response to NF. This mutant showed calcium spiking that was indistinguishable from the wild-type response: 38 of 38 cells on 8 plants showed calcium spiking (Fig. 2A; Table 1). It is interesting to note that 100% of the TRV25 cells showed calcium spiking, in contrast to 82.6% of wild-type cells. The size of the data set does not permit an assessment of whether this difference is significant. However, if the percent response proves reproducible in larger trials, it would imply that this mutant is affected either in sensitivity to NF or in feedback regulation of signal transduction from a downstream event.
Figure 2.
dmi3, nsp, and hcl mutants exhibit calcium spiking in response to NF. Root hair morphology was scored 16 h after application of 1 nM NF. Closed arrows indicate root hair branches. The open arrowhead indicates a swollen root hair. Traces represent the change in fluorescence intensity from one time point to the next (Xn+1 − Xn) of single root hairs injected with Oregon green–dextran. The amplitude of the fluorescence change is given in arbitrary units and cannot be compared from one cell to another, given that Oregon green is a nonratiometric dye. The traces represent individual cells and, although representative, each cell behaves slightly differently with regard to period and duration of calcium spiking. (A) dmi3 mutant TRV25; (B) nsp mutant B85; (C) hcl mutant B56.
Thus, of the three complementation groups that show similar mutant phenotypes and are proposed to act early in the NF perception/signal transduction pathway, two, dmi1 and dmi2, fail to induce calcium spiking in response to NF, whereas dmi3 shows calcium spiking equivalent to wild type.
Calcium Spiking in nsp and hcl Mutants.
We tested calcium spiking behavior in Nod− mutants that display root hair branching, denoting the ability not only to sense NF but also to alter their polar growth in response to the bacterial signal. It has been hypothesized that the genes corresponding to these mutant phenotypes act later in the pathway than do the DMI1, DMI2, and DMI3 loci. B85 (nsp-1), which carries a recessive mutation in the NSP locus, displays root hair branching akin to that of wild-type plants but differs in other NF and S. meliloti-induced responses (16). B85 displays greatly reduced RIP1 and ENOD11 expression, no cortical cell division, and no detectable ENOD40 expression in response to NF or S. meliloti (see Table 2; ref. 16). Of seven B85 plants tested, all showed some root hairs induced for calcium spiking: 13 of 24 cells showed calcium oscillations in response to NF (Fig. 2B). There was no significant difference in the lag before induction of spiking and the period between spikes in B85 as compared with wild type (Table 1).
We also assessed the calcium behavior of B56 (hcl-1), a mutant of the HCL locus. The B56 mutant shows root hair branching and wild-type expression of ENOD11 and RIP1 in response to NF. Additionally, this mutant is capable of limited cortical cell division when inoculated with S. meliloti (see Table 2; R. Catoira, T. Timmers, C.G., F. Maillet, C.G., R.V. Penmetsa, D.C., G.T. and J.D., unpublished results). Therefore, it has been hypothesized that this gene product acts after NSP in the activation of nodulation (R.C., T. Timmers, C.G., F. Maillet, C.G., R.V.P., D.C., G. Truchet and J.D., unpublished results). B56 initiated calcium spiking in response to NF (Fig. 2C). As shown in Table 1, all 10 of the B56 plants tested, with a total of 30 of 41 cells, displayed calcium spiking with characteristics indistinguishable from wild-type spiking. Thus, we conclude that the nsp and hcl mutants, which share the ability to respond to S. meliloti by root hair deformation, are able to activate calcium spiking in response to NF.
Discussion
In this study, we analyzed calcium ion spiking in a series of plant mutants with defects in early symbiotic responses to bacteria. All mutants fail to form nodules and show an early block in the developmental sequence leading to bacterial invasion. The five complementation groups studied can be distinguished to some extent by the degree of the root hair response to NF or bacteria. Mutants in three complementation groups, dmi1, dmi2, and dmi3, are characterized by root hair swelling but accomplish no further response: they do not display root hair branching, cortical cell division, or expression of early nodulin genes such as ENOD11, ENOD40, or RIP1 (16). Of these three complementation groups, dmi1 and dmi2 are completely or almost completely blocked in calcium spiking, whereas the third shows calcium spiking comparable to wild type. Mutants in two other complementation groups, nsp and hcl, show root hair branching, although the classical “shepherd's crook” structure that is required for infection does not form: both of these mutants are competent for calcium spiking. The observation that two genes are required in common for nodulation and calcium spiking establishes a strong correlation between these two behaviors. These two classes of mutants thus represent genes defined in an eukaryotic organism that are required for ligand-induced calcium spiking.
Correlation of calcium spiking with cellular signaling is widespread, but relatively few studies demonstrate specific output mechanisms for downstream effects. In two studies in cell culture lines derived from leukemic tissue, it was shown that calcium spiking may activate gene expression (5, 6). It has also been shown that calcium spiking is associated with changes in the rate of vesicle fusion in exocytosis (1). In Caenorhabditis elegans, calcium spiking has been linked genetically to rhythmic contractions of the intestine (29). Very little is known about the role of calcium spiking in plant systems. Indeed, calcium spiking has been observed in only two plant cell types: as a response to NF in legumes (27) and in response to the plant hormone abscisic acid and to elevated calcium levels in guard cells (30, 31). Considering the diverse roles hypothesized for calcium spiking in animal systems, it is particularly difficult to define how calcium spiking may be acting in the plants' response to NF. The unbiased nature of this genetic study may allow us to dissect the role of calcium spiking in the legume's response to NF.
The observation that dmi1 and dmi2 mutants are blocked for calcium spiking suggests that the corresponding genes act upstream in the pathway leading from NF perception. In principle, there should be a series of different cellular components that are required for calcium spiking, and mutations in any of them could abolish the response: the receptor(s) for NF perception or proteins that deliver, sequester, or degrade the NF ligand; components of the signal transduction pathway between the receptor and the calcium reservoir; the channels and/or pumps that control calcium release and reuptake; and the cellular functions that modulate feedback for these components. The phenotype of the dmi1 and dmi2 mutants, which show root hair swelling in response to NF, suggests that they have at least some component of the NF perception machinery. It is formally possible that they carry mutations in the NF receptor that weaken the perception or signal transduction domains, rendering them ineffective in activating downstream responses. However, the fact that both dmi1 and dmi2 mutants are defective both for the highly specific Sinorhizobium symbiosis and for the nonspecific mycorrhizal symbiosis makes it less likely that these shared components would be the receptors responsible for recognition itself.
Proceeding from the mutants defective in calcium spiking to those that are permissive, we need to consider theoretically possible roles of calcium spiking in the developmental sequence leading to nodulation. We observed calcium spiking in wild-type M. truncatula, in dmi3, which shows NF-induced root hair swelling, and in two Nod− mutants, in which the symbiotic response proceeds as far as root hair branching (nsp and hcl). It is possible that calcium spiking is part of the signal transduction chain in nodulation (Fig. 3A), or alternatively it may not be a signal transduction component but may be activated by components or steps that are shared with nodulation (Fig. 3B). Our finding that both of the mutants (nsp and hcl) that are able to progress to root hair branching are positive for calcium spiking suggests that the events leading to root hair branching lie downstream either of calcium spiking or of the last common signaling component that is shared by the two behaviors. Chemical inhibition of calcium spiking would more directly assess its role in nodulation, and this approach is currently being tested.
The dmi3 mutant is interesting in that it is phenotypically identical to the dmi1 and dmi2 mutants in all respects except calcium spiking. The calcium data therefore suggest that the three genes can be placed in a rough hierarchy, with DMI1 and DMI2 earlier than DMI3. If calcium spiking is a component in the direct signal transduction chain for NF signaling, then the position of DMI3 would be downstream of calcium spiking, potentially consistent with a gene product involved in transduction of the calcium-spiking signal to downstream targets.
In addition to the nodulation phenotypes, it is relevant to consider that dmi1, dmi2, and dmi3 mutants are also defective in the formation of mycorrhizal associations. Thus, mycorrhizal association and nodulation share common steps that appear to be early, in that two of the three lie upstream of calcium spiking. It is interesting to note that the genes shared between nodulation and mycorrhizal associations appear to span calcium spiking and, although there is no evidence to suggest a role for calcium spiking in mycorrhizal associations, this work suggests that an analysis of the induction of calcium spiking in mycorrhizal associations may be fruitful.
Calcium spiking is a relatively early response in the nodulation sequence. Other NF-induced responses occur within seconds to minutes after NF application, including rapid ion fluxes, membrane depolarization, transient extracellular and sustained cytoplasmic alkalinization, cytoplasmic calcium concentration increases, and cytoskeleton reorganization (reviewed in ref. 32). Additionally, calcium gradients are important in the tip growth of root hairs, and these are observed to change during the root hair response to NF (reviewed in ref. 21). It would be interesting to determine whether M. truncatula wild-type plants display early responses such as calcium or proton flux across the plasma membrane that have been reported for alfalfa (26) and whether dmi1 and dmi2 mutants, which are defective for calcium spiking, show any of these earlier NF-induced responses. Pharmacological evidence suggests involvement of a G-protein and phospholipase C in control of early plant nodulation gene expression (33). However, similar studies using mastoparan found no effect on the induction of calcium spiking in pea (34) or M. truncatula (R. Mustaphi and S.R.L., unpublished data). The cloning of the M. truncatula genes required for calcium spiking and nodulation should be informative for both the activation of signal transduction in plants and the induction of calcium spiking. These gene products may also provide information about the mechanisms of calcium spiking in diverse eukaryotic systems.
Supplementary Material
Acknowledgments
We thank G. Duc (Laboratoire des Légumineuses, Unité de Recherches de Génetique et d'Amélioration des Plantes, Institut National de la Recherche Agronomique, Dijon, France) for seed of TR25 and TRV25, D. Ehrhardt and S. Shaw for help with microscopy and for many useful discussions, K. Jennings for help with the analysis of the calcium spiking data, and J. A. Downie and S. Walker for communicating results before publication. S.R.L. is an investigator of the Howard Hughes Medical Institute. Additional support for this project was provided by International Human Frontier Science Program Award RG0327/1998-M, by the Institut National de la Recherche Agronomique and Centre National de la Recherche Scientifique Genome Program, France, and by Department of Energy Grant DE-FG03–90ER20010.
Abbreviations
- NF
Nod factor
- ENOD
early nodulin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230439797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230439797
References
- 1.Berridge M J, Bootman M D, Lipp P. Nature (London) 1998;395:645–648. doi: 10.1038/27094. [DOI] [PubMed] [Google Scholar]
- 2.Sanders D, Brownlee C, Harper J F. Plant Cell. 1999;11:691–706. doi: 10.1105/tpc.11.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsien R W, Tsien R Y. Annu Rev Cell Biol. 1990;6:715–760. doi: 10.1146/annurev.cb.06.110190.003435. [DOI] [PubMed] [Google Scholar]
- 4.Meyer T, Stryer L. Proc Natl Acad Sci USA. 1988;85:5051–5055. doi: 10.1073/pnas.85.14.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolmetsch R E, Xu K, Lewis R S. Nature (London) 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 6.Li W, Llopis J, Whitney M, Zlokarnik G, Tsien R Y. Nature (London) 1998;392:936–941. doi: 10.1038/31965. [DOI] [PubMed] [Google Scholar]
- 7.Dolmetsch R E, Lewis R S, Goodnow C C, Healy J I. Nature (London) 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 8.De Koninck P, Schulman H. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 9.Hirsch A M. New Phytol. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 10.Long S R. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadri A E, Bisseling T. In: The Rhizobiaceae. Spaink H P, Kondorosi A, Hooykaas P J J, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 403–416. [Google Scholar]
- 12.Schultze M, Kondorosi A. Annu Rev Genet. 1998;32:33–57. doi: 10.1146/annurev.genet.32.1.33. [DOI] [PubMed] [Google Scholar]
- 13.Denarie J, Debelle F, Prome J C. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 14.Downie J A. In: The Rhizobiaceae. Spaink H P, Kondorosi A, Hooykaas P J J, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 387–402. [Google Scholar]
- 15.Spaink, H. P., Kondorosi, A. & Hooykaas, P. J. J. (1998) (Kluwer, Dordrecht, The Netherlands), p. 566.
- 16.Catoira R, Galera C, de Billy F, Penmetsa R V, Journet E P, Maillet F, Rosenberg C, Cook D, Gough C, Dénarié J. Plant Cell. 2000;12:1647–1666. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penmetsa R V, Cook D R. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- 18.Schauser L, Roussis A, Stiller J, Stougaard J. Nature (London) 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 19.Albrecht C, Geurts R, Lapeyrie F, Bisseling T. Plant J. 1998;15:605–614. doi: 10.1046/j.1365-313x.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- 20.Szczyglowski K, Shaw R S, Wopereis J, Copeland S, Hamburger D, Kasiborski B, Dazzo F B, De Bruijn F J. Mol Plant–Microbe Interact. 1998;11:684–697. [Google Scholar]
- 21.Cardenas L, Holdaway-Clarke T L, Sanchez F, Quinto C, Feijo J A, Kunkel J G, Hepler P K. Plant Physiol. 2000;123:443–452. doi: 10.1104/pp.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrhardt D W, Atkinson E M, Long S R. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 23.Kurkdjian A C. Plant Physiol. 1995;107:783–790. doi: 10.1104/pp.107.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felle H H, Kondorosi E, Kondorosi A, Schultze M. Plant J. 1998;13:455–463. [Google Scholar]
- 25.Felle H H, Kondorosi E, Kondorosi A, Schultze M. Plant J. 1996;10:295–301. [Google Scholar]
- 26.Felle H H, Kondorosi E, Kondorosi A, Schultze M. Planta. 1999;209:207–212. doi: 10.1007/s004250050624. [DOI] [PubMed] [Google Scholar]
- 27.Ehrhardt D W, Wais R, Long S R. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 28.Keen N, Gelvin S, Long S. Mol Plant–Microbe Interact. 1999;12:835–838. [Google Scholar]
- 29.Dal Santo P, Logan M A, Chisholm A D, Jorgensen E M. Cell. 1999;98:757–767. doi: 10.1016/s0092-8674(00)81510-x. [DOI] [PubMed] [Google Scholar]
- 30.Staxen I P C, Montgomery L T, Gray J E, Hetherington A M, McAinsh M R. Proc Natl Acad Sci USA. 1999;96:1779–1784. doi: 10.1073/pnas.96.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAinsh M R, Webb A A R, Taylor J E, Hetherington A M. Plant Cell. 1995;7:1207–1219. doi: 10.1105/tpc.7.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Downie J A, Walker S A. Curr Opin Plant Biol. 1999;2:483–489. doi: 10.1016/s1369-5266(99)00018-7. [DOI] [PubMed] [Google Scholar]
- 33.Pingret J L, Journet E P, Barker D G. Plant Cell. 1998;10:659–672. doi: 10.1105/tpc.10.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker S A, Viprey V, Downie J A. Proc Natl Acad Sci USA. 2000;97:13413–13418. doi: 10.1073/pnas.230440097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.