Abstract
Reversible protein acetylation is controlled by the opposing actions of protein lysine acetyltransferases and deacetylations. Recent developments on the structure and biochemical mechanisms of histone acetyltransfers (HATs) have provided new insight into catalysis and substrate selection. Diverse families of HATs appear to perform a conserved mechanism of acetyl-transfer, where the lysine-containing substrate directly attacks enzyme-bound acetyl-CoA. The ability of HATs to form distinct multi-subunit complexes provide a means to regulate HAT activity by altering substrate specificity, targeting to specific loci, enhancing acetyltransferase activity, restricting access of non-target proteins, and coordinating the multiple enzyme activities of the complex. In the case of newly discovered Rtt109 HAT, association with distinct histone chaperones directs substrate selection between N-terminal lysines (H3K9, H3K23) and those (H3K56) within the histone fold domain. Moreover, the ability of some HATs to utilize longer chain acyl-CoA (i.e. propionyl-CoA) as alternative substrates suggests a potential direct link between the metabolic state of the cell and transcriptional regulation.
In 1964, Allfrey and coworkers showed that acetylation of histones was associated with more efficient RNA synthesis, suggesting a role for histone acetylation in the regulation of transcription [1]. Over the last decade, the enzymes responsible for site-specific histone acetylation were identified and their roles in replication, histone deposition, and other DNA-based processes have been established [2•,3]. Using acetyl-CoA as the acetyl donor, histone acetyltransferases (HATs) acetylate the ɛ-amine of lysine residues [4,5]. Acetylation removes the positive charge of lysine and alters interactions with DNA and other chromatin-associated proteins [3,6]. The dynamic interplay between HATs and histone deacetylases dictates the ultimate state of acetylation. The fact that the amino-acid sequence of histones are some of the most highly conserved among eukaryotes and that there is high sequence conservation of HATs, underscores the critical nature of histone acetylation in genome regulation [2•,3,7].
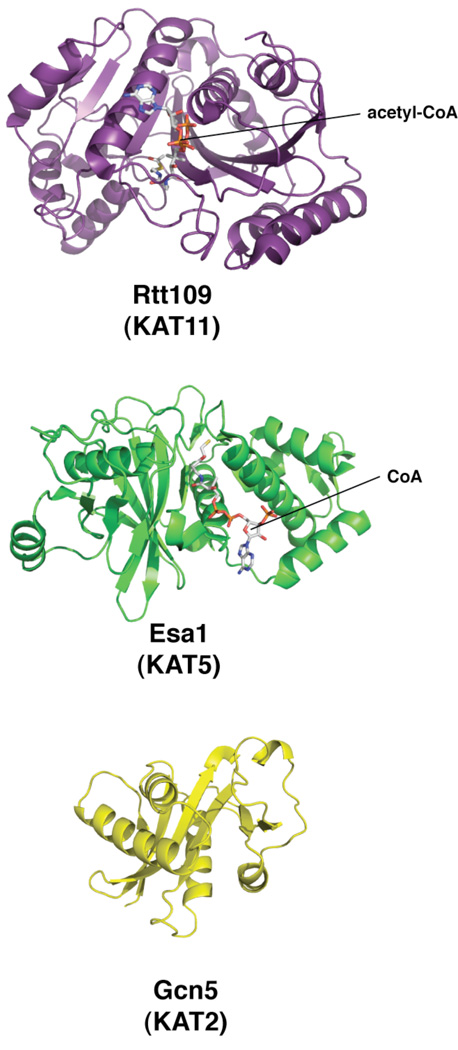
HATs can be categorized into three major families: the MYST family containing MOZ, Ybf2/Sas3, Sas2, Tip60, the Gcn5 related N-acetyltransferase or GNAT family, and the more recently characterized p300 family (Table 1)[3,5,8*]. In addition, there are a number of putative acetyltransferases (i.e. Spt10), which contain motifs similar to those found in HATs but their acetyltransferase activity remains to be confirmed [9]. Although this review will focus on their histone acetylation activity, it is important to note that many of these enzymes are capable of acetylating non-histone proteins, and thus could be generally termed protein lysine acetyltransferases [3,10,11]. The MYST and GNAT families are largely unrelated in primary sequence, but most HATs have an acetyl-CoA binding domain or Motif A, and a similar active site structure (Figure 1) [4,9]. However, not all HATs harbor a recognizable Motif A, as is the case for Rtt109 [9,12•–14]. The apparent diversity among known HATs has lead to speculation that these differences reflect diverse catalytic mechanisms and substrate specificity. In this review, we will highlight the most recent structural and biochemical investigations on the molecular mechanisms of histone acetylation. As an aid to the reader, Table 1 lists the HATs discussed in this review according to their respective families, a brief summary of their histone specificity, and major protein complexes associated with each HAT.
Table 1.
The Major Families of Histone Acetyltransferases
| Common/Gene Namea |
KAT Designationb |
Histone Specificity |
Known Complexes |
|---|---|---|---|
| GNAT family of Histone Acetyltransferasesc | |||
| Gcn5 | 2 | H3K9, 14, 36 | SAGA, Gcn5/Ada2/Ada3, ATAC, TFTC |
| p/CAF | 2B | H3K14 | STAGA |
| MYST family of Histone Acetyltransferases | |||
| Esa1 (Tip60 in H. sapiens) |
5 | H4K5, K8, K12, K16; Htz1K14 |
NuA4, Piccolo NuA4 |
| Sas2 (MOF in H. sapiens) |
8 | H4K16 | SAS-I |
| Sas3 | 6 | H3K14, K23 | NuA3 |
| MOZ | 6A | H3K14 | MOZ |
| p300 family of Histone Acetyltransferases | |||
| CBP | 3A | H2AK5; H2B | Numerous |
| p300 | 3B | H2AK5; H2B | Numerous |
| Rtt109 | 11 | H3K56, K9, K23 |
Rtt109•Vps75, Rtt109•Asf1 |
Designations used in this review
K (lysine) Acetyltransferase nomenclature proposed by Allis, et al., 2007 to simplify the identification and classification of new histone acetyltransferases [8•]
GNAT = Gcn5-related N-acetyltransferase; MYST = MOZ, Ybf2/Sas3, Sas2, Tip60
Figure 1. Representative structures of protein/histone lysine acetyltransferases from S. cerevisiae.
X-ray crystal structures of Rtt109 bound to acetyl-CoA (purple, 3D35), Esa1 bound to CoA (green, 1FY7) and Gcn5 (yellow, 1YGH) [15,19,27•].
CONSERVED MECHANISM OF ACETYLATION
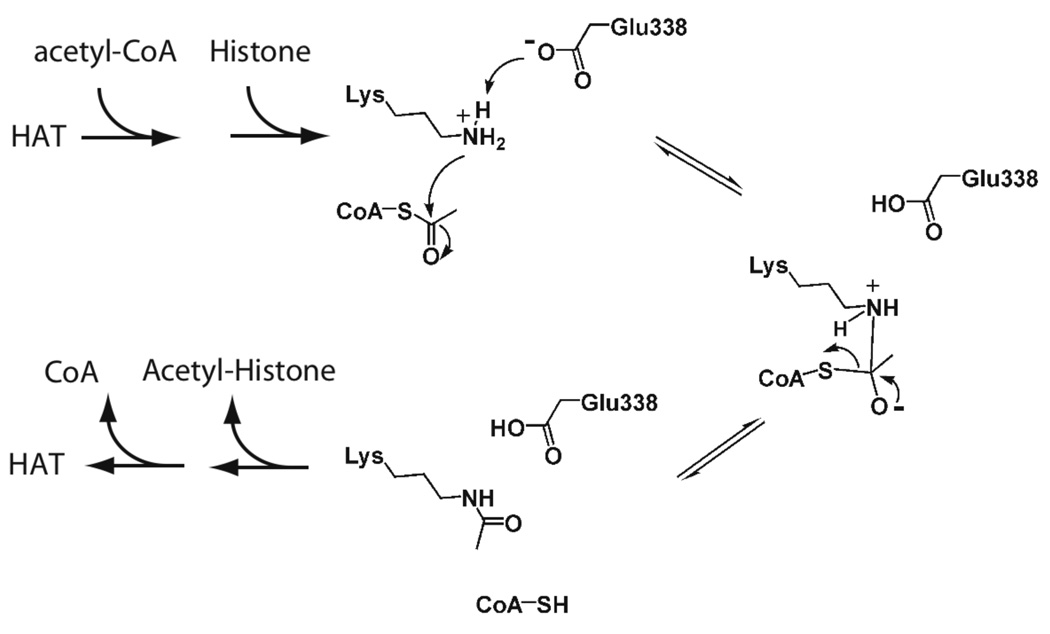
How HATs achieve acetyl-transfer has been the subject of much debate and investigation. For the GNAT family of HATs, initial biochemical and structural work revealed a sequential mechanism of acetyl transfer [15–18]. In this mechanism, acetyl-CoA and the protein substrate bind to form a ternary complex before any chemical catalytic step (Figure 2). An active-site glutamate (base) assists in deprotonating the lysine, allowing nucleophilic attack on the carbonyl carbon of acetyl-CoA. A putative tetrahedral intermediate forms, which then collapses to the reaction products, CoA and the acetylated protein (Figure 2). This direct attack mechanism is similar to that described for the glycoside acetyl-transferases, serotonin acetyltransferases, and amino acid acetyltransferases (reviewed in [7]). Although the original structure of MYST family member Esa1 suggested a similar catalytic mechanism to that for Gcn5-like enzymes, a subsequent study from the same group reported the structure of Esa1 with an acetylated active site cysteine [19,20]. In contrast, this structure was presented as evidence for a two step acetyl-transfer mechanism [20]. In this proposed mechanism, Esa1 binds acetyl-CoA followed by attack of the Cys-304 sulfhydryl on the acetyl-moiety, yielding an acetylated enzyme intermediate (Ac-S-Cys304). Dissociation or release of CoA would permit binding of the lysine-containing substrate. The lysine nucleophile is then deprotonated by an active site base (E338 in Esa1) for attack on the acetylated-Cys, yielding acetylated peptide. This mechanism is distinct from the direct attack mechanism established for Gcn5. The possibility of a two-step chemical mechanism for Esa1 was surprising given the strong structural homology between Esa1 and Gcn5-like HATs [19,21,22]. However, a more recent study provided compelling biochemical data to support a one step catalytic mechanism, where lysine directly attacks the acetyl-CoA from the ternary complex enzyme•acetyl-CoA•histone-peptide [23•] (Figure 2). The major distinction between these two studies was the version of Esa1 employed [20,23•]. Yan, et al. used a truncated version of Esa1, whereas Berndsen, et al. used full-length Esa1 alone and Esa1 in complex with two accessory proteins, known as the piccolo NuA4 complex [24]. Mutagenesis of Cys-304 to alanine yielded a piccolo NuA4 complex (and full-length Esa1 alone) that was catalytically indistinguishable from wild-type Esa1 enzymes harboring cysteine at position Cys-304 [23•]. Thus, efficient catalysis by Esa1 does not require Cys-304 nor the formation of an acetylated cysteine intermediate. Analogous to Gnc5, Glu338 of Esa1 functions as a base to abstract the proton from the bound substrate [23•]. This new evidence supports the idea that MYST and Gcn5 family HATs utilize a universal mechanism for acetyl transfer, as depicted in Figure 2.
Figure 2. Propose kinetic and catalytic mechanism of acetyl transfer.
Proposed sequential mechanism of acetylation for histone/protein acetyltransferases. After binding acetyl-CoA and peptide substrate to form a ternary complex, an active site glutamate (e.g. Glu338 from Esa1) deprotonates the ɛ-amine of substrate lysine. Lysine attacks the carbonyl carbon of the acetyl moiety of acetyl-CoA forming a tetrahedral intermediate, which then collapses to form CoA and acetylated product. The recent p300 structure did not reveal a putative base, although a backbone amide may assist in removing a proton from the ɛ-amine [26•].
The catalytic mechanism for the HAT p300 has remained unsettled for nearly a decade. Initial studies suggested an acetylated intermediate in the mechanism mechanism [25], however, recent biochemical and structural efforts revealed that p300 utilizes a special type of sequential mechanism with a short-lived ternary complex, known as a Theorell-Chance mechanism [26•]. The X-ray structure (at 1.7 Å) of the p300 HAT domain suggested that the backbone amide of Trp1436 aids in deprotonating the substrate lysine for attack on the carbonyl carbon of bound acetyl-CoA [26•]. An active site Tyr residue is proposed to serve as a general acid and protonate the sulfhydryl of CoA after acetyl-transfer. Recently, Rtt109, a putative HAT from yeast, was found to be a distant structural homolog of p300, however whether Rtt109 uses a similar mechanism of catalysis is unclear [27•]. The active site does not harbor any obvious nucleophilic or acid/base residues to suggest the type mechanism used by this unique class of enzymes. Given the fact that Rtt109 requires a histone chaperone for efficient catalysis [12•,13,28•,29•], the solved Rtt109 structure likely represents an inactive form of the enzyme [27•]. Deletion of a loop that is critical for chaperone binding was removed to aid in crystallization [27•]. It is tempting to speculate that chaperone binding may reorient the active site and position a base for an efficient direct-attack mechanism. Though the mechanism of Rtt109-like HATs remains to be determined, there is now compelling evidence to suggest that known HATs employ a common direct-attack mechanism without the need for a covalent acetyl-enzyme intermediate.
HAT COMPLEXES
In many respects, protein acetylation is a post-translational modification that rivals phosphorylation. However, protein/histone acetylation appears to be carried out by a surprisingly limited number of protein acetyltransferases [5,8•]. Although more protein acetyltransferases may yet be discovered, the ability of a limited number of HAT catalytic subunits to exist in diverse multi-subunit complexes might explain this apparent conundrum. Their interactions with other proteins serve to regulate the catalytic activity of the acetyltransferase subunit and to help coordinate the multiple (enzymatic and binding) functions within the protein complex. Targeting the HAT activity of multi-subunits complexes is often a means to restrict acetylation to specific regions or loci of the genome, as appears to be the case with Gcn5 (recently reviewed in [30–33•]). Additionally, the existence of multiple enzyme activities within the same complex appears to be a way to coordinate the many enzymatic processes required to mediate transcriptional activation. The SAGA complex contains the histone ubiquitin protease Ubp8 and the histone acetyltransferase Gcn5 [34]. In yeast, the coordinated activities of Gcn5 and Ubp8 are needed for efficient transcription of SAGA regulated genes such as GAL1 and ADH2, as deletion of both cripples growth more severely than either single deletion [35]. The human NuA4 complex contains the HAT, Tip60, but also ATM, a DNA damage related kinase. In response to DNA damage, Tip60 acetylates ATM allowing for phosphorylation of Chk2 and p53 by ATM [36,37]. Rtt109 associates with the histone chaperones Vps75 or Asf1 [12•,13,28•,29•]. Association of Rtt109 with Asf1 is critical for H3K56 acetylation in vivo, while Rtt109•Vps75 has recently been shown to function in H3K9 and H3K23 acetylation in vivo [28•,29•]. Moreover, the activities of both subunits appear to be interconnected, as both chaperones enhance the HAT activity of Rtt109, and Rtt109 stimulates histone deposition by Vps75 and Asf1 [12•,28•,29•,38,39](C.E. Berndsen and J.M. Denu, unpublished data). Thus, HAT interacting partners appear to regulate HAT activity by a.) altering substrate specificity, b.) targeting to specific loci, b.) enhancing acetyltransferase activity, c.) restricting access of non-target proteins, and d.) coordinating the multiple enzyme activities of the complex.
CATALYTIC ACTIVATION OF ACETYL-TRANSFER
Kinetic studies comparing Esa1 and Rtt109 as individual subunits with their respective physiological complexes indicates that other subunits enhance the catalytic efficiency by greater than 50-fold, with only minor effects on the Michaelis constant (Km) [12•,27•,29•,40]. Although there are ~10 additional subunits of NuA4, Esa1 minimally requires Yng2 and Epl1 (the piccolo NuA4 complex) for full catalytic activity and nucleosome recognition [24,40]. The existence of two complexes containing Esa1•Epl1•Yng2 suggests separate functions. Piccolo NuA4 is proposed to function as a genome-wide H4 HAT complex, while the additional subunits of NuA4 assist in targeting the HAT complex to specific loci [24]. For the Rtt109 HAT, stimulation of activity by Vps75 results in ~50-fold increase of the kcat value, though the dissociation constant (Kd) for histone H3 and the Km for H3 peptide were not enhanced for the Rtt109•Vps75 complex [27•,29•] (Table 2). As was the case with Esa1, binding of these “helper” proteins to Rtt109 does not appear to alter overall affinity for substrate [29•,41], but rather induces the active conformation of the HAT domain, leading to enhanced rates of catalysis. An alternative explanation for the stimulatory effect of histone chaperones on Rttt109 activity would be that the chaperone binds and presents the substrate in the proper orientation for the direct attack on bound acetyl-CoA. From the individual structures of Rtt109 [27•] and Vps75 [29•,41], we present a conceptual model for their functional interaction (Figure 3). Additional experiments will be needed to explore these mechanisms in detail.
Table 2.
Comparison of kinetic constants for HAT catalyzed acetyl transfer
| HAT | kcat, s−1 | Km AcCoA, µM | Km peptide, µM | Reference |
|---|---|---|---|---|
| p300 | 4.1 ± 0.1 × 100 | 40 ± 0.6 | 1.6 ± 0.1 × 102a | [26•] |
| Gcn5 | 1.7 ± 0.1 × 100 | 2.5 ± 1.4 | 4.9 ± 0.8 × 102 b | [18] |
| Rtt109 | 1.7 ± 0.1 × 10−3 | 0.3 ± 0.1 | 8.3 ± 2.9 × 101 c | [29•] |
| Rtt109•Vps75 | 1.3 ± 0.4 × 10−1 | 1.0 ± 0.2 | 7.5 ± 1.5 × 101 c | [29•] |
| Esa1 | 1.9 ± 1.0 × 10−3 | 1.0 ± 0.4 | 1.4 ± 1.0 × 103 d | [40]; C.E.B. and J.M.D., unpublished data |
| Piccolo NuA4 | 1.6 ± 0.1 × 100 | 2.5 ± 0.3 | 2.2 ± 0.3 × 102 d | [23•] |
Calculated using a peptide of H4 residues 4 to 15
Calculated using a peptide of H3 residues 1 to 20 plus a C-terminal cysteine
Calculated using a peptide of H3 residues 1 to 20
Calculated using a peptide of H4 residues 1 to 20
Figure 3. Conceptual model for the interaction of histone acetyltransferase Rtt109 with histone chaperone Vps75.
The structures of Vps75 and Rtt109 have been solved separately. The basic patches of Vps75 (shown in light blue) were manually aligned with the acidic patches of Rtt109 (shown in red) in MacPyMOL (DeLano Scientific LLC, San Francisco) [27•,29].
In stark contrast, Gcn5, p/CAF and p300 appear to be constituitive HATs that do not require “helper” proteins to exhibit full catalytic activity. These HATs show catalytic activity on a similar scale to that of the Esa1 and Rtt109 complexes (Table 2). From our current understanding of HAT catalysis, this distinction might point to the existence of two major regulatory mechanisms among HATs. In one case, Esa1 and Rtt109 represent low-activity HATs that are stimulated by regulatory “helper” proteins, Yng2-Epl1 and Vps75/Asf1, respectively. With Gcn5, p/CAF and p300, which present constuitively-active HATs, accessory proteins may function to restrict access to non-target substrates, either through direct targeting or through inhibition [30,33,42]. Although the cellular control of HAT activity will likely entail multiple layers involving post-translation modifications and changes in the pools of the short-chain-CoA available to acetyltransferases (see below), the current data certainly suggest these two general mechanisms may exist.
SUBSTRATE SPECIFICITY OF HATS
Molecular understanding for substrate selection by HAT domains remains elusive. Though a number of studies have provided new insight, it is unclear how important the active site and catalytic domains are in specific lysine selection. The catalytic domains of Gcn5 and p/CAF have been crystallized with a number of peptide substrates including sequences from histone and p53 [21,22,43]. Early work on Tetrahymena Gcn5 had shown that the sequence G-K-X-P within histone H3, which includes the primary Gcn5 substrate K14, made several key contacts within the active site that are conserved with other GNAT members including p/CAF [44]. As more substrates of Gcn5 were identified, further studies were unable to define a clear consensus sequence, which lead to the proposal that the non-catalytic subunits of the Gcn5 complex may define the substrate specificity [22,43]. However, it was noted that the interactions between the HAT and the side chains in the +2 and +4 positions relative to the substrate lysine appear to be similar across several divergent substrates [43]. These data suggest that selection of H3K14 and sites within p53 is based on context-specific contacts between substrate and the active sites of Gcn5 and p/CAF.
Among all known HATs, p300/CBP appear to exhibit the broadest protein specificity [3,26•,42]. The recent crystal structure of the catalytic domain revealed an electronegative pocket near to the active site, fitting with the previous observation that p300 prefers histone acetylation sites with a positive charge in the −3 or +4 position [25]. In the Theorell-Chance kinetic mechanism of p300, the central ternary complex noted for other HATs (Figure 1) is transiently formed, suggesting a “looser” association with the peptide substrate [27•]. These collective observations indicate that p300 may not have the intimate peptide substrate contacts, which are observed in other HAT structures. Substrate specificity and targeting may be driven by interactions outside of the catalytic domain. Though other protein factors such as E1A and Nap1 can modulate p300/CBP HAT activity, it remains unclear whether this regulation alters site or substrate specificity, as appears to be the case for the HAT activity of Rtt109 (described below) [45,46].
For Gcn5, p/CAF, and p300, only 3 to 5 residues on either side of the substrate lysine are required for efficient binding and catalysis by the catalytic domain. Recent studies on the MYST family HATs, Esa1 and MOZ indicate that efficient acetylation is influenced by interactions with more distal regions of the substrate [40,47]. Although piccolo NuA4 is largely an H4 tail specific enzyme, the complex acetylates tail peptides (up to the first 20 amino acids) of H3 and H4 at slow rates [22,25,40]. Strikingly, full-length histone H4 is acetylated 2000-fold faster than histone tail peptides, indicating that interactions with the histone-fold domain of H4 is critical for the high catalytic efficiency and the ability of the enzyme to acetylated four lysine residues with the same H4 tail [40,48]. In another interesting example, DNA was found to enhance the acetyltransferase activity of MOZ [47]. Based on the presented biochemical data, a model was proposed where MOZ is directed to acetylate specific loci based on direct interaction with DNA, however it remains to be seen how DNA sequence specificity may regulate MOZ HAT activity [47].
Lastly, HAT specificity can be modulated through interactions with other accessory proteins. Very recently, two groups identified H3K9 and H3K23 as targets of Rtt109 in vitro and in vivo [28•,29•]. Prior to these studies, Rtt109 had been linked to H3K56 acetylation and histone deposition from newly synthesized H3 [49]. Acetylation of the H3 tail was only apparent when Rtt109 was paired with the histone chaperone Vps75, but not with the distinct chaperone Asf1, which specifies H3K56 acetylation [28•,29•,49]. Thus, the chaperone associated with Rtt109 dictates the acetylation site specificity. These studies provide direct evidence that recognition of acetylation sites can be influenced by association with a single accessory protein. While we have presented data covering three distinct mechanisms for substrate selection, it is important to note that these mechanisms are not mutually exclusive, and reflect remaining gaps in our molecular understanding of HAT regulation between and among HAT families.
USE OF LONGER CHAIN ACYL-COA BY PROTEIN ACETYLTRANSFERASES
To date, the use of longer chain acyl-CoAs by protein lysine acetyltransferases has been an underdeveloped research area. A recent MS analysis of protein acylation in humans suggests the presence of lysine propionylation and butyrylation on histone H4 and p53, however the proteins that perform these modifications in vivo are unknown [50•]. In vitro, Esa1 and p/CAF can transfer the propionyl group from propionyl-CoA with slightly lower efficiency (5–10-fold lower in kcat/Km) compared with acetyl-CoA, providing direct evidence that some HATs are endowed with the ability to act as more general acyltransferases [23•,51]. A cellular connection between HATs and longer chain acylation of lysine has not been established and the functional implications of protein propionylation and butyrylation are unclear. In Salmonella, N-propionylation of propionyl-CoA synthetase downregulates enzyme activity, similar to the regulation of bacterial and mammalian acetyl-CoA synthetase activity by reversible acetylation [52–55•]. The ability of diverse acyl-CoA pools to act as substrates of protein acetyltransferases is an exciting future directing that might provide a direct link between the metabolic state of the cell and transcriptional regulation. Moreover, use of longer chain acyl-CoAs offers the possibility of discovering new functions for HATs.
CONCLUSIONS AND PERSPECTIVES
Recent structural and enzymic studies have provided tremendous insight into the molecular mechanisms of HATs. Many of these in vitro studies have employed truncated proteins for reasons of practicality and obtaining suitable quantities for biochemical and structural experiments. However, the intricacy of nucleosome recognition and the complex regulation of HAT activity gleaned from the current literature, indicates that such in vitro studies are just a starting point for understanding the fascinating array of mechanisms which control HAT activity. In particular, future investigations on alternate acyl-CoA substrates and on protein substrate selection by shuttling HATs between different protein complexes will be necessary to advance our current knowledge.
References (•denotes paper of high significane published in the last 2 years)
- 1.Allfrey VG, Faulkner R, Mirsky AE. Acetylation And Methylation Of Histones And Their Possible Role In The Regulation Of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. •An excellent review of the biological implications associated with site-specific histone acetylation.
- 3.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 5.Hodawadekar SC, Marmorstein R. Chemistry of acetyl transfer by histone modifying enzymes: structure, mechanism and implications for effector design. Oncogene. 2007;26:5528–5540. doi: 10.1038/sj.onc.1210619. [DOI] [PubMed] [Google Scholar]
- 6.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 7.Vetting MW, LP SdC, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 8. Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. •This paper suggests a new consistent nomenclature for many chromatin-modifying enzymes.
- 9.Neuwald AF, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 10.Fu M, Wang C, Zhang X, Pestell RG. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem Pharmacol. 2004;68:1199–1208. doi: 10.1016/j.bcp.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 11.Yang XJ, Gregoire S. Metabolism, cytoskeleton and cellular signalling in the grip of protein N-ε- and O-acetylation. EMBO Rep. 2007;8:556–562. doi: 10.1038/sj.embor.7400977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. •A number of papers, including references 12, 13, 14, 28, 29, 38, 39, 40 & 41, demonstrated that yeast Rtt109 is a bona fide HAT, whose activity is controlled by two histone chaperones Asf1 and Vps75.
- 13.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 16.Tanner KG, Langer MR, Denu JM. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry. 2000;39:15652. doi: 10.1021/bi005121q. [DOI] [PubMed] [Google Scholar]
- 17.Lau OD, Courtney AD, Vassilev A, Marzilli LA, Cotter RJ, Nakatani Y, Cole PA. p300/CBP-associated factor histone acetyltransferase processing of a peptide substrate. Kinetic analysis of the catalytic mechanism. J Biol Chem. 2000;275:21953–21959. doi: 10.1074/jbc.M003219200. [DOI] [PubMed] [Google Scholar]
- 18.Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 19.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat Struct Biol. 2002;9:862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, Fletcher CM, Zhou J, Allis CD, Wagner G. Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature. 1999;400:86–89. doi: 10.1038/21922. [DOI] [PubMed] [Google Scholar]
- 22.Clements A, Poux AN, Lo WS, Pillus L, Berger SL, Marmorstein R. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol Cell. 2003;12:461–473. doi: 10.1016/s1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- 23. Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46:623–629. doi: 10.1021/bi602513x. •This study demonstrated that the highly efficient catalytic mechanism of MYST HAT Esa1 is similar to that of the GCN5-like enzymes, and does not involve an acetyl-cysteine enzyme intermediate.
- 24.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J Biol Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- 26. Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. •This paper describes the long-awaited structure determination for the catalytic domain of p300. Overall, the structure resembles other known HAT structures, even though sequence homology is minimal. Kinetic data suggest a GCN5-like sequential mechanism, where the ternary complex is transiently formed known as a Theorell-Chance mechanism.
- 27. Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verrault A, Cole PA, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1448. •This study presents the structure determination of the newly discovered HAT Rtt109. Though homology is poor, the catalytic domains of Rtt109 and p300/CBP are similar in overall architecture.
- 28. Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone Control of the Activity and Specificity of the Histone H3 Acetyltransferase Rtt109. Mol Cell Biol. 2008 doi: 10.1128/MCB.00182-08. •This paper and reference 29 demonstrated that Rtt109 and Vps75 function together as a histone H3 tail acetyltransferase.
- 29. Berndsen CE, Tsubota T, Lindner SE, Lee S, Holton JM, Kaufman PD, Keck JL, Denu JM. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1459. •This paper and reference 41 present the structure of histone chaperone Vps75 and describe its unique biochemical role in activating the Rtt109 HAT.
- 30.Baker SP, Grant PA. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pillus L. MYSTs mark chromatin for chromosomal functions. Curr Opin Cell Biol. 2008;20:326–333. doi: 10.1016/j.ceb.2008.04.009. •A good review of the biological functions of MYST family HATs.
- 32.Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- 33.Nagy Z, Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- 34.Sanders SL, Jennings J, Canutescu A, Link AJ, Weil PA. Proteomics of the eukaryotic transcription machinery:identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol Cell Biol. 2002;22:4723–4738. doi: 10.1128/MCB.22.13.4723-4738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 39.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 40.Berndsen CE, Selleck W, McBryant SJ, Hansen JC, Tan S, Denu JM. Nucleosome recognition by the Piccolo NuA4 histone acetyltransferase complex. Biochemistry. 2007;46:2091–2099. doi: 10.1021/bi602366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park YJ, Sudhoff KB, Andrews AJ, Stargell LA, Luger K. Histone chaperone specificity in Rtt109 activation. Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 43.Poux AN, Marmorstein R. Molecular basis for Gcn5/PCAF histone acetyltransferase selectivity for histone and nonhistone substrates. Biochemistry. 2003;42:14366–14374. doi: 10.1021/bi035632n. [DOI] [PubMed] [Google Scholar]
- 44.Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 45.Perissi V, Dasen JS, Kurokawa R, Wang Z, Korzus E, Rose DW, Glass CK, Rosenfeld MG. Factor-specific modulation of CREB-binding protein acetyltransferase activity. Proc Natl Acad Sci U S A. 1999;96:3652–3657. doi: 10.1073/pnas.96.7.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asahara H, Tartare-Deckert S, Nakagawa T, Ikehara T, Hirose F, Hunter T, Ito T, Montminy M. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol Cell Biol. 2002;22:2974–2983. doi: 10.1128/MCB.22.9.2974-2983.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holbert MA, Sikorski T, Carten J, Snowflack D, Hodawadekar S, Marmorstein R. The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting. J Biol Chem. 2007;282:36603–36613. doi: 10.1074/jbc.M705812200. [DOI] [PubMed] [Google Scholar]
- 48.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol Cell Biol. 2005;25:8179–8190. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. •A mass spectroscopy study that provided compelling evidence that histones can be modified by propionylation and butyrylation of lysine residues. References 23, 51 & 52 provide additional evidence that such modifications can be performed through enzyme-dependent mechanisms.
- 51.Leemhuis H, Packman LC, Nightingale KP, Hollfelder F. The human histone acetyltransferase P/CAF is a promiscuous histone propionyltransferase. Chembiochem. 2008;9:499–503. doi: 10.1002/cbic.200700556. [DOI] [PubMed] [Google Scholar]
- 52.Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J Biol Chem. 2007;282:30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- 53.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 55. Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. •This study along with references 53 and 54 provided the first direct evidence that metabolic enzymes from both bacteria and mammals are regulated by reversible protein acetylation.