Summary
The sudden availability of DNA sequencing technologies that rapidly produce vast amounts of sequence information has triggered a paradigm shift in genomics, enabling massively parallel surveying of complex nucleic acid populations. The diversity of applications to which these technologies have already been applied demonstrates the immense range of cellular processes and properties that can now be studied at the single-base resolution. These include genome resequencing and polymorphism discovery, mutation mapping, DNA methylation, histone modifications, transcriptome sequencing, gene discovery, alternative splicing identification, small RNA profiling, DNA-protein and possibly even protein-protein interactions. Thus, these deep sequencing technologies offer plant biologists unprecedented opportunities to increase the understanding of the functions and dynamics of plant cells and populations.
Keywords: deep sequencing, genome sequencing, transcriptome, methylation, epigenetics, small RNA, natural variation
Introduction
The application of genomic techniques to plant research has yielded a multitude of discoveries concerning plant cellular biology, development and evolution. Now, the sudden rise of relatively low cost and rapid “next-generation” DNA sequencing technologies is dramatically advancing our ability to comprehensively interrogate the nucleic-acid based information in a cell at unparalleled resolution and depth. Already this technology has been employed to study genome sequence variation, ancient DNA, cytosine DNA methylation, protein-DNA interactions, transcriptomes, alternative-splicing, small RNA populations and mRNA regulation (Figure 1), with a number of these applications being effectively applied to plant systems. Current deep sequencing technologies produce many gigabases of single-base resolution information and can perform multiple genome-scale experiments in a single experimental run, thus being effective in the analysis of many plant genome equivalents. However, it should be noted that some significant challenges remain in the employment of this new technology, most evident are informatics and data processing issues that arise from the generation of such large (terabytes per run) volumes of data. Here we discuss several applications of these “now-generation” DNA sequencing technologies and the insights they have yielded into the diversity of plant genome regulation.
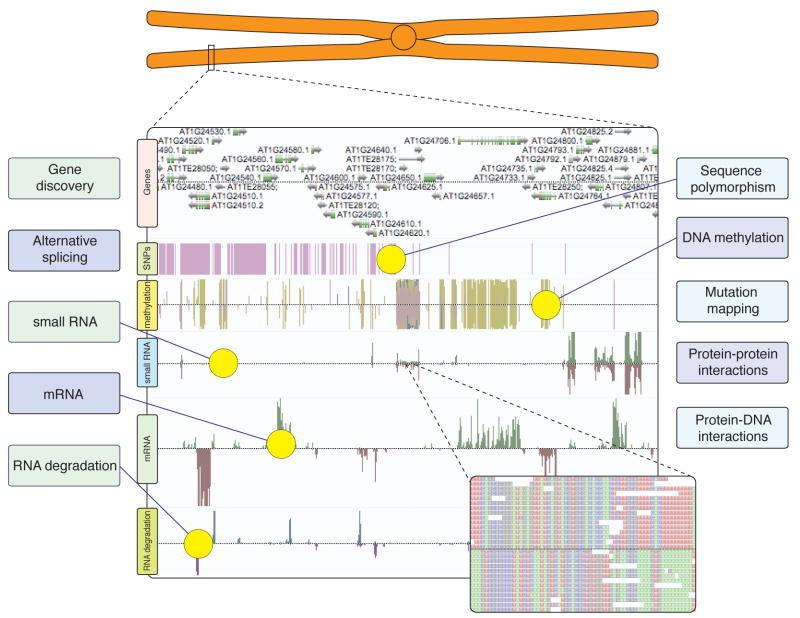
Figure 1. Advanced DNA sequencing technologies underly diverse approaches to unravel plant cellular activities.
Massively parallel DNA sequencing of complex nucleic acid populations now enables numerous subsets of genomic and cellular information to be rapidly characterized at unprecedented resolution and breadth. The AnnoJ genome browser (www.annoj.org) excerpt shown above represents approximately 100 kilobases of Arabidopsis thaliana chromosome 1. Single nucleotide polymorphisms between Col-0 and Ler-1 ecotypes (Lister, O’Malley, Ecker, unpublished), single-base DNA methylation maps, strand-specific smRNA and mRNA components of the transcriptome, and RNA-degradation products from Arabidopsis thaliana flower buds, all generated by ultra high-throughput DNA sequencing, have been integrated to illustrate the holistic views of genomic and transcriptional regulation and variation that can now be routinely captured [41,48].
Currently, there are three widely deployed deep sequencing platforms in hundreds of research labs and in some core facilities worldwide, the Genome Sequencer FLX from 454 Life Sciences/Roche, Illumina Genome Analyzer, and Applied Biosystems SOLiD. Each instrument essentially massively parallelizes individual reactions, sequencing hundreds of thousands to hundreds of millions of distinct, relatively short (50 to 400 bases) DNA sequences in a single run. The technical details of the operation and chemistries of each sequencer have been reviewed in detail recently ([1, 2]). Here, we will briefly outline the quantity and constitution of sequence data produced by each platform. It should be noted that each of these platforms have seen dramatic and rapid increases in total yield, sequence quality and read length, such that the figures quoted will likely be rapidly surpassed by the time of publication of this review. The Genome Sequencer FLX from 454 Life Sciences is capable of producing over a million reads of up to 400 bases per 10 hour run, for a total yield of 400 – 600 megabases. The Illumina Genome Analyzer will yield over one hundred million high-quality short reads (up to 76 bases) per 3–5 day run, totaling several gigabases of aligned sequence. Finally, the Applied Biosystems SOLiD system will also produce hundreds of millions of short reads (up to 50 bases) per flow cell in a similar time frame to yield an equivalent quantity of sequence as the Illumina instrument. Furthermore, all three platforms offer the paired-read sequencing technique, where sequence is produced from both ends of a long DNA molecule, increasing the unambiguous mapping of sequence reads by spanning repetitive regions and anchoring one repetitive read to a distinct genomic location by its unique partner sequence. The base-calling error rates observed with the new sequencing technologies are on average ten times greater than capillary based Sanger sequencing, and the type of error varies between the different platforms [2]. However, the massive increase in sequence output affords the possibility to generate multiple passes of the same sequence, thereby greatly reducing error rates.
Applications for genome analysis
Genome sequencing and polymorphism discovery
Identification of sequence polymorphisms in related but phenotypically distinct individuals or groups within a species is an essential step in elucidation of the causative genetic differences that give rise to observed phenotypic variation. Furthermore, the distribution of genetic polymorphism is informative of population structure and evolutionary history. Hybridization of genomic DNA to high-density oligonucleotide arrays has successfully been used to identify genetic polymorphisms in several organisms including human, mouse and Arabidopsis thaliana [3–5]. However, utilization of tiling microarrays to identify genetic polymorphisms is limited to genomic regions that are highly similar to the reference strain sequence upon which the tiling array is designed, as efficient probe hybridization is necessary for deconvolution of the sequence in the other strains. Consequently, the analysis of genomic sequence variation is confined to these highly similar sequences, while regions containing small to large insertions or deletions, or a high density of polymorphisms cannot easily be interrogated.
The recent development of deep sequencing technologies is a major boon for the aforementioned areas of investigation, in which interrogating the genomic sequence of a wide range of individuals, strains or species is essential to generating highly informative datasets. The ability to generate vast amounts of sequence data from any organism enables the rapid discovery of much greater sequence variation than has been identified previously. Through a recent study in Arabidopsis thaliana, Ossowski et al. (2008) reported the resequencing of two naturally occurring and geographically distinct strains of Arabidopsis thaliana (Bur-0 and Tsu-1) with short reads generated by the Illumina sequencing technology [6•]. Furthermore, the study details the development of a new computational mapping tool, ShoRe, which enables identification of both SNPs and 1–3 bp indels at high sensitivity and specificity. Within these two studied strains, over 800,000 non-redundant single-nucleotide polymorphisms (SNP) were identified relative to the reference strain Col-0, constituting a dramatic increase in SNP discovery relative to previous array-based experiments [3]. Furthermore, over 79,000 1–3 bp indels were identified in the genomes of these two strains, resulting in 1,839 potential frame shifts, and regions that showed significantly higher coverage than expected were identified, likely indicating duplicated regions. Finally, 3.4 megabases of the Bur-0 and Tsu-1 genomes that was identified as highly dissimilar, duplicated or deleted relative to the reference Arabidopsis thaliana genome was targeted for de novo sequence assembly, resulting in the generation of 10,921 high-confidence contigs of up to 408 bp. Clearly, a wide assortment of polymorphism information can be gleaned from limited short read sequencing of divergent Arabidopsis thaliana accessions, and the sequence of Bur-0 and Tsu-1 will be highly informative for ongoing research into extant Bur-0/Tsu-0 recombinant inbred lines. The study by Ossowski et al. marks the first data release of the international cooperative endeavor to sequence the genomes of 1,001 distinct strains of Arabidopsis thaliana (http://1001genomes.org), which will provide a vast resource for the comprehensive study of global polymorphism, population structure, and analysis of the genetic basis of natural phenotypic variation.
New developments in sequencing technology, such as significantly longer reads and paired reads separated by multiple kilobases, must to be applied to enable true de novo assembly of the complete plant genomes. Application of these technological advances will enable significantly more comprehensive detection of the genetic diversity such as large structural variation within related genomes, and consequently aid elucidation of the polymorphisms that dictate phenotypic variation.
Mutation mapping by deep sequencing
Screening of populations subjected to mutagenesis and identification of the causative genetic lesions of mutant phenotypes is a fundamental approach in the discovery of gene function. Forward genetic screens have proven extremely powerful in Arabidopsis thaliana for assigning genes to specific biological pathways [7]. The success of this approach is, in part, due to the highly accurate sequence of its compact genome [8], facile genetics, and extensive collection of mapping markers [9]. However, identifying the causative mutation commonly takes several months to years after generating a mapping population, so approaches to expedite this step will be highly valuable. In a modification of an approach termed bulked segregant analysis [10, 11], deep sequencing of a pool of F2 individuals containing only mutant plants from a mapping population enables rapid mapping of the mutation. Every sequenced SNP between the two parental strains of the mapping population acts as a marker (Figure 2), and hundreds of thousands of SNPs can now be routinely detected with relatively low genome coverage [6•]. Tracts homozygous for the genotype of the mutagenized strain are indicative of no recombination events occurring within that region, and thus are within physical proximity of the mutation. Furthermore, the sequence within this region can be scoured for potential mutations to rapidly identify the exact location of the genetic lesion, although sequencing errors and accumulated non-causative polymorphisms in the mutant population compared to the reference sequence may contribute to false-positive identification. Recently, using a “sequencing with prior mapping” approach, Sarin et al. (2008) reported the use of the Illumina platform to sequence the genome of the C. elegans mutant lsy-12 to identify the causative mutation [12•]. Notably, for organisms with genomes of moderate size such as Arabidopsis thaliana, a 76 base read/paired-end sequencing run that yields ~40x coverage currently takes only two weeks and costs a few thousand dollars, both factors that will see continual, rapid and dramatic improvement based upon the progression in the last two years. While this mutation-mapping approach offers great potential and is already being applied in a number of plant laboratories, both statistical predictions and empirical testing of the size of the mutant pool and the required coverage are necessary to determine the most effective experimental strategies. Looking ahead, mutation mapping will likely soon undergo even more dramatic advances. Recent studies have already demonstrated the identification of specific mutations by deep sequencing without inter-strain crosses to generate a mapping population [13•, 14•]. Thus, with the rapid increases in sequence output it is now conceivable to directly identify mutations in plant genomes, effectively taking the Mendel (genetic crosses) out of mutation mapping.
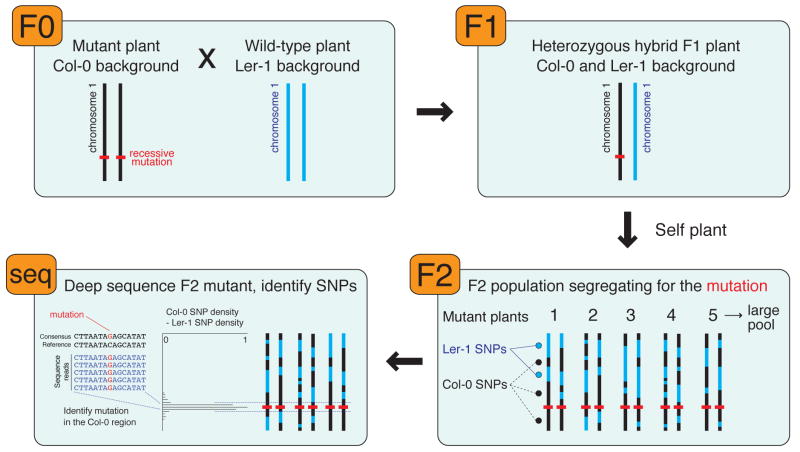
Figure 2. Identification of mutations by deep sequencing.
A plant with Col-0 background that harbors a recessive mutation leading to a mutant phenotype is crossed to a wild-type Ler-1 plant. The heterozygous F1 hybrid plant is allowed to self-fertilize to produce a large pool of F2 plants that are segregating for the mutation. A large number of F2 plants that display the mutant phenotype are pooled and their gDNA subjected to deep sequencing. The density of single nucleotide polymorphisms (SNPs) inherent in the Ler-1 strain is subtracted from the density of SNPs indicative of the Col-0 background, identifying a discrete region on the chromosome in which only Col-0 marker SNPs are present. The deep sequencing data in this interval is then scoured for the potential causative mutation.
Analysis of DNA-protein interactions through ChIP-seq
DNA-protein interactions mediate innumerable critical nuclear processes that govern genome organization, replication and interpretation of the inherent underlying information. Chromatin structures such as nucleosome composition and position, and post-translational modifications of histones influence chromatin compaction and interactions with transcription machinery, thus affecting proximal transcriptional activity [15–18]. Therefore, comprehensive genome-wide maps of such chromatin composition and state, and more broadly the full range of DNA-protein interactions, are essential to generate a more complete understanding of genome and transcriptional regulation. While these interactions were historically gradually revealed by analysis of interactions at a small number of genomic loci, more recent studies have utilized genomic tools such as high-density oligonucleotide arrays to interrogate the sites of interaction throughout entire genomes. The ChIP-chip method involves immunoprecipitation of specific chromatin through its interaction with a protein of interest that is crosslinked to proximal genomic DNA in the context of its in vivo interactions [19, 20]. Purification, labeling and hybridization of the immunoprecipitated genomic DNA to arrays enables identification of the genomic sites at which interaction of the protein with the genomic DNA occurred [21, 22]. ChIP-chip has been used extensively to produce comprehensive maps of DNA-protein interactions in plants and animals [23–27]. With the availability of new sequencing technologies, the chromatin immunoprecipitation technique has rapidly been coupled to shotgun sequencing to generate even higher resolution maps of protein-DNA interactions, an approach dubbed “ChIP-seq”, revealing distinct patterns of transcription factor binding, RNA polymerase II, and histone modifications in human and mouse lineage-committed, differentiating, as well as pluripotent and induced-pluripotent stem cells [28••–32]. With several gigabases of sequence generated in each sequencing run, ChIP samples are perfectly suited for analysis with deep sequencing technology, generally requiring only a fraction of the total output of a single run to saturate detection of sites of protein-DNA interaction. In fact, the rapidly increasing output of the DNA sequencers such as the Illumina Genome Analyzer and Applied Biosystems SOLiD likely already provides a cost-benefit over array hybridization for analysis of ChIP samples, particularly in organisms that possess large genomes that are distributed over several arrays. Sample barcoding, by addition of a short unique sequence tag to all sequenced molecules within one library, and subsequent multiplexing will further decrease the cost [33•]. Further advantages over ChIP-chip are evident in the higher resolution of the interactions that can be observed through the distribution of ChIP-seq short read tags. While there are no publications of the utilization of ChIP-seq in plant systems, numerous laboratories are currently employing this technique to gain new insights into DNA-protein interactions in plant cells and a flurry of papers utilizing this new method is expected soon.
Genome-wide detection of sites of DNA methylation
Methylation of cytosines in the nuclear genomes of diverse eukaryotic lineages is an epigenetic modification that is required for numerous cellular processes, including transposon silencing, genomic imprinting, embryogenesis and gene regulation [34–39]. Several distinct molecular pathways control the deposition of DNA methylation in plants, so clearly the comprehensive detection of these sites at single-base resolution is necessary to gain an understanding of the pathways involved in its patterning and how it affects the underlying genetic information.
Single-base resolution analysis of sites of DNA methylation can be achieved by sodium bisulfite (BS) treatment of genomic DNA, which converts cytosines, but not methylcytosines, to uracil [40]. Subsequent sequencing of PCR-amplified bisulfite-converted DNA allows determination of the methylation state of the cytosines in the sequenced region of the genome, as methylcytosine will be sequenced as cytosine, and unmethylated cytosine as thymine. While historically this approach was limited to analysis of a small number of loci, deep sequencing technologies have recently enabled two groups to conduct shotgun bisulfite sequencing of the entire Arabidopsis thaliana genome with a technique dubbed BS-seq or methylC-seq, offering an unprecedented view of the DNA methylome [41•, 42•]. Using the Illumina Genome Analyzer, Cokus et al. and Lister et al. generated 2–3 gigabases of uniquely aligned bisulfite sequence to comprehensively identify sites of DNA methylation throughout the Arabidopsis thaliana genome at single base-resolution, including previously unidentified sites of cytosine methylation, and local sequence motifs associated with DNA methylation. The relationship with small RNA abundance, downstream effects upon transcription of modifying methylation patterns, and dynamics of DNA demethylation were also uncovered [42•].
Application of methylC-seq to study distinct cell types, related but genetically distinct natural populations, and organisms exposed to various biotic and abiotic stresses will provide an unparalleled assessment of the extent to which cytosine methylation patterns vary within and between organisms.
Applications for transcriptome sequencing
Deep sequencing of small RNA populations
RNA silencing represents a pathway that controls expression of specific genes transcriptionally and post-transcriptionally [43]. In RNA silencing, small RNAs (smRNAs) comprise the sequence-specific effectors of RNA silencing pathways that direct the negative regulation or control of genes, repetitive sequences, viruses, and mobile elements [44, 45].
To gain insights into the total population and gain a better understanding of smRNA function in plants a number of groups turned to sequencing the smRNA component of the plant transcriptome (smRNAome). Numerous groups have recently employed Genome Sequencer FLX from 454 Life Sciences and Illumina Genome Analyser sequencing technologies to look at the smRNAome of various plant species [42•, 46–60]. Putting these two technologies to work, the sequencing of smRNAomes from plants containing various genetic lesions has resulted in the elucidation and categorization of millions of smRNAs, as well as the identification of biogenesis factors and regulators of specific smRNA populations [42•, 48–51, 53, 55, 57]. For instance, sequencing the smRNAomes of Arabidopsis thaliana plants harbouring lesions in genes encoding DNA methyltransferases in conjunction with single-base resolution DNA methylation analysis (see above) revealed a strong correlation between the location of smRNAs and DNA methylation, a disruption in biogenesis of specific smRNA size classes upon loss of CpG DNA methylation, and the potential of smRNAs for directing strand-specific DNA methylation in regions of RNA-DNA homology [42•]. In another study, sequencing experiments using Arabidopsis thaliana rdr2 and maize mop1-1 mutant plants, which lack a homologous RNA-dependent RNA polymerase, revealed loss of this protein results in a significant decrease in the 24 nt smRNA population of the smRNAome. This loss of 24 nt smRNAs was accompanied in the sequencing experiments by an increase in sequencing of those that were 21 nt in length, which through subsequent analysis resulted in the identification of numerous unidentified miRNAs throughout the Arabidopsis thaliana (rdr2) and maize (mop1-1) genomes. Furthermore, these studies revealed that 24 nt smRNAs, which are mostly associated with repetitive elements and heterochromatic regions of the genome, comprise the bulk of the Arabidopsis thaliana and maize smRNAome complexity [53, 55].
With accessibility to these technologies becoming increasingly available, the number of plant species with sequenced smRNAomes is ever increasing [46, 47, 52, 54–60]. So far this collection of sequence data has elucidated that smRNAomes are not statically maintained between all species. More specifically, the distribution of smRNAs amongst various size classes has been found to differ between plants. This differential distribution of smRNA lengths is hypothesized to reflect a disparity in the maintenance of genomic organization between plant species that have dramatic variations in the quantity of their genetic material [54, 61].
Ultimately, with millions of sequence reads generated in each run, and the ability to determine specific nucleotide length of all identified smRNAs machines such as the 454 sequencer, Illumina Genome Analyser, and Applied Biosystems SOLiD provide ideal platforms for complete indexing of the plant smRNAome. Additionally, the increased use of barcoding of numerous smRNA samples [51], and subsequent multiplexing will result in the sequencing of smRNAomes from an even greater variety of plant species. With the ensuing flood of smRNA sequencing data from an immense collection plant species, a clearer view of the dynamic nature of plant smRNAomes will emerge. Additionally, these datasets will aid in elucidating how these small regulatory RNA molecules have evolved between plant species to regulate genomes with such disparity in size.
mRNA sequencing for transcript discovery and profiling
As the astounding and unexpected complexity of eukaryotic transcriptomes has become apparent over the last few years [24, 62–68], so the requirement has grown for techniques that allow broad but accurate characterization of the dynamic cellular complement of transcripts. Ideally such approaches will incorporate highly specific, sensitive and quantitative measurements over a large dynamic range with a flexibility to identify unanticipated novelties in transcript structures and sequences.
A number of studies have recently used deep sequencing to perform surveys of the mRNA component of the transcriptome in various organisms, enabling parallel quantification and annotation of cellular transcripts. While sequencing of cDNA pools is a well established technique, for example the sequencing of EST libraries [69], the ability to rapidly and cheaply generate diverse cDNA sequence datasets will allow the transcriptional activity of a vast array of different cell types, mutants and environmental conditions to be analyzed. Deep sequencing of cDNA, referred to as RNA-seq, overcomes several shortcomings of microarray-based detection of transcripts, including probe cross-hybridization [70], restricted signal dynamic range, and low sensitivity and specificity, which often lead to difficulties in detection of low abundance transcripts and discrimination between similar sequences. Sequence-level transcript information has much greater power to distinguish between paralogous genes, better detection of low abundance transcripts, and allows replicable digital quantification based upon counting of sequence reads [71–75]. Furthermore, RNA-seq can identify transcript sequence polymorphisms, novel trans-splicing and splice isoforms, and there is no strict-requirement for a reference genome sequence. Whilst approaches such as SAGE, CAGE and MPSS have enabled parallel sequencing of short reads from many transcripts, they suffer from a poor coverage of each transcript and potentially ambiguous mapping due to the short read length [76–78]. In contrast, RNA-seq can produce complete coverage of transcripts, providing information about the sequence, structure and genomic origins of the entire transcript.
Several strategies have been employed to perform shotgun sequencing of cellular mRNAs, but they can be broadly categorized as either “stranded” RNA-seq, yielding strand-specific data that informs about transcript directionality, or “strandless” RNA-seq, where sequencing of double-stranded cDNA fragments loses the strand of origin information [79•]. The first papers reporting RNA-seq of plant transcripts with one of the new deep sequencing technologies utilized the 454 sequencer, generating strandless RNA-seq data from double stranded cDNA of Medicago truncatula, Arabidopsis thaliana and maize [80–82]. Cheung and colleagues [81] sequenced adapter-ligated fragments of a normalized Medicago truncatula cDNA library, assembling the reads into contigs representing thousands of previously unobserved and rare transcripts. In Arabidopsis thaliana seedlings, Weber et al. [81] generated reads mapping to 17,449 genes, accounting for ~90% of the transcripts estimated to be expressed in the sample, identifying reads from previously unannotated transcripts and predicted genes with no prior EST support. Finally, Emrich and colleagues [81] sequenced cDNA from maize shoot apical meristem cells isolated by laser-capture microdissection, identifying over 25,000 genomic sequences, including nearly 400 orphan transcripts with no homology to sequences from any other species and which appeared to be expressed in a cell-type specific manner. Clearly, the sensitivity of the shotgun sequencing is applicable for characterization of the transcript complement of individual cell types.
Several recent publications have utilized the Illumina Genome Analyzer and Applied Biosystems SOLiD instruments to generate vast datasets of short expressed tags in Arabidopsis thaliana, human, mouse and yeast [42•, 71–75, 83]. Essentially, these instruments yield vastly more transcriptome sequence per run than the 454 Life Sciences instrument, typically over one hundred million individual reads, however the length of these reads is significantly shorter than those from the 454 instrument. Thus, while many more unique sequence tags are generated, the shorter read length of the Illumina and Applied Biosystems machines provide a challenge to perform transcript assembly, identification of multiple splicing events within the same mRNA molecule, and unambiguous read alignment to some transcripts with highly similar sequences. However, the vast quantity of short read sequence is extremely powerful for transcript quantification, gene discovery, correction of transcriptional unit structure annotation, and detection of alternative splicing [72••, 74].
In a recent study, Lister et al. [42•] utilized a strand-specific RNA-seq technique to sequence the transcriptome from flower buds of wild-type and DNA methyltransferase or DNA demethylase deficient mutant Arabidopsis thaliana plants. By overlaying the RNA-seq data with the single-base resolution detection of DNA methylation in the same tissues, Lister and colleagues identified hundreds of genes that displayed altered transcript abundance upon perturbation of proximal DNA methylation patterns. Importantly, the stranded RNA-seq data was essential for identification of the strand from which the intergenic transcripts originated and unambiguous identification of repetitive transposon sequences reactivated upon loss of the repressive methylation modifications and alteration of proximal smRNA abundance (Figure 3).
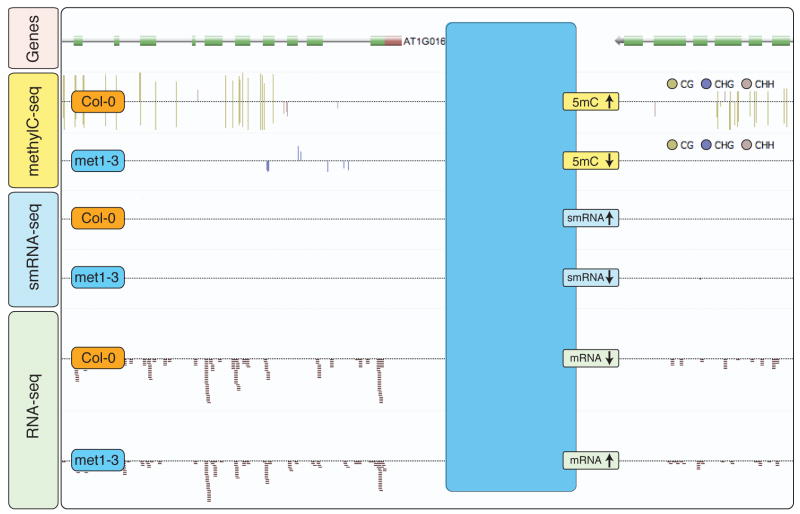
Figure 3. Integration of multiple deep-sequencing datasets for identification of DNA methylation-repressed transcripts.
Shotgun sequencing was used to generate single-base resolution maps of DNA methylation and the smRNA and mRNA components of the transcriptome in wild-type (Col-0) and DNA methyltransferase-deficient mutant (met1-3) plants. Integration of these diverse datasets and visualization in the Anno-J deep-sequencing browser (www.annoj.org) revealed hundreds of intergenic transcribed regions that were normally suppressed in wild-type plants, where loss of DNA methylation in the met1-3 mutant was accompanied by a decrease in smRNA abundance and an increase in transcriptional activity [41].
While RNA-seq offers previously unparalleled means to characterize cellular transcriptional activity, numerous methodological advances that are now being pursued offer to greatly enhance its effectiveness. Paired-read sequencing can be used assess the splicing patterns of multiple distal exons within a single transcript to be studied, while with single short reads it is generally only possible to assess one splice event. With increases in read length constantly being pursued eventually it will be feasible to sequence and assemble an entire transcript, thus revealing the precise splicing pattern. Such a development would also greatly facilitate an understanding of the transcriptome of plant species that do not yet possess high quality reference sequences, allowing identification of novel transcripts where shorter reads at this point may preclude effective contig assembly. It will be essential for RNA-seq techniques to be refined to require significantly less starting material, so as to enable the sequencing of single cells to characterize their transcriptional complement and identify cell-type specific transcripts. Together, such developments will greatly improve the value of RNA-seq, providing researchers with a more comprehensive understanding of the composition and dynamics of plant cell transcriptomes.
Recently, more specialized RNA-seq approaches have been developed to sample the 3′ cleavage fragments produced by endonucleolytic cuts, and in so doing captured a global snapshot of degraded RNAs [49•, 84•, 85•]. These “degradome” sequencing approaches exploit the 5′-RACE principle but ignore the 5′ mRNA cap and selectively clone mRNA molecules with a 5′ monophosphate [49•, 84•, 85•]. Analysis of the degradome sequencing data revealed that the vast majority of expressed genes had sequencing reads that mapped to them, the majority mapping specifically to the 3′ ends of mRNA molecules, suggesting that some level of endonucleolytic cleavage mostly targeted to the 3′ end of mRNAs and subsequent turnover is the norm for most expressed transcripts [49•, 84•, 85•]. Additionally, this type of sequence information, which is riddled with sequenced miRNA-directed cleavage sites, has been used to identify known and previously unidentified miRNA target mRNAs [84•, 85•]. Overall, these recent studies illustrate how high-throughput sequencing technologies can be utilized to gain insights into global RNA dynamics within plants.
Future prospects and concluding remarks
The advent of widely available new or now-generation sequencing technologies has spawned a remarkable array of applications to study genomic and cellular dynamics and features with unprecedented precision and breadth. Many of these new sequence-enabled techniques have been applied to plant systems, producing intriguing insights into cellular function, and genome and population dynamics that could not previously have been obtained. Widespread adoption of these new sequencing technologies will allow researchers to characterize a vast assortment of plant processes in both model and non-model species. The many varied techniques will inevitably be applied to generate detailed temporal and spatial maps of cellular states and activities, profiling not only different cell types within an organism but, with suitable advances in sample preparation and amplification methods, perhaps also single cells. A tantalizing goal is the effective integration of the many complex and rich sequencing datasets to yield cohesive views of cellular activities and dynamics, yet clearly there are substantial bioinformatic challenges that lie ahead on the path to this objective.
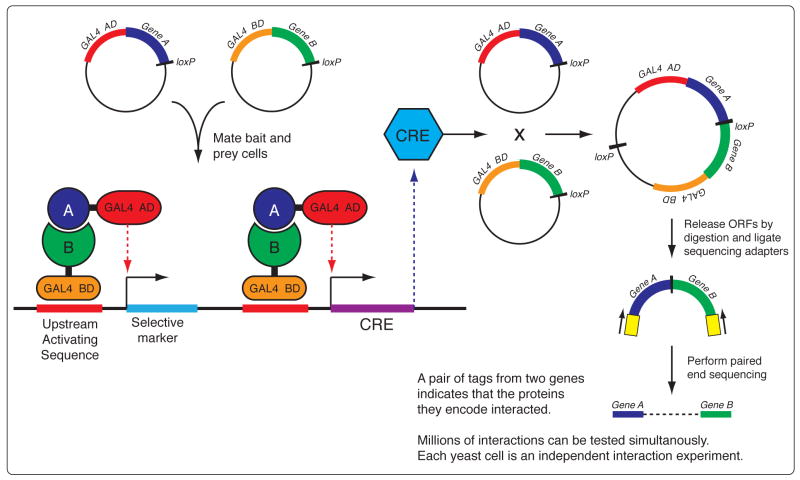
Theoretically, any cellular process or experimental assay for which the output is in nucleic acid form can be comprehensively interrogated, providing an opportunity for the development of a wide assortment of novel applications. For example, it should be possible to combine the yeast two-hybrid screening method [86] with deep sequencing to perform a massively parallel protein-protein interaction experiment, interrogating every pairwise permutation of the full protein-coding complement of an organism’s genome to generate a complete direct-interaction network. In this proposed technique (Figure 4) interaction of bait and prey constructs results in the activation of the CRE recombination system and expression of a selective marker gene. loxP sites situated at the end of each gene in the bait and prey constructs will be recombined to form a chimeric DNA molecule containing the two gene ORFs that encode the interacting proteins. Restriction digestion to release the chimeric molecule followed by paired-end sequencing of its two ends will yield a pair of sequences, one from each of the genes, thus identifying the two proteins that directly interacted. Two complex pools of yeast cells, each one containing the full complement of an organism’s gene ORFs fused to either the bait or the prey domain, would be mixed and allowed to mate. Deep sequencing performed on the complex pool of resulting chimeric DNA molecules would reveal every pairwise interaction that took place, interrogating the hundreds of millions of possible interactions between every protein encoded in a eukaryotic genome, Such a parallelized approach will be the only possible avenue through which to test the 784 million possible interactions of the 28,000 proteins encoded in the Arabidopsis thaliana genome.
Figure 4. Massively parallel interrogtion of all pairwise protein interactions for all proteins encoded by a genome by bait-prey recombination and deep-sequencing.
Interaction of bait and prey constructs results in the activation of the CRE recombination system and expression of a selective marker gene. Recombination at loxP sites located at the end of each gene forms a chimeric DNA molecule containing the two genes that encode the interacting proteins. Digestion to release the chimeric ORFs followed by paired-end sequencing of its two ends will produces one sequence tag from each of the genes, thus identifying the two proteins that directly interacted. Two complex pools of yeast cells, each one containing the full complement of an organism’s genes fused to either the bait or the prey domain, would be mixed and allowed to mate. Sequencing of the complex pool of chimeric ORFs would reveal all pairwise interaction that occurred, interrogating the hundreds of millions of possible interactions between any two proteins encoded in a eukaryotic genome.
As enabling as this leap in technology has been, several companies already claim to soon deliver momentous increases in sequence read length and output (e.g. Pacific Biosciences, http://www.pacificbiosciences.com; Complete Genomics, http://www.completegenomics.com; Visigen Biotechnologies, http://visigenbio.com). With such advances it may soon be possible to apply these new technologies to the study of plants with much larger genomes, and to survey a wide range of plant species, thus dramatically increasing the understanding of the diversity of plant life.
Acknowledgments
We thank Dr. Robert Schmitz for valuable input in the manuscript preparation. R.L. is supported by a Human Frontier Science Program Long-term Fellowship. B.D.G. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1909-06). This work was supported by grants from the National Science Foundation, the Department of Energy, the National Institutes of Health, and the Mary K. Chapman Foundation to J.R.E.
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest related to this publication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mardis ER. Next-generation DNA sequencing methods. Annual review of genomics and human genetics. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 2.Shendure JHJi. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 3.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. Common Sequence Polymorphisms Shaping Genetic Diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 4.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 5.Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294:1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 6•.Ossowski S, Schneeberger K, Clark R, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008 doi: 10.1101/gr.080200.108. The authors perform resequencing of two strains of Arabidopsis thaliana with the Illumina Genome Analyser and develop new computational tools to align the sequence reads and identify different types of genetic polymorphisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Page DR, Grossniklaus U. The art and design of genetic screens: Arabidopsis thaliana. Nat Rev Genet. 2002;3:124–136. doi: 10.1038/nrg730. [DOI] [PubMed] [Google Scholar]
- 8.AGI. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 9.Jander G. Gene identification and cloning by molecular marker mapping. Methods Mol Biol. 2006;323:115–126. doi: 10.1385/1-59745-003-0:115. [DOI] [PubMed] [Google Scholar]
- 10.Giovannoni JJ, Wing RA, Ganal MW, Tanksley SD. Isolation of molecular markers from specific chromosomal intervals using DNA pools from existing mapping populations. Nucleic Acids Research. 1991;19:6553–6558. doi: 10.1093/nar/19.23.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Sarin S, Prabhu S, O’Meara MM, Pe’er I, Hobert O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nature Methods. 2008;5:865–867. doi: 10.1038/nmeth.1249. This paper (and [13•], [14•]) provides an example of identification of an EMS mutation by resequencing with the Illumina Genome Analyser. In this study the authors located the mutation in a strain of C. elegans starting from rough mapping of the lesion to a 4 megabase region. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 14.Srivatsan A, Han Y, Peng J, Tehranchi A, Gibbs R, Wang J, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 16.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 17.Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- 18.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 19.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 20.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 21.Kim TH, Ren B. Genome-wide analysis of protein-DNA interactions. Annual review of genomics and human genetics. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- 22.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nature Protocols. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernatavichute Y, Zhang X, Cokus S, Pellegrini M, Jacobsen S, Dilkes B. Genome-Wide Association of Histone H3 Lysine Nine Methylation with CHG DNA Methylation in Arabidopsis thaliana. PLoS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang ZD, Paccanaro A, Fu Y, Weissman S, Weng Z, Chang J, Snyder M, Gerstein MB. Statistical analysis of the genomic distribution and correlation of regulatory elements in the ENCODE regions. Genome Res. 2007;17:787–797. doi: 10.1101/gr.5573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. The authors performed ChIP-seq to generate high resolution maps of 20 histone methylation modifications, the histone variant H2A.Z, RNA polymerase II and the insulator binding protein CTCF throughout the human genome, identifying characteristic patterns linked to gene transcriptional activity and regulatory elements. [DOI] [PubMed] [Google Scholar]
- 29.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 30.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Research. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nature Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. This paper details methods for incorporating “barcode” sequence tags before the read sequence for multiplexing many samples with the new sequencing technologies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 35.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 36.Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 37.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 40.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. The authors performed shotgun bisulfite sequencing of the Arabidopsis thaliana genome to identify sites of cytosine DNA methylation at single base resolution, identifying sequence motifs associated with DNA methylation, distinct periodicity of the modification, and its alteration in a range of mutant plants deficient in DNA methyltransferase enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. This study used the Illumina sequencing technology to create single-base DNA methylation maps, characterise the smRNA component of the transcriptome, and perform strand-specific RNA-seq in wild-type and DNA methyltransferase and demethylase mutant Arabidopsis thaliana plants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 44.Almeida R, Allshire RC. RNA silencing and genome regulation. Trends Cell Biol. 2005;15:251–258. doi: 10.1016/j.tcb.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 46.Barakat A, Wall K, Leebens-Mack J, Wang YJ, Carlson JE, Depamphilis CW. Large-scale identification of microRNAs from a basal eudicot (Eschscholzia californica) and conservation in flowering plants. Plant J. 2007;51:991–1003. doi: 10.1111/j.1365-313X.2007.03197.x. [DOI] [PubMed] [Google Scholar]
- 47.Barakat A, Wall PK, Diloreto S, Depamphilis CW, Carlson JE. Conservation and divergence of microRNAs in Populus. BMC Genomics. 2007;8:481. doi: 10.1186/1471-2164-8-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc Natl Acad Sci U S A. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu C, Kulkarni K, Souret F, Muthuvalliappan R, Tej S, Poethig R, Henderson I, Jacobsen S, Wang W, Green P, Meyers B. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci U S A. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pandey SP, Gaquerel E, Gase K, Baldwin IT. RNA-directed RNA polymerase3 from Nicotiana attenuata is required for competitive growth in natural environments. Plant Physiol. 2008;147:1212–1224. doi: 10.1104/pp.108.121319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L) Genome Biol. 2007;8:R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dolgosheina EV, Morin RD, Aksay G, Sahinalp SC, Magrini V, Mardis ER, Mattsson J, Unrau PJ. Conifers have a unique small RNA silencing signature. Rna. 2008;14:1508–1515. doi: 10.1261/rna.1052008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 63.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 64.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 65.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 66.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 67.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 68.Laubinger S, Zeller G, Henz S, Sachsenberg T, Widmer C, Naouar N, Vuylsteke M, Scholkopf B, Ratsch G, Weigel D. At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 2008;9:R112. doi: 10.1186/gb-2008-9-7-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 70.Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Research. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cloonan N, Forrest A, Kolle G, Gardiner B, Faulkner G, Brown M, Taylor D, Steptoe A, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 72••.Mortazavi A, Williams B, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. In this study the authors utilize the RNA-seq technique to sequence the transcriptome of various mouse tissues, identifying a multitude of transcript splice junctions, exploring various technical issues associated with the approach, and developing a powerful RNA-seq computational analysis platform. [DOI] [PubMed] [Google Scholar]
- 73.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 75.Wilhelm B, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett C, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 76.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 77.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 79•.Cloonan N, Grimmond S. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008;9:234. doi: 10.1186/gb-2008-9-9-234. The authors developed a strand-specific RNA-seq technique and explored technical issues related to this new methodology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheung F, Haas B, Goldberg SM, May G, Xiao Y, Town CD. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7:272. doi: 10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weber A, Weber K, Carr K, Wilkerson C, Ohlrogge J. Sampling the Arabidopsis Transcriptome with Massively Parallel Pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marioni J, Mason C, Mane S, Stephens M, Gilad: RNA-seq Y. An assessment of technical reproducibility and comparison with gene expression arrays. Genome Research. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84•.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. This paper (and [49•], [85•]) describe a massively parallel adaptation of the 5′ RACE technique to investigate RNA degradation dynamics and miRNA-mediated transcript cleavage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.German M, Pillay M, Jeong D, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis L, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 86.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]