SUMMARY
Deciphering the multiple layers of epigenetic regulation that control transcription is critical to under-standing how plants develop and respond to their environment. Using sequencing-by-synthesis technology we directly sequenced the cytosine methylome (methylC-seq), transcriptome (mRNA-seq), and small RNA transcriptome (smRNA-seq) to generate highly integrated epigenome maps for wild-type Arabidopsis thaliana and mutants defective in DNA methyltransferase or demethylase activity. At single-base resolution we discovered extensive, previously undetected DNA methylation, identified the context and level of methylation at each site, and observed local sequence effects upon methylation state. Deep sequencing of smRNAs revealed a direct relationship between the location of smRNAs and DNA methylation, perturbation of smRNA biogenesis upon loss of CpG DNA methylation, and a tendency for smRNAs to direct strand-specific DNA methylation in regions of RNA-DNA homology. Finally, strand-specific mRNA-seq revealed altered transcript abundance of hundreds of genes, transposons, and unannotated intergenic transcripts upon modification of the DNA methylation state.
INTRODUCTION
Methylation of cytosines in nuclear DNA is an epigenetic modification found in diverse eukaryotic organisms that imparts an additional layer of heritable information upon the DNA code. In higher eukaryotes, DNA methylation is involved in myriad essential processes, including embryogenesis, genomic imprinting, and tumorigenesis in mammals, and in transposon silencing and gene regulation in plants (Bestor, 2000; Li et al., 1992; Lippman et al., 2004; Rhee et al., 2002; Zhang et al., 2006; Zilberman et al., 2007). DNA methylation patterns are established and perpetuated through DNA replication by DNA methyltransferases, which in eukaryotes catalyze the transfer of a methyl group to cytosine, forming 5-methylcytosine. The flowering plant Arabidopsis thaliana is an exceptionally tractable organism in which to conduct genomic studies of the biology of DNA methylation, due to the high-quality sequence of its compact genome (119 Mb) and a diverse collection of viable null DNA methyltransferase mutants. Whereas methylation at CpG dinucleotides predominates in animals, in plant cells distinct pathways govern the methylation of cytosines throughout all sequence contexts (Bernstein et al., 2007; Henderson and Jacobsen, 2007). DNA methylation is established in all contexts by DRM1/2, homologs of the mammalian DNMT3a/b de novo DNA methyltransferases (Cao et al., 2003; Cao and Jacobsen, 2002). A DNA methylation targeting system termed RNA-directed DNA methylation (RdDM) operates in plant cells, whereby 21–24 nt small RNA (smRNA) molecules generated by DICER-LIKE3-dependent endonuclease activity are incorporated into ARGONAUTE4, presumably to guide DRM1/2 activity to the corresponding genomic DNA (Zilberman et al., 2004; Li et al., 2006; Qi et al., 2006). Methylation at CpG sites is maintained through genome replication by the DNA methyltransferase MET1, a homolog of mammalian DNA methyltransferase 1 (Finnegan and Dennis, 1993; Kankel et al., 2003; Saze et al., 2003), while the plant-specific DNA methyl-transferase CMT3 primarily methylates in the CHG sequence context (where H = A, C, T) (Jackson et al., 2002). Furthermore, the recent characterization of the DNA demethylases ROS1, DME, DML2, and DML3 in Arabidopsis suggests that subsets of genomic DNA methylation patterns are the products of antagonistic methylation-demethylation activity (Gong et al., 2002; Penterman et al., 2007). It remains to be determined how DNA demethylase activity is regulated, and a precise understanding of the genomic targets of methylation and demethylation is essential to deconvolute how these opposed activities forge the methylation landscape that is observed.
Immunoprecipitation-ChIP studies with a methylcytosine-specific antibody have provided a map of the regions of the Arabidopsis genome that contain methylated DNA (Zhang et al., 2006; Zilberman et al., 2007). However, this approach suffers from low resolution and an inability to identify the precise sequence context of the methylation site(s). The regulatory potential of altering the methylation state of single cytosines has been established (Weaver et al., 2004), so clearly, genome-wide determination of DNA methylation status at the single-base resolution is the essential precursor for unraveling how this ubiquitous epigenetic modification regulates the underlying genomic information.
The gold-standard technique for determining the methylation state of any cytosine in a DNA sequence is treatment of genomic DNA with sodium bisulfite, which under denaturing conditions converts cytosines, but not methylcytosines, into uracil (Frommer et al., 1992), which can subsequently be distinguished by sequencing. This approach is conventionally applied to only a small set of genomic locations. Here we have combined novel methods with a next-generation sequencing by synthesis technology to enable direct sequencing of the entire cytosine methylome of Arabidopsis at single-base resolution (methylC-seq). This revealed extensive, previously undetected, DNA methylation, enabled both the context and level of methylation at each site to be assessed, and identified effects of the local sequence composition upon DNA methylation state. Deep sequencing of the cytosine methylomes of mutant plants defective in methylation maintenance (met1-3), establishment (drm1-2 drm2-2 cmt3-11), and demethylation (ros1-3 dml2-1 dml3-1) identified the subsets of genomic targets upon which the different classes of enzymes act. Hundreds of discrete regions of dense demethylation were identified that overlapped significantly with promoters and 3′UTRs, and a subset of transposons proximal to protein-coding genes was also identified that are consistently demethylated. Deep sequencing of the cellular smRNA component of the transcriptome (the “smRNAome”) exposed a direct relationship between the location and abundance of smRNAs and DNA methylation, showed a tendency for smRNAs to direct strand-specific DNA methylation in the region of RNA-DNA homology, and demonstrated DNA methylation-dependent amplification of proximal smRNA abundance. Finally, strand-specific mRNA-seq revealed changes in the transcript abundance of hundreds of genes, trans-posons, and unannotated intergenic transcripts upon altering their DNA methylation state. Altogether, these comprehensive and highly integrated data sets reveal previously uncharted subsets of the epigenome and provide deep insights into the complex interplay between DNA methylation and transcription.
RESULTS AND DISCUSSION
Bisulfite Sequencing of the Arabidopsis Genome
Genomic DNA was isolated from Arabidopsis (ecotype Col-0) immature floral tissue, fragmented, and ligated to adaptor oligo-nucleotides in which every cytosine was methylated. We used floral tissue, as it has previously been shown to contain a more diverse population of smRNAs (Lu et al., 2005), and more abundant DNA methylation (J. Yazaki, H. Shiba, J.R.E., unpublished data). Subsequent treatment with sodium bisulfite under denaturing conditions was performed to convert unmethylated cytosines into uracil, after which the converted gDNA was sequenced with the Illumina Genetic Analyzer (GA). Four different methods of bisulfite conversion were compared to assess the most effective for conversion efficacy while minimizing template degradation (Table S1; Supplemental Experimental Procedures). Reads aligning to the unmethylated chloroplast genome that is isolated and sequenced in conjunction with the nuclear genome were used to calculate the frequency of cytosine conversion (Fojtova et al., 2001). Two consecutive bisulfite treatments yielded optimal results, with a conversion rate of 99.14% (Table S1) and minimal template degradation. Therefore, this method was used for all subsequent bisulfite-sequencing library construction.
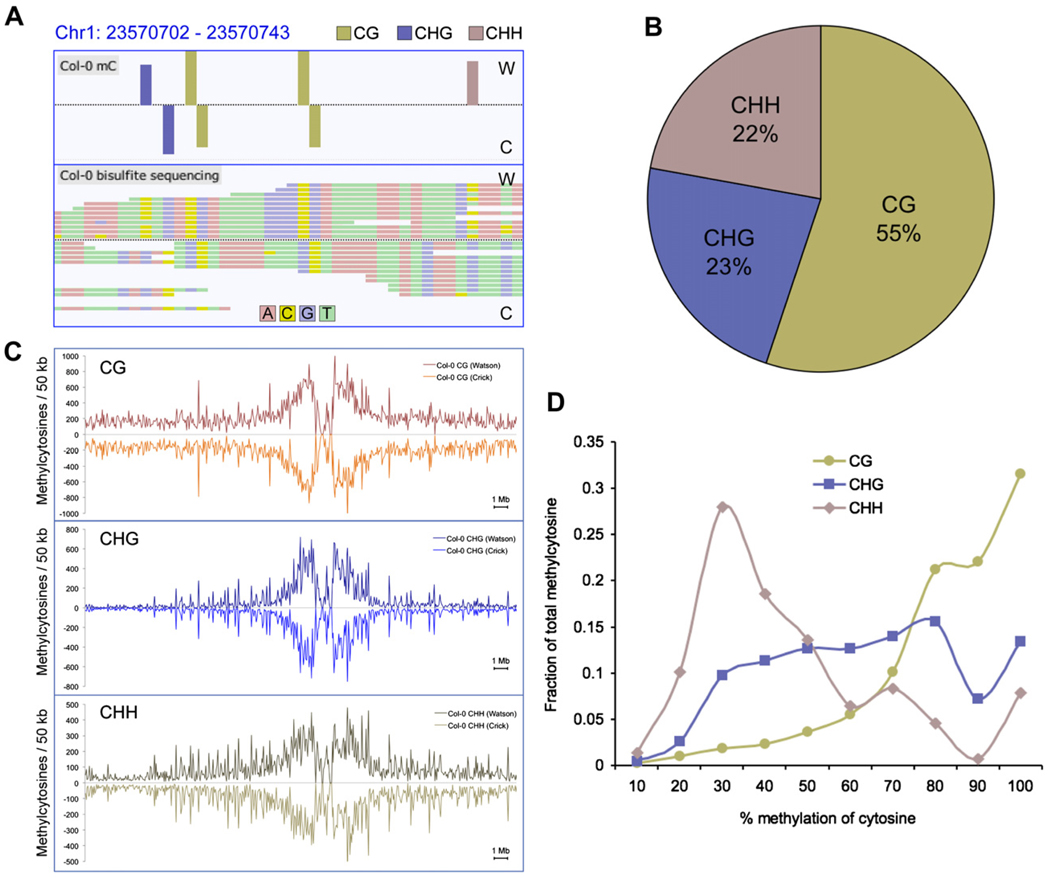
Sequence reads were filtered as described (Supplemental Experimental Procedures), only retaining reads that mapped uniquely to the Col-0 reference genome. To abolish any con-founding effects due to clonal duplication during library preparation, we removed all but one read in cases where multiple reads shared the same start coordinate. A statistical analysis of the incidence and effect of clonal reads is provided in the Supplemental Experimental Procedures. From 55,805,931 aligned reads, 39,113,599 were unique and nonclonal, yielding an average depth of 8.0 read coverage per base for each DNA strand, and overall unique read coverage of 78.5% of all cytosines in the genome with at least two reads (Figure S1; Table S2). Sequencing of cytosines at cytosine positions in the chloroplast reference genome in each bisulfite-treated library provided a measure of the sum of the rates of nonconversion and thymidine to cytosine-sequencing errors. Using this value as a measure of the false methylcytosine discovery rate, a binomial probability distribution was used to calculate the minimum sequence depth at a cytosine position at which a methylcytosine could be called while maintaining a false positive rate below 5%. Applying this algorithm, we identified 2,267,447 methylated cytosines in the nuclear genome of Col-0 flower buds, accounting for 5.26% of all genomic cytosines, or 6.70% of the cytosines for which sufficient sequencing depth (more than one read) was generated (Table S3). Notably, reads that contained several cytosines were not removed from the data set, so as to avoid skewing the detection of DNA methylation in highly methylated regions, providing an unbiased assessment of DNA methylation throughout the genome. Viewing of the methylC-seq reads in our custom built genome browser (http://neomorph.salk.edu/epigenome.html) clearly shows the strand-specific identification of unconverted cytosines, indicative of DNA methylation in each context (Figure 1A). The relative prevalence of DNA methylation in each sequence context throughout the genome was assessed, revealing that 55% were in CG context, while 23% and 22% were in the CHG and CHH contexts, respectively (Figure 1B).
Figure 1. DNA Methylation Context and Chromosomal Distribution in Wild-Type Col-0.
(A) Methylcytosines identified (top panel) in Col-0 from bisulfite-converted sequencing reads (bottom panel) for a region of chromosome 1, as represented in the AnnoJ browser.
(B) The fraction of methylcytosines identified in each sequence context for Col-0, where H = A, C, or T.
(C) The density of methylcytosines identified in each context, on each strand throughout chromosome 1, counted in 50 kb bins.
(D) Distribution of the percentage methylation at each sequence context. The y axis indicates the fraction of the total methylcytosines that display each percentage of methylation (x axis), where percentage methylation was determined as the fraction of reads at a reference cytosine containing cytosines following bisulfite conversion. Fractions were calculated within bins of 10%, as indicated on the x axis.
To validate the methylC-seq methylcytosine predictions in the same gDNA sample, we performed methylcytosine immunoprecipitation (mCIP) and hybridization to whole-genome tiling arrays (Zhang et al., 2006). The mCIP-array approach identified 13,166 methylated regions encompassing ~13.6 Mb of the nuclear genome. Comparison to the methylcytosines identified by methylC-seq revealed that 98.6% of the regions identified by mCIP contained one or more methylcytosines in the overlapping sequence data, but these accounted for only 51.7% of the total methylcytosines discovered by the bisulfite sequencing (Table S4). The predicted methylation density from methylC-seq was found to be 8-fold higher in mCIP regions than in non-mCIP regions, indicating an mCIP bias for heavily methylated regions and demonstrating the higher sensitivity achieved with the methylC-seq approach. Analysis of the context of the methylation showed that the mCIP regions contained 44.6%, 70.1%, and 50.7% of the total CG, CHG, and CHH methylcytosines, respectively, indicating that mCIP tends to be biased toward discovery of regions containing CHG methylation. Thus, the demonstrated higher sensitivity, increased coverage, and reduced bias of the methylC-seq approach allows for the discovery of a previously uncharted segment of the Arabidopsis DNA methylome.
While methylation in all sequence contexts is present at a higher density in the pericentromeric regions of Arabidopsis nuclear chromosomes (Figure 1C and Figure S2), CHG methylation appeared most enriched in the pericentromeric regions, likely due to its preference for methylation of transposon-related sequences (Tompa et al., 2002; Kato et al., 2003). In contrast, CG and CHH methylation, although most dense in the pericentromeric regions, is commonly observed throughout the euchromatic chromosome arms. As we obtained appreciable read depth for a large proportion of the genome, we were able to estimate the level of methylation at each methylcytosine, calculated from the number of cytosines sequenced divided by the total read depth. Interestingly, each methylation context showed distinct profiles of methylation level. CG methylation sites, maintained by MET1 after genome replication, were predominantly highly methylated, consistent with the highly pervasive maintenance nature of this methylation type (Figure 1D). In stark contrast, CHH sites that were found to contain methylcytosines tended to manifest a higher fraction of unmethylated cytosines, perhaps indicating that the methylation was only present in a subset of the cell types of the floral tissue, or that CHH methylcytosine is more variable even within the same cell type. Interestingly, CHG sites were found across a broad range of methylation levels (Figure 1D).
Local Sequence Effects upon DNA Methylation State
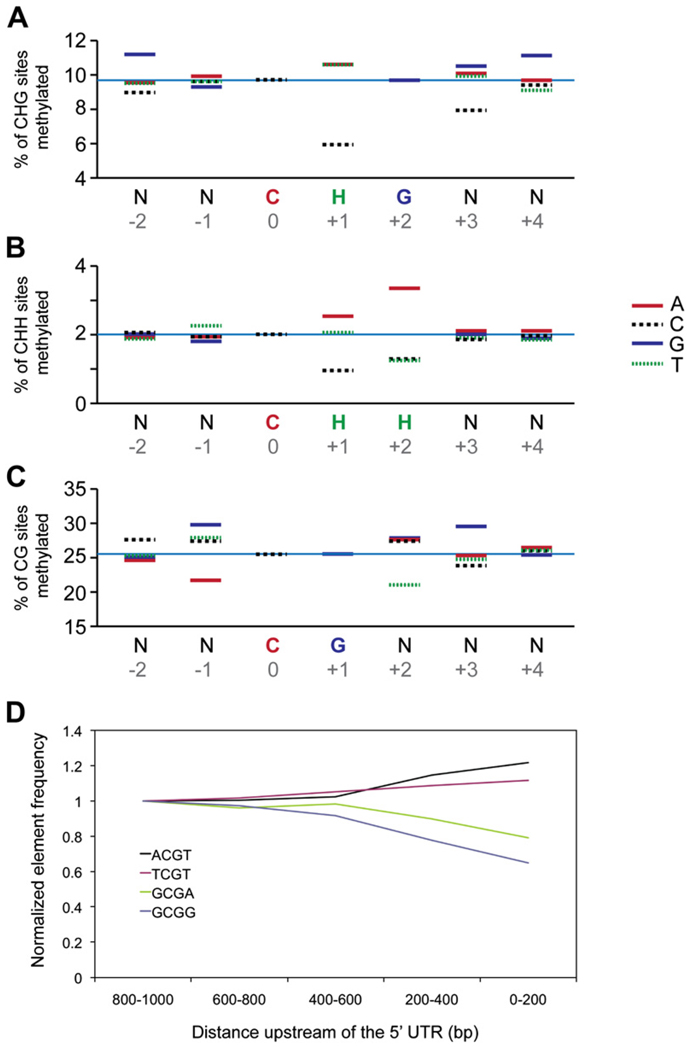
An analysis of sequence content within the general classes of CG, CHG, and CHH revealed additional local sequence effects on cytosine methylation (Figure 2; Table S5). In both the CHH andCHG contexts, a cytosine immediately followed by another cytosine has a significantly lower tendency to be methylated than a cytosine neighboring an adenine or thymine. This is clearly illustrated in the CHG context in which the CTG and CAG sites are found to be methylated at twice the frequency of a CCG, and similarly in the CHH context, where methylation at CTH and CAH are two and three times more likely than at a CCH site, respectively (Figures 2A and 2B). As opposed to the repressive effect of cytosine, adenines in 3′ positions of the CHH context are associated with an increase in the cytosine methylation frequency. This effect is strongest in the +2 positions where a CHA is methylated 3-fold more often than either CHC or CHT (Figure 2B). Finally, in the CG context, an adenine at −1 and thymidine at +3 are both 25% less likely to be methylated than when any other bases are in these positions (Figure 2C). In combination, these positional effects produce large differences in methylation states, such as in the CHG context where CTGG is 6.5 times more likely to be methylated than CCGC, or in the CG context, where GCGG is twice as likely to be methylated as the palindromic ACGT. As each methylation context shows unique sequence effects, it is more likely that these differences are due to each enzyme’s substrate preferences, as opposed to sequence-specific steric effects influencing 5-cytosine availability.
Figure 2. The Effect on Cytosine Methylation Frequency by Neighboring Bases.
(A–C) The methylation frequency of cytosines in the (A) CHG, (B) CHH, and (C) CG contexts are shown as a function of proximal base composition. Base composition effects two positions upstream (−2) to four bases downstream (+4) of a particular cytosine are interrogated by dividing the number of methylcytosines to total cytosines of each base at each position.
(D) The percent contribution of each CG tetramer (−1 to +2) was calculated in five 200 bp pair sections in the 1 kb region immediately upstream of all genes in the genome. For each tetramer, the percent contribution in the 800–1000 bp region was used to normalize all other regions.
To ensure that these local sequence effects are not due to an overrepresentation of specific sequences in heavily methylated genomic regions, we examined sequence-context trends exclusively in densely methylated locations. In these regions, the repression of methylation in the CHH and CHG context by a +1 cytosine and the increase in methylation associated with a +2 adenine are still clearly seen, albeit at a higher baseline level of methylation. For the CG context, the repressive effects of −1 adenine and +2 thymine observed in the whole genome are not visible in the highly methylated regions, though this may be in part due to the fact that these regions are nearly saturated, with 75% of all cytosines methylated.
One potential repercussion of cytosine methylation sequence preferences could be enrichment or depletion of certain sequences in regions divergent in methylation content, such as a gene promoter. To test for this we calculated the percent content of each CG tetramer (−1 to +2) in adjacent 200 bp regions from 1 kb upstream up to the 5′UTR of genes. The percent content of each region was then normalized to the percent content in the 800–1000 bp region (Figure 2D). We observed a depletion of the most highly methylated tetramers (CGCA and GCGC) and a corresponding enrichment of the most lowly methylated tetramers (ACGT and TCGT) as we moved toward the start of the gene. This result suggests that MET1 sequence preference may directly affect sequence content in gene promoters.
The Cytosine Methylomes in DNA Methyltransferase and Demethylase Mutants
In order to better understand the regulatory pathways that fashion the observed patterns of DNA methylation, methylC-seq was performed for a set of highly informative mutant plants deficient in CG maintenance DNA methylation (met1-3 mutant, referred to as met1 henceforth) (Saze et al., 2003), non-CG maintenance, and de novo DNA methylation (drm1-2 drm2-2 cmt3-11 triple mutant, termed ddc) (Chan et al., 2006), or a triple mutant that eliminates nearly all DNA demethylation activity (ros1-3 dml2-1 dml3-1 triple mutant, termed rdd) (Penterman et al., 2007). As with Col-0, deep methylC-seq was performed on gDNA isolated from immature floral tissue harvested from plants of each mutant, and yielded a similar conversion rate and percent of genomic cytosines covered by two or more independent reads in every genotype (Figure S1; Table S2 and Table S3). As the Col-0 data set contained more aligned read sequence than the mutant data sets (Table S2) and read depth may be affected by the methylation state of the bisulfite converted DNA, a subset of direct comparisons were conducted that only involved interrogation of the bisulfite sequencing reads where the total read depth was 6- to 10-fold coverage for each DNA strand (12- to 20-fold sequence coverage in total) for every genotype examined, to ensure unbiased comparisons between genotypes.
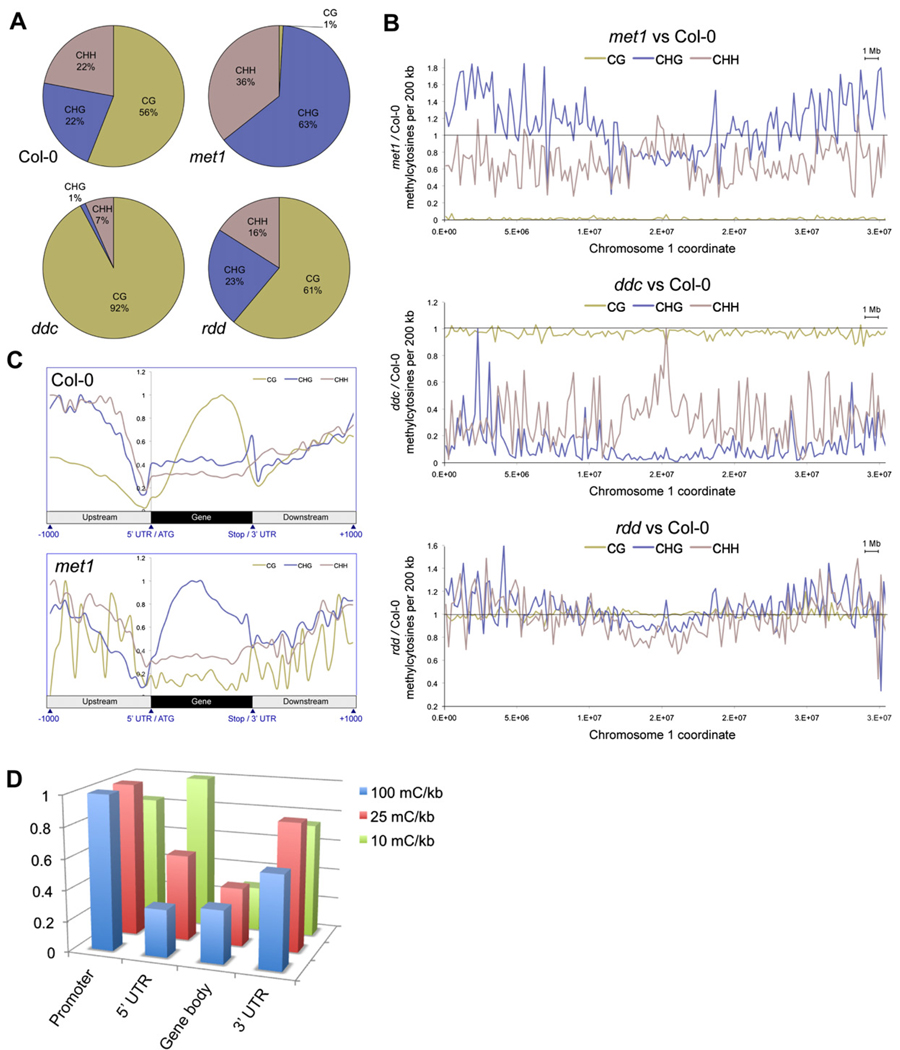
Overall, CHG methylcytosine numbers showed little change in met1, while CHH methylation decreased by approximately half (Figure S3). CG methylation was dramatically reduced to only 1% of the total methylcytosines (Figure 3A), 0.5% of the number of methylcytosines present in the CG context in Col-0 (Table S3). Interestingly, higher levels of CHG methylation were evident in the euchromatic regions of the nuclear chromosomes in met1 compared to Col-0 (Figure 3B and Figure S4), indicating immediate recruitment of non-CG DNA methyltransferase activity in first-generation mutant plants homozygous for the met1 null allele. This is in contrast to a recent report that new CHG methylation appears only after several generations of the absence of a functional MET1 allele (Mathieu et al., 2007). Calculation of the difference in percentage methylation at every methylcytosine identified in Col-0 or met1 mutant over chromosome 1 showed that met1 displays significant CHG hypermethylation in the euchromatic regions of the chromosome, whereas CHH methylation is slightly reduced and CG methylation almost abolished (Figure 3B). The new CHG methylation was found to be widespread and present in the bodies of over two thousand genes, of which 78% contain CG methylation in Col-0 (e.g., Figure S5). The density of DNA methylation in each context was calculated over each gene, in 1 kb upstream/downstream, and the profile normalized for all genes. In Col-0 this genic profile clearly showed the abundant CG body methylation that tends to be distributed toward the 3′ of the gene and depleted at the 5′ and 3′ (Figure 3C), as reported previously (Zhang et al., 2006; Zilberman et al., 2007). Interestingly, the genic profile for met1 revealed that the average profile of gene body CHG methylation adopts a pattern that is very similar to the wild-type CG body methylation (Figure 3C), suggesting that the met1 CHG body methylation may perform a compensatory role to accommodate for the loss of CG methylation.
Figure 3. Methylation in DNA Methyltransferase and DNA Demethylase Mutant Plants.
(A) The fraction of methylcytosines in each sequence context for each genotype. Positions were compared only where all genotypes had a sequence read depth between 6 and 10.
(B) Ratio of the number of methylcytosines in each mutant versus Col-0 per 200 kb, where read depth was 6–10. The horizontal line represents Col-0, the plotted line represents percentage methylation in the mutant versus Col-0.
(C) Average distribution of DNA methylation in each context throughout genes and 1 kb upstream and downstream in Col-0 and met1. Values are normalized to the highest point within each profile.
(D) Density of sites of hypermethylation in rdd. The number of sites of DNA methylation present in rdd and absent in Col-0 was tabulated for promoters (1 kb upstream), 5′UTRs, gene bodies, and 3′UTRs, and the density of methylation in each normalized to the length of each feature. The number of features that contained hypermethylation above different lower thresholds of density was calculated, and the number of features of each type normalized relative to the feature with the most counts, for each lower threshold.
CG methylation patterns and abundance in ddc were generally similar to Col-0 (Figure 3B and Figure S4); however, CHG methylation was reduced to only 1% of the total methylcytosines in ddc, just 3% of the number identified in Col-0 at similar read depths, likely due to the absence of CMT3 (Figure 3A, 3B, Figure S3, and Figure S4). Interestingly, CHH methylation, thought to be maintained by DRM1 and DRM2 and persistent smRNA signals, was reduced by 80% in ddc mutant plants, indicating that there is likely another enzyme that can perform de novo DNA methylation in Arabidopsis (Figure 3A, 3B, Figure S3, and Figure S4).
The DNA demethylase triple mutant rdd showed similar overall numbers and context distribution of methylation to Col-0 when surveyed at a read depth of 6–10 (Figure 3A and Figure S3), unlike the methyltransferase mutants that effect nearly complete loss of methylation in their respective contexts. However, though the total number of methylcytosines identified in the demethylase mutant was similar to wild-type, measurement of the methylcyto-sine density in 1 kb segments, with a 500 bp overlap, tiling across the whole genome identified hundreds of discrete regions in which the density of DNA methylation was at least 2-fold greater (Figures S6A–S6D). An even distribution of these hypermethylated regions throughout nuclear chromosomes was evident, except for locations proximal to the centromere (Figure S6E). Individual sites of hypermethylation present in rdd but absent in Col-0 were identified, where both genotypes had sufficient read depth to enable interrogation of the methylation status. The density of hypermethylation sites located in four features was calculated: promoters (1 kb upstream), 5′UTRs, coding sequence (gene bodies) and 3′UTRs. The number of features that contained hypermethylation densities above lower thresholds of 10, 25, and 100 methylcytosines per kilobase were counted, and the number of features of each type normalized relative to the feature with the most counts, for each lower threshold (Figure 3D). It was evident that the most highly hypermethylated regions were most frequently located in promoters and 3′UTRs, with a relative depletion in 5′UTRs and gene bodies, indicating that demethylation was most active in these regions flanking the gene. Clearly, these DNA demethylases have widespread activity throughout the genome, actively removing methylcytosines in a variety of gene level contexts for reasons that are currently undetermined.
Sequencing of the smRNAome from DNA Methylation- and Demethylation-Deficient Mutants
In order to further characterize the cellular forces that govern the landscape of DNA methylation, we performed ultradeep sequencing of the smRNAome for Col-0 and mutant plants lacking DNA methyltransferases or demethylases. We were interested in the smRNAome because it has been previously demonstrated that a subset of the cellular smRNA pool targets DNA methylation through RdDM (Qi et al., 2006), an essential process for the establishment of DNA methylation and its maintenance in asymmetric contexts.
The 15–30 nt subset of total RNA was isolated from immature floral tissue from wild-type plants, and molecules possessing a 5′ monophosphate were ligated with specific oligonucleotide adapters to generate a library amenable to high-throughput sequencing with the Illumina GA (see Supplemental Experimental Proceduers). Identification of the adapters in the resulting sequences allowed precise determination of the lengths of each of the smRNA molecules. A total of 2,625,243 smRNAs were identified that aligned perfectly to the genome, of which 1,479,577 aligned uniquely to 737,606 locations and were termed “unique mapping smRNAs.” The remaining 1,145,666 sequences each aligned to multiple locations and encompassed 431,949 distinct smRNAs aligning to 2,104,289 genomic locations, and they were referred to as “multiple mapping smRNAs” (Table S6). As far as we are aware, this constitutes the largest set of distinct small RNA sequences from a single population reported to date (Matzke et al., 2007). The most abundant species of smRNA were 24 nt in length, followed by 21-mers (Figure S7A). Whether an smRNA that is homologous to multiple sequences in the genome is able to direct DNA methylation at each location is an unresolved question. However, we found that 52.4% of the genomic regions to which any sequenced smRNA aligned did not contain methylcytosines, whereas only 14.6% of locations to which unique mapping smRNAs aligned did not contain DNA methylation (Figures S7B and S7C). This indicates that multiple mapping smRNAs do not act at all homologous loci. The genomic positions of all smRNAs encompassed 39% of the Col-0 methylcytosines and the unique smRNAs only 28%, suggesting that multiple mapping smRNAs are responsible for a considerable subset of RdDM. However, we focused our subsequent analyses toward identifying the causal relationships between DNA methylation and smRNAs by using only the unique mapping smRNA subset to reduce occlusion of associations caused by incorrect assumptions of the true location of smRNA activity.
All smRNA loci were searched for the presence of methylcytosines on either strand of the nuclear genome. In Col-0, 85.4% of smRNA loci contained at least one methylcytosine. To further quantify the association between methylation and the presence of smRNAs, we calculated the odds ratio for the correspondence of these two genomic features. To calculate this ratio, we first determined that the odds of cytosine methylation versus cytosine nonmethylation at smRNA loci is 1 to 1.02 (408,769 mC to 417,271 C), and the odds of cytosine methylation versus cytosine nonmethylation at non-smRNA loci is 1 to 26.4. The ratio of these two odds is 25.9, that is, a 25.9 greater odd of finding a methylcytosine at a smRNA locus than at a non-smRNA locus. These data provide strong evidence supporting an important role for smRNAs in targeting of DNA methylation. It is worth noting, however, that smRNAs are only associated with approximately a third of all genomic cytosine methylation. Potentially, epigenetic marks such as histone modifications are involved in directing methylation at these other cytosines.
Whereas a previous report did not find appreciable evidence of trans-acting siRNAs (tasiRNAs) directing DNA methylation (Zhang et al.,2006),we observe abundant DNA methylation dependent on MET1, DRM1, DRM2, and CMT3 overlapping the smRNA generating regions of five of the six trans-acting siRNA generating loci (TAS), TAS1A, TAS1B, TAS1C, TAS2, and TAS3(Figure S8). Furthermore, an increase in DNA methylation proximal to the tasiRNA clusters can be observed in the DNA demethylation triple mutant, rdd, indicating that without the demethylase activity the DNA proximal to the tasiRNAs is being targeted for de novo methylation.
Interconnection of the smRNAome and the DNA Methylome
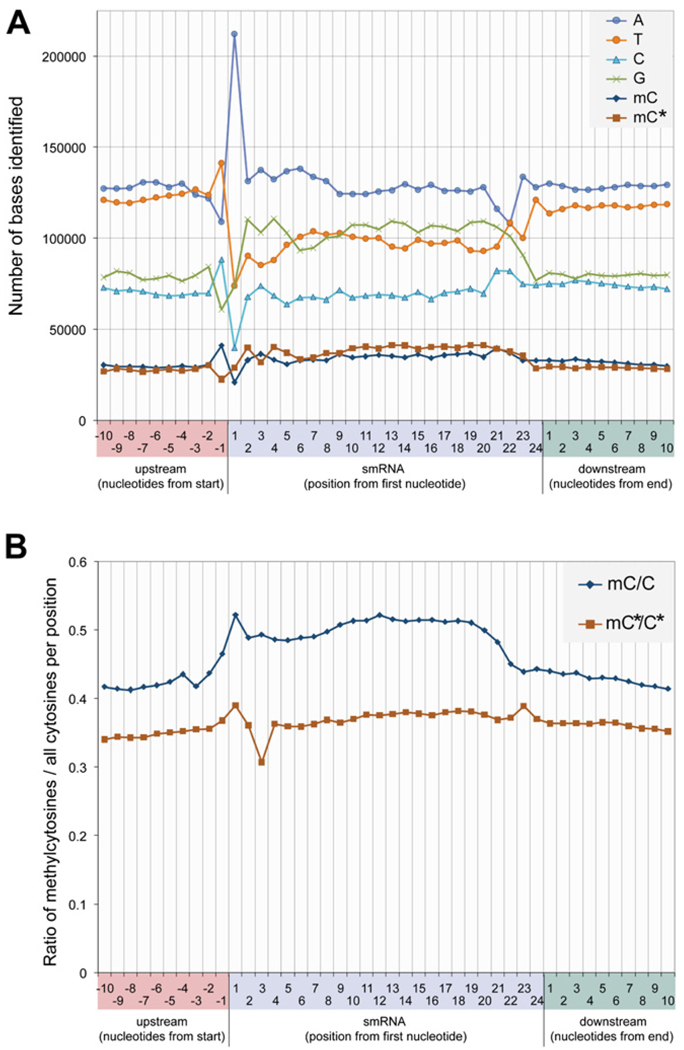
The base composition and strand-specific DNA methylation state of bisulfite converted genomic DNA sequence was analyzed within the region homologous to all 21–24 nt unique smRNAs sequenced from wild-type floral tissue, and the 10 nt immediately flanking them on both sides. The base composition is displayed for the strand identical to the smRNA sequence, termed the sense strand, in Figure 4A. Interestingly, strong biases in the base composition directly within the sequence matched by the smRNA are evident, as exemplified by an increased propensity for guanine and a decreased representation of thymine. In the case of 24 nt smRNAs, adenine is the most common first base of the sequenced smRNAs, followed by a lower and approximately equal distribution throughout the other 3 nt. Additionally, thymine is highly overrepresented at position −1 in relation to 24 nt smRNA sequences, which is consistent with the tendency for endonucleases to favor cleaving after uracil. Thus, we observe a tendency for a TA dinucleotide that would provide an optimal motif for cleavage of a double-stranded RNA species by the DICER-LIKE3 endonuclease (Huesken et al., 2005; Reynolds et al., 2004). Curiously, the other major size classes of smRNAs manifest conspicuously different sequence patterns. For instance, 21 nt smRNAs are enriched for adenine at positions 1 and 21, whereas 22 and 23 nt smRNAs most commonly have adenine as their last nucleotide (Figure S9). Methylcytosines on the sense and antisense strand were identified at similar frequencies (Figure 4A). However, because guanine is more abundant on the sense strand, the frequency at which a cytosine is methylated on the sense strand is greater than on the antisense strand. This tendency for methylation to be found on the sense strand relative to the smRNA is clearly observed in the ratio of methylcytosines to all cytosines at each position underlying an aligned smRNA on each strand (Figure 4B). On the sense strand, the ratio of methylated to unmethylated cytosines is higher specifically from nt 1 to 21 of the sequence that the smRNA matches, whereas on the antisense strand no such enrichment in the region of gDNA-smRNA homology is observed. The tendency to find smRNAs overlapping one another in clusters may partially occlude the detection of highly localized effects on underlying DNA methylation. There-fore, the clear enrichment of methylcytosines on the sense strand in the precise location to which the smRNA aligns strongly indicates both that smRNAs target DNA methylation in their region of genomic homology, and that the deposition of methylation has strand specificity, more frequently targeting the sense strand. This suggests that the smRNA may be directing DNA methylation on the strand opposite the one to which it can hybridize.
Figure 4. Distribution of smRNA Lenghts and Overlap between DNA Methylation and 24-mer smRNAs.
(A) Nucleotide frequency and distribution flanking and within uniquely aligning 24-mer smRNAs.
(B) Methylcytosine distribution presented as the ratio of methylcytosines to all cytosines located at each position flanking and within uniquely aligning 24-mer smRNAs. Abbreviations: mC, methylcytosine on the sense strand relative to the smRNA sequence; C, total cytosine on the sense strand; mC*, methylcytosine on the antisense strand; C*, total cytosine on the antisense strand.
DNA Methylation-Associated Amplification of smRNA
In light of the strong correlation between smRNAs and the presence of underlying DNA methylation, the smRNA populations of met1, ddc, and rdd mutant plants were sequenced to investigate whether the cellular smRNA pool changes in response to alterations in DNA methylation. Again, for each mutant the number of distinct smRNA sequences discovered supersedes any previous report.
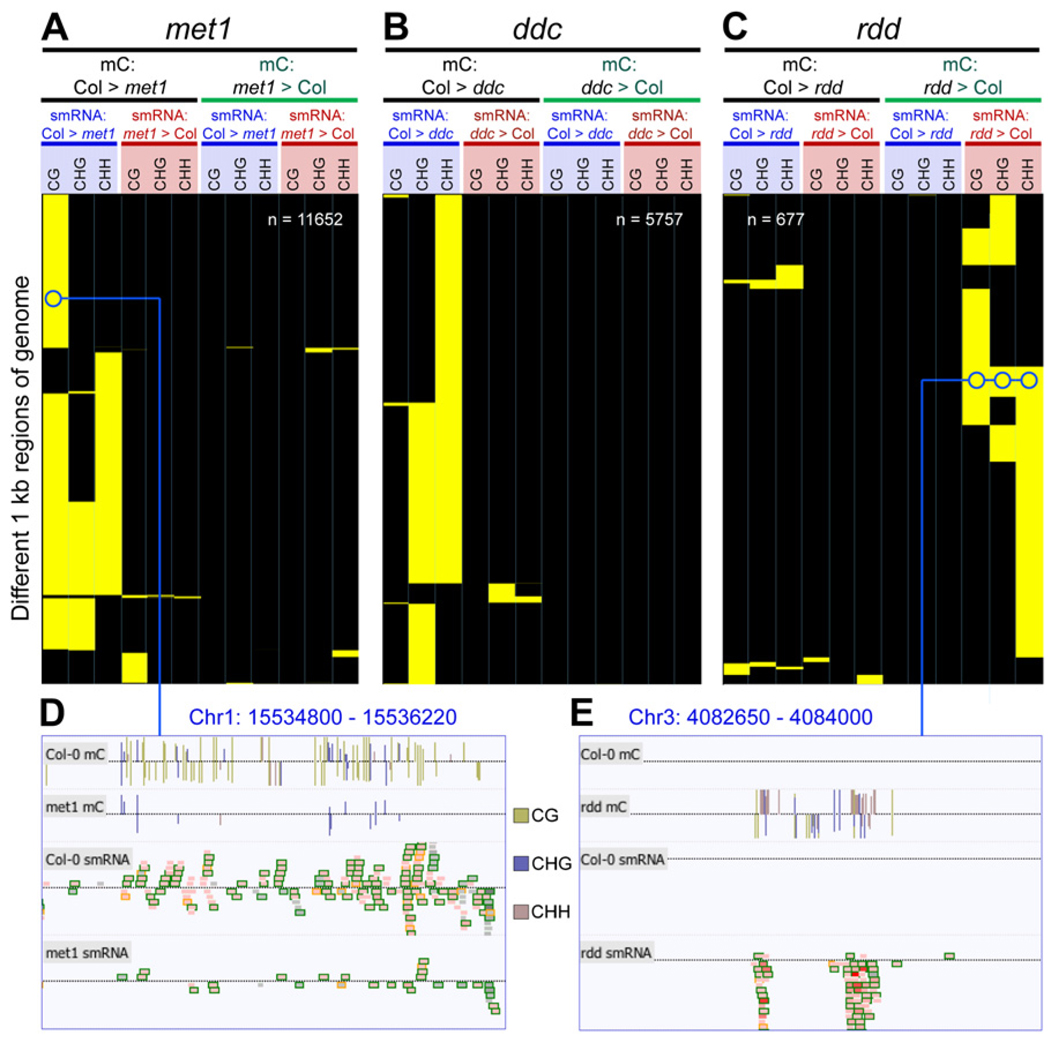
To examine the effect on smRNA populations resulting from disruption of the DNA methyltransferase and demethylase activities, we tabulated the methylcytosine content and smRNA abundance for 1 kb regions with a 500 bp overlap tiling across the entire genome for wild-type and each of the mutants. A methylcytosine was counted only if there was sufficient methylC-seq read depth in every genotype to interrogate the position (>1 depth). By a pairwise comparison between wild-type and each of the mutants (met1, ddc, and rdd), we identified 1 kb regions that displayed more than a 3-fold difference in methylcytosine density and a 5-fold difference in smRNA density (see Experimental Procedures). From this analysis, a profile of the coincident changes in DNA methylation and smRNA abundance was generated for each region. Hierarchical clustering was performed upon the patterns of DNA methylation and smRNA changes in regions that displayed any significant difference (Figure 5). We identified 11,652 regions that showed coincident changes in DNA methylation and smRNA abundance between wild-type and met1 based on these parameters (Figures 5A and 5D). Approximately 92% of the regions had both higher DNA methylation in every combination of sequence context and higher smRNA density in wild-type relative to met1. The loss of non-CG methylation in met1 at locations where smRNAs align suggests a role for MET1-dependent CG methylation in maintaining RdDM.
Figure 5. Methylcytosine Density Correlates with smRNA Abundance.
(A–C) Hierarchical clustering of all 1 kb regions of the nuclear genome that have significantly different methylcytosine and smRNA density between Col-0 and (A) met1, (B) ddc, and (C) rdd (see Experimental Procedures). Yellow indicates >3-fold change in methylcytosine density and >5-fold difference in number of sequenced bases of smRNA; black indicates no change above fold change thresholds. Minimum methylcytosines: 8 per kilo-base, minimum smRNA: 300 sequenced bases per kilo-base.
(D) DNA methylation and smRNAs in Col-0 and met1.
(E) DNA methylation and smRNAs in Col-0 and rdd. Tracks are shown for DNA methylation sites and smRNA. smRNAs are colored internally by their uniqueness (red: maps to a single location, greyscale: maps to multiple locations), and a surrounding box indicates the size class (orange:21-mer, black: 24-mer), and the shading represents the copy number (darker: more copies, lighter: fewer copies). Abbreviations: mC, methylcytosine.
A similar trend of coincident higher DNA methylation and smRNA abundance in wild-type relative to ddc was observed, with 95% of the 5757 altered regions displaying less non-CG methylation and smRNA abundance in the mutant (Figure 5B). The remaining 5% of the regions had a decrease in non-CG methylation but an increase in the smRNA abundance. Clearly there is a strong tendency for a decrease in DNA methylation to be accompanied by a reduction in the number of proximal smRNAs. The same analysis was undertaken for rdd, where we observed the complementary effect, that when DNA methylation density increased in the absence of demethylase activity, the proximal smRNA population was enlarged (Figures 5C and 5E). Approximately 90% of the 677 altered regions displayed this relationship, while the remaining 10% showed a higher density of DNA methylation and smRNAs in wild-type. The dual directionality of this relationship illustrates that the presence of DNA methylation is highly associated with an increase in steady-state smRNA levels in the vicinity and supports a model in which DNA methylation at a smRNA generating locus can effect an increase in the production of smRNAs. Coupled with the evidence supporting a role for smRNA in directing over a third of the DNA methylation, these data indicate that at a subset of genomic loci, DNA methylation and smRNAs may act in a self-reinforcing positive feedback loop.
Interestingly, in met1 we noted that the loss of DNA methylation at some transposons resulted not in an overall decrease in smRNA abundance, but the sudden appearance of abundant 21-mer smRNAs, where in every other genotype 24-mers pre-dominate (Figure S10). Notably, this increase in 21-mer abundance is clearly reflected in the proportion of total smRNAs in met1 that are 21-mers (Figure S7A). In many cases, however, these smRNA do not result in de novo methylation, either due to their ectopic nature or perhaps the lack of a functional MET1 isoform. Thus, it appears that the loss of DNA methylation at CG sites in a subset of transposons results in disruption and redirection of smRNA biogenesis at those loci, significantly altering the overall smRNAome composition.
Distinct Patterns of DNA Methylation and Abundance of smRNAs Are Controlled by Different DNA Methyltransferases
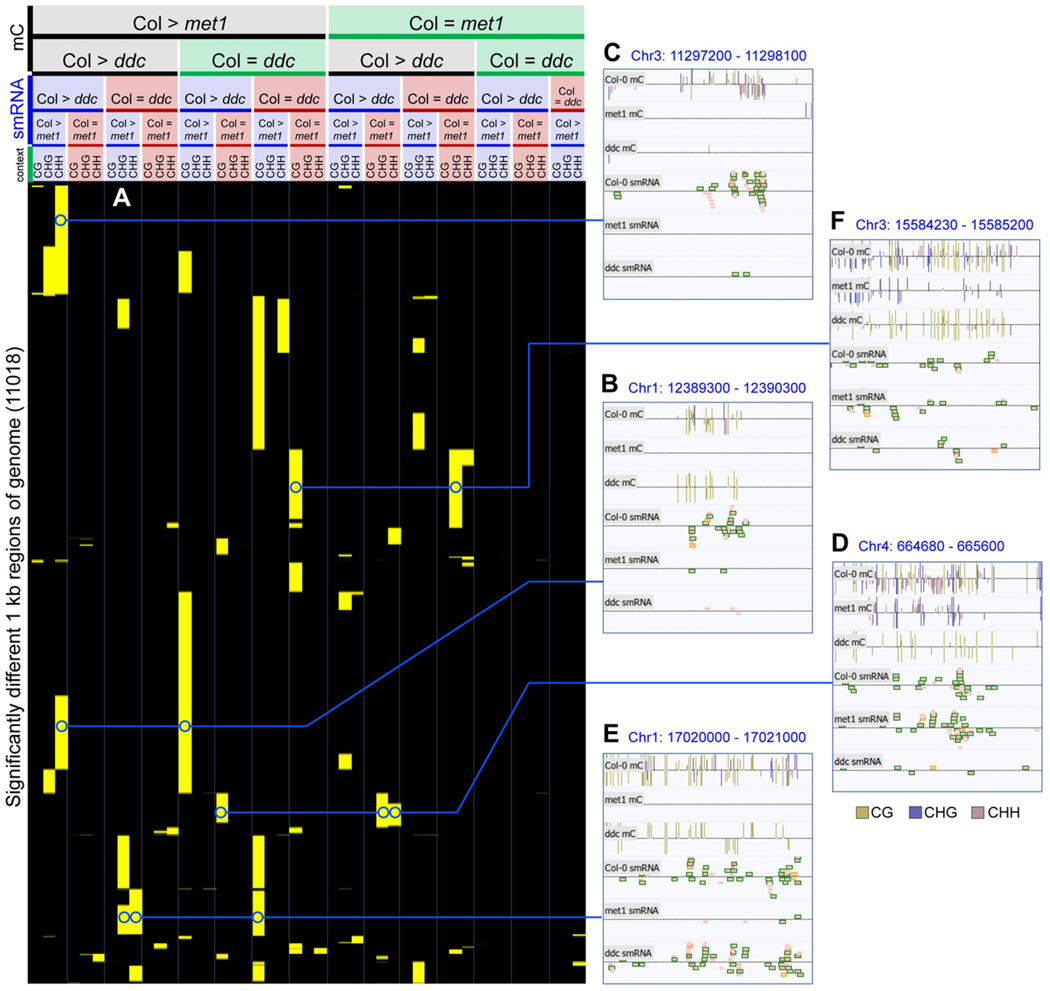
The coincident reduction of smRNAs and DNA methylation in any context and the observation that loss of MET1 activity often resulted in a decrease in CHG and CHH methylation (Figure 5A) prompted us to examine whether there were regional differences in the control of DNA methylation and smRNA abundance governed by the different DNA methyltransferases. In order to fully capture the diversity of responses in both met1 and ddc, we expanded the pairwise analysis of altered DNA methylation and smRNAs to cluster regions of wild-type, met1, and ddc together in a single comparison. Similar to the profiling described above, overlapping 1 kb regions tiling across the entire genome were interrogated for whether they contained equivalent (<2-fold difference) or lower (>3-fold change) densities of DNA methylation and/or smRNAs in both met1 and/or ddc relative to wildtype (see Experimental Procedures). The 11,018 regions that had altered DNA methylation or smRNA abundance in at least one mutant were subjected to hierarchical clustering to identify subsets of genomic regions that displayed a similar response in both DNA methylation and smRNA changes in each of the DNA methyltransferase mutants (Figure 6). All possible combinations were observed, reflecting a large diversity of control of DNA methylation and smRNAs and the variable overlap of different DNA methyltransferases on a genome-wide scale, reported only anecdotally previously. Examples are shown for subsets of loci for which smRNA production is significantly reduced upon loss of met1 and ddc (Figures 6B and 6C), ddc only (Figure 6D), and met1 only (Figure 6E), while a subset of the regions display a reduction in DNA methylation without loss of smRNAs (Figure 6F). The varied patterns suggest that different hierarchies of establishment of DNA methylation in each sequence context may be under the control of smRNAs at distinct genomic locations. Interestingly, the presence of numerous regions where MET1 activity, but not DRM1/2 or CMT3, is essential for the establishment of DNA methylation and the accumulation of smRNAs (Figure 6E) raises the possibility that the maintenance of CG methylation at that locus is required for the activity of DRM1/2 or CMT3, or that MET1 itself can act as a de novo DNA methyltransferase, as has been suggested previously (Aufsatz et al., 2004).
Figure 6. Diverse Patterns of Interaction between DNA Methylation and smRNAs.
(A) Hierarchical clustering of all 1 kb regions of the nuclear genome that have significantly different DNA methylation and/or smRNA density between Col-0 and met1 or ddc, and regions that show no difference (see Experimental Procedures). Yellow indicates >3-fold mC density and/or >5-fold smRNA difference;black indicates no difference above thresholds (<2-fold change in mC density or sequenced bases of smRNA). Minimum Col-0 mC: 8 per kilobase, minimum Col-0 smRNA: 300 sequenced bases per kilobase.
(B–F) Typical examples from various clusters visualized in the AnnoJ browser. Tracks are shown for DNA methylation sites and smRNA. smRNAs are colored as in Figure 5. Abbreviations: mC, methylcytosine.
Whole Transcriptome Sequencing
A primary effect of epigenetic marks such as methylcytosines is the regulation of transcription, as evidenced by previous reports of DNA methylation controlling the downregulation and silencing of transposons and some genes (Lippman et al., 2004; Zhang et al., 2006). As a final stage in our effort to develop a comprehensive and integrative map of epigenetic regulation governed by DNA methylation and small RNAs in Arabidopsis, we have developed a novel method (mRNA-seq) to sequence the transcriptome of wild-type, met1, ddc, and rdd plants, in order to identify subsets of the transcriptome that are regulated by DNA methylation.
In an effort to maximize the resolution and reduce the bias of our map of transcript space, we developed a method that enables strand-specific transcript sequencing and has no selection for polyadenylated transcripts (see Supplemental Experimental Procedures). Total RNA was isolated from the immature floral tissue, and highly abundant rRNAs removed using specific LNA oligonucleotides, after which the remaining RNA was fragmented by metal hydrolysis and ligated to specific 5′ and 3′adapters. This method is highly strand specific, as 95.7% of transcriptome reads, when aligned to the entire Col-0 reference genome (TAIR7), were found to map to annotated exons in the correct strand orientation, while only 0.8% of reads map to the antisense of annotated exons. The remaining 3.5% of the reads aligned to regions of the genome annotated as intergenic. For the most part, reads aligning to the intergenic regions cluster into discrete patches, suggesting that these may constitute new, unannotated transcripts (e.g., Figure S11).
For wild-type, met1, ddc, and rdd, 247–354 Mb of transcript sequence was generated per genotype (Table S7). In an effort to quantify transcript levels we counted the number of reads mapping within each AGI annotated gene model. To allow for equal comparisons between each genotype, for each data set we normalized the number of sequenced bases per gene model by the factor of the total sequenced bases in the data set to the data set with the most sequence generated. A 2-fold difference in total sequenced bases of a transcript was used as the minimum threshold for identification of genes that had large changes in transcript abundance between wild-type and each mutant. Comparisons were not made for any gene that in both genotypes being compared contained less than one sequenced base per base of the gene model, to eliminate variability due to lowly expressed genes. To reduce the effect of sample variability upon prediction of altered transcript abundance, each genotype was compared to two others, having to display a 2-fold difference in sequenced bases of transcript to both genotypes (met1 was compared to Col-0 and rdd, ddc to Col-0 and rdd, and rdd to Col-0 and ddc).
A comparison of the genes predicted as upregulated in met1 by a tiling array-based approach (Zhang et al., 2006) to our mRNA-seq results revealed a high concordance between the two data sets. It is noteworthy that though the two studies utilized distinct sources of cellular material (floral tissue here versus all above-ground tissue by Zhang et al. [2006]), we identified 42.6% of the 319 upregulated genes in the array study, accounting for 23.1% of the 589 genes we predicted here as upregulated. We identified as upregulated six of the nine genes that Zhang et al. (2006) validated by northern blot. A closer inspection of the remaining three, which were pseudogenes, revealed reads present only in met1, but the number of reads fell below our significance threshold of one sequenced base per base of the cDNA sequence (AT5G32430: 2 reads, AT1G38194: 11 reads, AT4G06730: 6 reads). Only mRNA-seq reads that mapped unambiguously to the reference sequence were included in this analysis, whereas reads that mapped to multiple identical genomic sequences, as are common in pseudogenes, were not included in this analysis. Therefore, mRNA-seq provides an effective means to identify changes in transcript abundance that can be unambiguously assigned to the original genomic location, avoiding crosshybridization issues that may confound array-based studies. In addition to the genes upregulated in the methylation mutants, a smaller set of genes were found to be downregulated, primarily in the met1 background. Interestingly, in a subset of these met1-downregulated genes, novel body CHG methylation was established upstream of the repressed gene. This could indicate that the compensatory CHG body methylation in met1 may cause proximal transcriptional perturbation.
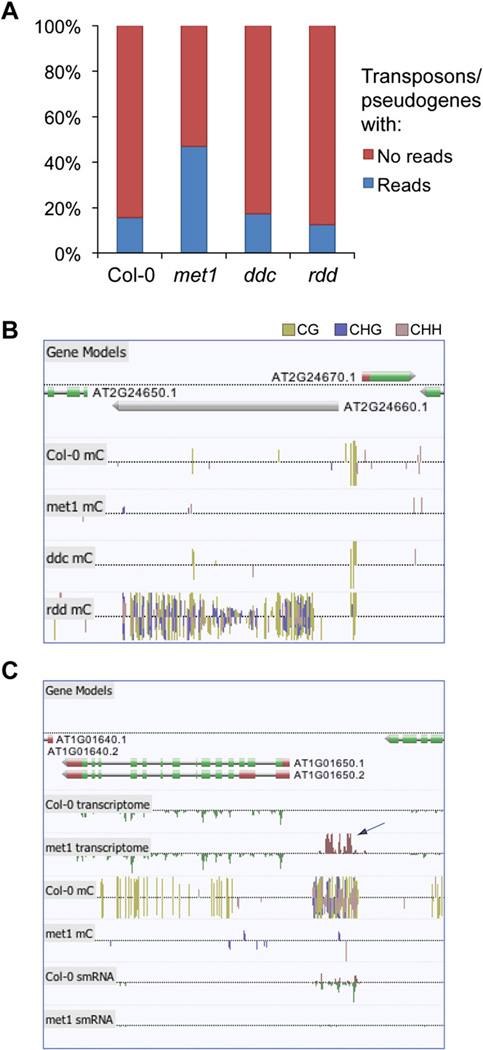
Using stringent conditions, we identified 3.04%, 1.22%, and 0.53% of genes with altered transcript abundance in met1, ddc, and rdd, respectively (Table S7). Notably, we found 281 transposons and pseudogenes that were upregulated in met1, accounting for ~46% of the upregulated genes, consistent with the role of methylation in genome defense through silencing of transposon transcription. Whereas crosshybridization prevented past array-based studies from reliably detecting changes in the transcript abundance of most transposons, mRNA-seq enables unambiguous assignment of reads to the unique sequences within the transposon. Indeed, we found that 47.0% of transposons/pseudogenes (henceforth referred to collectively as transposons) had at least one read in met1, as opposed to just 15.7% in wild-type (Figure 7A), with 3.7-fold more bases of transposons sequenced in met1 than wild-type following normalization to total bases sequenced. Particular classes of transposable elements have the ability to excise and translocate genomic sequences in their proximity and, consequently, are potentially powerful engines of genetic diversity. To determine whether certain lineages of transposons are more likely to be expressed after loss of methylation, we selected all 233 members of the Mutator-like element (MULE) (Yu et al., 2000) transposon family, and generated a phylogenetic tree from the alignment of the DNA sequences (Figure S12). Annotation of the individual MULEs that we found to be upregulated in met1 through mRNA-seq indicated that half of the MULEs comprising two closely related groups contained 46 of the 51 reactivated elements. This dramatic enrichment suggests either that these groups share sequences that are essential for reactivation, or that the nonreactivated set is suppressed by an alternative silencing mechanism.
Figure 7. Identification of Upregulated Transposons and Intergenic Transcripts Regulated by DNA Methylation through mRNA-Seq.
(A) The percentage of transposons/pseudogenes in the Arabidopsis genome for which mRNA-seq-based evidence of transcription was present or absent in each genotype.
(B) Hypermethylation of transposons in rdd. Browser visualization of methylcytosines located within and proximal to the copia-like retrotransposon At2g24660. Dense methylation is present within the retrotransposon only in rdd.
(C) An upregulated intergenic transcript identified in the DNA methyltransferase mutant met1 (indicated by arrow). Sites of DNA methylation are shown, and the color reflects the methylation context, as indicated.
In contrast to met1, for ddc we identified 10% more transposons with associated mRNA-seq reads than wild-type, but interestingly in rdd we found that 20% fewer transposons contained at least one read (data not shown). Furthermore, we observed some transposons that were hypermethylated in rdd yet depleted of DNA methylation in wild-type plants (Figure 7C and Figure S13), as observed previously in ros1 mutant plants (Zhu et al., 2007). Interestingly, these demethylated transposons were frequently found close to protein-coding genes and were often accompanied by an increase in smRNA abundance. Taken together, these data indicate that rdd actively maintains a subset of transposons in a demethylated state, perhaps partly to protect neighboring genes from the effects of a local increase in smRNA abundance and potential silencing.
Alignment of unique mRNA-seq reads to the genomic DNA sequence enabled us to scour the intergenic space for reads originating from unannotated genes. A custom algorithm was used to identify intergenic adjacent reads located within the average Arabidopsis intron size + 1 standard deviation to each other, from which hypothetical transcript units were generated (see Supplemental Experimental Procedures). A total of 250 upregulated intergenic transcripts were identified in met1, of which 206 displayed coincident decreases in DNA methylation, and 61% of which displayed reduced smRNA abundance (Figure 7D). In ddc, 74 novel intergenic transcripts were detected, of which 40 displayed changes in DNA methylation state. Together, with the identification of hundreds of derepressesd transposons, these results indicate that mRNA-seq, in conjunction with smRNA-seq and sequencing of the single base cytosine methylome, provide an unprecedented view of the composition and dynamics of the DNA methylation-suppressed transcriptome (Figure S14).
Permutation tables have been generated for all combinations of changes and equivalence in DNA methylation and smRNA and mRNA levels for every genotype in every gene or new intergenic transcript in the genome. This format enables the integration of all three data sets in a format that conveys the relative frequency of different combinations of epigenetic and transcriptional change throughout the genome. The hyperlinked permutation tables can be accessed for all genes (http://neomorph.salk.edu/genes.html) and intergenic transcripts (http://neomorph.salk.edu/intergenic.html).
Conclusion
In this study, we have described novel methodologies developed to produce a comprehensive integrated map of the genomic distributions of methylcytosines, smRNAs, and transcripts in Arabidopsis at unprecedented resolution. Through the simultaneous study of these three interrelated phenomena in wild-type plants and in informative mutant backgrounds, we have helped to illuminate, genome-wide, the scope and sophistication of the interactions that exist between methylation and smRNA, and their ultimate effect on transcriptional regulation. As a resource for the larger community we have made available all the data sets to GenBank, and we have displayed them in our powerful and easy-to-use genome browser, AnnoJ (http://neomorph.salk.edu/epigenome.html), which was developed for the purpose of this study. The methods we have developed and the highly informative data sets we have made available will contribute positively to the important work of unmasking the role of these powerful epigenetic regulatory mechanisms in eukaryotes.
EXPERIMENTAL PROCEDURES
Supplemental Information
Further details on the plant materials, experimental procedures, high-through-put sequencing, processing, and mapping of Illumina GA sequence reads are provided in the Supplemental Experimental Procedures. A thorough analysis of the incidence of duplicate reads is provided.
MethylC-Seq Library Generation
Five micrograms of purified genomic DNA was sonicated and ligated to Illumina methylated DNA adapters (San Diego, CA), after which adapter-ligated molecules of 120–170 bp were enriched by 18 cycles of PCR with primers complementary to the adaptor sequences. See Supplemental Experimental Procedures for more detail.
smRNA-Seq Library Generation
RNA of 15–30 nt length was gel-purified from total RNA, after which specific 5′ and 3′ Illumina RNA adapters were sequentially ligated to the smRNA molecules. Adapter-ligated molecules were reverse transcribed and enriched by 15 cycles of PCR with primers complementary to the adaptor sequences. See Supplemental Experimental Procedures for more detail.
mRNA-Seq Library Generation
Twenty micrograms of total RNA was depleted of 18S and 28S rRNA, after which the remaining RNA was decapped, fragmented by metal hydrolysis, dephosphorylated, and the 3′ end ligated to the Illumina 3′ smRNA adaptor. 3′ adaptor-ligated RNA was phosphorylated and ligated to the Illumina 5′ smRNA adaptor at the 5′ end. Adaptor-ligated molecules were reverse transcribed and enriched by 20 cycles of PCR with primers complementary to the adaptor sequences. See Supplemental Experimental Procedures for more details.
Dissection of the Genome into Region Profiles
Multiple profile tables were constructed for categories of defined genomic regions, including moving windows of various sizes and various step sizes (e.g., 1000/500 bp and 400/200 bp) and annotation categories (e.g., gene models). Each profile table describes each genomic region in terms of total expression level, smRNA abundance, and methylation content.
Hierarchical Clustering of Regions with Altered DNA Methylation and smRNA Levels
One kilobase regions displaying greater than 3-fold difference in methylcyto-sine density and greater than 5-fold difference in smRNA density between wild-type and mutant were regarded as having altered DNA methylation and smRNA levels, while less than 2-fold difference was deemed equivalent. For a region to be considered, a minimum density of nine methylcytosines of the context being interrogated and 300 sequenced bases of smRNA in one of the two genotypes in each pairwise comparison were required. A pairwise comparison of wild-type and mutant was performed for every region, interrogating all possible permutations of DNA methylation and smRNA differences. If a difference was detected, the region was attributed a positive score for the change permutation (for example, lower DNA methylation and higher small RNA levels), generating a profile for each 1 kb region of all coincident changes. These were subjected to hierarchical clustering with Gene Cluster 3.0, using an uncentered correlation similarity metric and complete linkage clustering method. The resulting .cdt file was visualized in TreeView 1.1.1.
Permutation Tables
Permutation tables were constructed using the profile tables to cluster regions and genes by common DNA methylation, smRNA, and mRNA changes between mutants. Lower thresholds for calling a significant change were as follows: DNA methylation, 1.4-fold difference in number of methylcytosines, >1% of available cytosines in the region methylated; smRNA, 2-fold difference, sequenced bases >60% of the total number of bases in the region; mRNA, 2-fold difference, sequenced bases >100% of the total number of bases in the region. If these thresholds were not met, the region was determined as equivalent for the parameter being compared. If a region did not contain the minimum number of methylcytosines or sequence coverage of smRNA or mRNA in all genotypes being compared, it was considered unchanged. The number of sequenced bases of mRNA-seq within each profile region was adjusted by a single factor for each genotype, based on the relative total number of mRNA-seq reads obtained per genotype, so as to equalize all data sets to an equivalent global total mRNA signal. To reduce variability in the mRNA-seq comparisons, the level of mRNA in each mutant was compared to both wild-type and one other mutant: met1 was compared to Col-0 and rdd, ddc to Col-0 and rdd, and rdd to Col-0 and ddc).
Intergenic Transcript Definition
Coarse expression region boundaries for novel intergenic transcripts were determined by joining overlapping and proximal mRNA-seq reads, tolerating a maximum gap of the mean intron size in Arabidopsis (167 bp) + 1 standard deviation. Each subsequent region was then profiled and used to identify a set of unannotated genes.
AnnoJ Genome Browser
AnnoJ is a REST-based genome annotation visualization program built using Web 2.0 technology. Licensing information and documentation are available at http://www.annoj.org.
Supplementary Material
Supplemental Data include 17 figures, 13 tables, and Supplemental Experimental Procedures and can be found with this article online at http://www.cell.com/cgi/content/full/133/3/523/DC1/.
ACKNOWLEDGMENTS
We thank Dr. Junshi Yazaki and Dr. Hiroshi Shiba for sharing immunoprecipitation data regarding DNA methylation in floral tissue, and Huaming Chen for assistance with microarray data. We gratefully acknowledge Dr. Robert L. Fischer for providing the ros1-3 dml2-1 dml3-1 mutant. R.L. is supported by a Human Frontier Science Program Long-term Fellowship. B.D.G. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1909-06). J.T.-F. is supported by Hackett and Ernest and Evelyn Havill Shacklock Scholarships from the University of Western Australia. The development of the visualization and profiling tool AnnoJ was supported by the Australian Research Council through its Centres of Excellence Scheme (CE0561495, DP0771156). This work was supported by grants from the National Science Foundation, the Department of Energy, the National Institutes of Health, and the Mary K. Chapman Foundation to J.R.E.
Footnotes
ACCESSION NUMBERS
Data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE10877 and the NCBI Short Read Archive accession numbers SRA000284 (methylC-seq), SRA000285 (smRNA-seq), and SRA000286 (mRNA-seq).
REFERENCES
- Aufsatz W, Mette MF, Matzke AJ, Matzke M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol. Biol. 2004;54:793–804. doi: 10.1007/s11103-004-0179-1. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the arabidops is DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006;2:e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fojtova M, Kovarik A, Matyasek R. Cytosine methylation of plastid genome in higher plants. Fact or artefact? Plant Sci. 2001;160:585–593. doi: 10.1016/s0168-9452(00)00411-8. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 2003;13:421–426. doi: 10.1016/s0960-9822(03)00106-4. [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gameto-genesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Tompa R, McCallum CM, Delrow J, Henikoff JG, van Steensel B, Henikoff S. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr. Biol. 2002;12:65–68. doi: 10.1016/s0960-9822(01)00622-4. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Yu Z, Wright SI, Bureau TE. Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics. 2000;156:2019–2031. doi: 10.1093/genetics/156.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include 17 figures, 13 tables, and Supplemental Experimental Procedures and can be found with this article online at http://www.cell.com/cgi/content/full/133/3/523/DC1/.