Summary
Mammalian telomeres are associated with shelterin, the telomere specific protein complex that solves the end-protection problem. The telomeric shelterin binding sites, TTAGGG repeats, are maintained by telomerase, which solves the end-replication problem. We report that the TTAGGG repeat arrays of human and mouse telomeres challenge the DNA replication machinery, giving rise to replication-dependent defects that resemble those of the aphidicolin-induced common fragile sites. Conditional gene deletion experiments showed that efficient duplication of telomeric DNA requires the shelterin component TRF1. In the absence of TRF1, telomeres activate the ATR kinase in S phase and show a fragile site phenotype in metaphase. SMARD showed that TRF1 promotes efficient replication of TTAGGG repeats and prevents fork stalling. Two helicases that can remove G4 DNA structures, BLM and RTEL1, were required to repress the fragile telomere phenotype. These results identify a second telomere replication problem that is solved by the shelterin component TRF1.
Keywords: TRF1, ATR, Telomere, Replication, Fork stalling
INTRODUCTION
Human and mouse telomeres are composed of very long arrays (2–100 kb) of TTAGGG repeats that are maintained by telomerase. These repeats serve as binding sites for shelterin, the telomere specific protein complex that protects chromosome ends from the DNA damage response (de Lange, 2005). Most of the telomeric DNA, however, is maintained by semi-conservative DNA replication. Here we show that TTAGGG repeats cause replication problems and that telomeres resemble the aphidicolin-induced common fragile sites.
Fragile sites represent specific chromosomal regions that challenge replication, especially under conditions of limiting nucleotide pools or partial inhibition of DNA polymerases (Durkin and Glover, 2007). Examples are the common fragile sites, which are prone to display abnormal features in metaphase chromosomes when cells experience replication stress. Specifically, treatment with low levels of the DNA polymerase inhibitor aphidicolin induces site-specific breaks or gaps in metaphase chromosomes (Glover et al., 1984). The molecular basis of this replication dependent instability is not known. Common fragile sites are large and sequence motifs that might explain their behavior have not been identified. The occurrence of breaks or gaps at common fragile sites is enhanced when replication stress is combined with deficiency in the ATR kinase pathway, which responds to stalled replication forks (Casper et al., 2002). Similarly, inhibition of homology-directed repair, which facilitates replication restart after replication fork collapse, exacerbates the expression of common fragile sites (Arlt et al., 2004). The idea that common fragile sites represent regions where replication forks stall and collapse is consistent with the increased rate of recombination at these loci (Feichtinger and Schmid, 1989; Glover and Stein, 1987). Indeed, common fragile sites are hotspots for deletions and other chromosome rearrangements in cancer (Yunis and Soreng, 1984; LeBeau and Rowley, 1984).
Our data identify telomeres as aphidicolin-induced fragile sites and establish that the shelterin protein TRF1 is required to prevent telomere replication problems. TRF1 is one of the six distinct proteins that make up shelterin ((Chong et al., 1995); reviewed in (de Lange, 2005)). TRF1 and its paralog TRF2 bind to double-stranded TTAGGG repeats of the telomere with high fidelity. Both proteins are abundant at telomeres, binding throughout the telomeric DNA tract. TRF1 and TRF2 interact with TIN2, which also recruits TPP1 and POT1 to chromosome ends. POT1 also binds telomeric DNA but unlike TRF1 and TRF2, it interacts with the single-stranded TTAGGG repeats in the 3’ overhang. TRF2 and POT1 contribute to the protection of chromosome ends by repressing DNA damage signaling by the ATM and ATR kinases, respectively (Lazzerini Denchi and de Lange, 2007). TRF2 and POT1 also repress the two main DNA repair pathways, non-homologous end-joining (NHEJ) and homology-directed repair (HDR) (Celli and de Lange, 2005; Celli et al., 2006; Palm et al., 2009). Although TRF1 has a similar architecture as TRF2 (Fairall et al., 2001), it has distinct domains and a different set of interacting partners ((Chen et al., 2008); reviewed in (Palm and de Lange, 2008)). TRF1 has been shown to contribute to telomere length regulation (van Steensel and de Lange, 1997; Smogorzewska et al., 2000) but its role in telomere protection had not been established. Because TRF1 deletion in the mouse is lethal (Iwano et al., 2004; Karlseder et al., 2003), we generated a conditional allele to examine the role of TRF1 in telomere biology.
RESULTS
Conditional deletion of TRF1
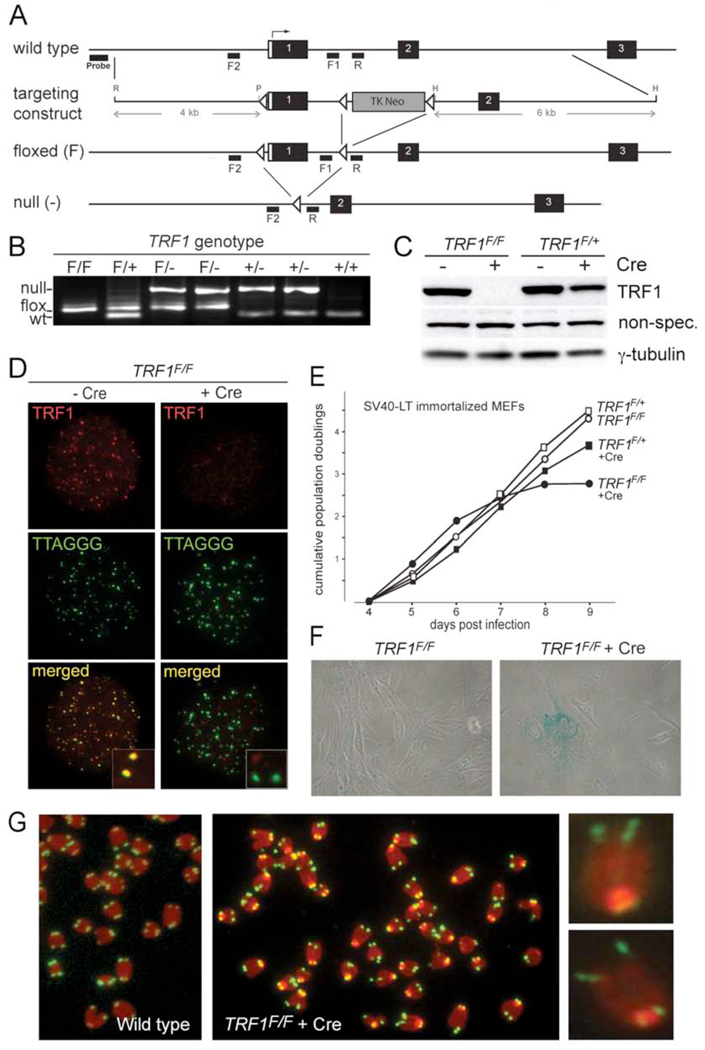
We generated a conditional allele of the mouse TRF1 gene that allows Cre-mediated deletion of exon 1, which contains the translation start site (TRF1F; Fig. 1A,B). Introduction of Cre into TRF1F/F mouse embryo fibroblasts (MEFs) resulted in the expected loss of TRF1 protein within 72 hours (Fig. 1C and D). Consistent with previous reports on the lethality of TRF1 deletion (Iwano et al., 2004; Karlseder et al., 2003), loss of TRF1 induced a growth arrest and senescence in primary and SV40-LT immortalized MEFs (Fig. 1E, F). As deletion of TRF1 was better tolerated in immortalized MEFs, they were used for these studies unless indicated otherwise. The cell cycle arrest and other phenotypes of Cre-mediated TRF1 deletion were suppressed by exogenous TRF1 (Suppl. Fig. 1 and see below), demonstrating that they are indeed a consequence of TRF1 loss.
Figure 1. Conditional deletion of mouse TRF1.
(A) Schematic of the mouse TRF1 locus on chromosome 1 (NCBI locus ID 21749), the targeting construct, and the altered alleles of TRF1. R: EcoRI, P: PvuII, H: HindIII. F1. F2 and R: PCR primers.
(B) TRF1 PCR on tail DNA from mice of the indicated genotypes using the F1 and F2 forward primers and the R reverse primer. PCR products: wild type, 100 bp; flox, 152 bp; null allele, 500 bp.
(C) Immunoblot monitoring loss of TRF1 upon Cre-treatment of TRF1F/F MEFs. TRF1 (Ab1449) was detected 3 days after Cre treatment of TRF1F/F and TRF1F/+ MEFs. -Cre: mock infection, γ-tubulin: loading control.
(D) IF-FISH to monitor TRF1 at telomeres of TRF1F/F MEFs at day 3 post Cre. TRF1 IF, red; telomeric FITC PNA probe, green.
(E) Graph representing proliferation of TRF1-deficient MEFs.
(F) Phase-contrast microscopic images of primary TRF1F/F MEFs before and after Cre treatment. Cells were stained for SA-β-galactosidase at day 7 post Cre.
(G) Metaphase spreads showing the fragile telomere phenotype in TRF1 null cells. Telomeres highlighted by FISH (green) and DNA was stained with DAPI (red) at day 4 post Cre.
TRF1-deficient cells did not show a strong telomere fusion phenotype, as fewer than 2% of the metaphase chromosomes became joined and genomic DNA analysis showed that both the telomeric restriction fragment lengths and the telomeric 3’ overhang were unaltered (Suppl. Fig. 2A, B). These results contrast the phenotype of TRF2 deletion, which induces telomere fusions and a concomitant loss of the 3’ overhang (Celli and de Lange, 2005). Deletion of TRF1 also did not lead to the strong increase in telomeric overhang signals observed upon deletion of POT1b, nor did the cells show the endoreduplication phenotype associated with the loss of POT1a (Suppl. Fig. 2B, C) (Hockemeyer et al., 2006). Furthermore, TRF1 deletion did not change the expression levels of Rap1, POT1a, or TRF2 (Suppl. Fig. 2D, E and data not shown) and there were only moderate effects on the association of Rap1, TRF2, TPP1, and POT1a with telomeric DNA as measured by ChIP (Suppl. Fig. 2F).
TRF1 deletion results in aberrant telomeres in metaphase
The most notable phenotype of TRF1 deletion was a high incidence of telomeres with an aberrant structure in metaphase (Fig. 1G and Fig. 2A). The telomeric FISH signal at individual chromatid ends is normally represented as a single signal with an intensity that is roughly equal to the telomeric signal of the sister chromatid end. After TRF1 deletion, a large fraction of chromatids had multiple telomeric signals (Fig. 2A, B). In some cases, the multiple signals were spatially separated from the chromatid end, as if the telomeric DNA had failed to condense or was broken. We refer to these various abnormal telomeric patterns as fragile telomeres.
Figure 2. Effects of TRF1, aphidicolin, and ATRsh.
(A) Examples of the fragile telomeres in TRF1F/F MEF metaphases at day 4 post Cre, Telomeric DNA: FITC PNA probe (green); DNA: DAPI (blue).
(B) Quantification of fragile telomeres induced by deletion of TRF1 with or without treatment with 0.2 µM aphidicolin or ATR shRNA. Bars represent mean values of three independent experiments with SDs. Asterisks: P < 0.01 based on a two-tailed Student's t-test.
(C) Quantification chromosome breaks/gaps. Experimental conditions as in (B).
(D) Quantification of long arms telomere associations in response to the indicated treatments. Experimental conditions as in (B). Sister telomere associations were only scored on long arms. Sister associations were significantly reduced (P < 0.05 based on a two-tailed Student's t-test) by treatment of TRF1 null cells with ATR shRNA but not by aphidicolin treatment or absence of DNA ligase IV (lig4−/−).
Up to 20% of the telomeres showed this aberrant structure in TRF1 null cells whereas fragile telomeres were less frequent (<5%) in control cells (Fig. 2B). Chromosome-orientation FISH (CO-FISH) showed that both sister telomeres were roughly equally prone to display the fragile phenotype (Suppl. Fig. 3A and data not shown). Aberrant telomeric structures (often referred to as telomere doublets) resembling the fragile telomeres documented here, were previously reported in several settings including ES cells lacking TRF1 (Philippe et al., 1999; Undarmaa et al., 2004; Iwano et al., 2004; van Overbeek and de Lange, 2006; Okamoto et al., 2008), but the underlying telomeric defect had not been identified.
In addition to the fragile telomeres, we observed frequent associations of sister telomeres in TRF1 null cells (Fig. 2D). These telomere associations did not result from NHEJ since they also occurred upon deletion of TRF1 from cells lacking DNA ligase IV (Fig. 2D). The molecular nature of these associations remains to be determined but given the data presented below, it is tempting to speculate that the sister-telomere associations represent the recently described sister-chromatid bridges at fragile sites (Chan et al., 2009).
Fragile telomeres are induced by aphidicolin and respond to inhibition of ATR
In order to test whether the fragile telomere phenotype resembles that of the common fragile sites, we examined metaphases of wild type MEFs treated with low concentrations of aphidicolin (0.2 µM). Consistent with previous data (Glover et al., 1984), aphidicolin induced breaks in ∼8% of chromosomes (Fig. 2C, Suppl. Fig. 3B, C). Importantly, aphidicolin induced a striking increase in the frequency of fragile telomeres (Fig. 2B). The effect of aphidicolin was additive with deletion of TRF1, resulting in ∼28% of telomeres showing this phenotype (Fig. 2B). Aphidicolin did not affect the sister telomere associations seen after TRF1 deletion (Fig. 2D).
In order to determine the effect of ATR on the fragile telomere phenotype, TRF1F/F MEFs were treated with Cre and subsequently with ATR shRNA (Suppl. Fig. 3D). As for the common fragile sites (Casper et al., 2002), ATR inhibition strongly enhanced the fragile telomeres phenotype of TRF1-deficient cells (Fig. 2B, Suppl. Fig. 3B–E). In contrast, inhibition of ATR did not increase the sister telomere association phenotype of TRF1 null cells (Fig. 2D). Collectively, the data obtained with cells treated with aphidicolin or ATR shRNA demonstrate that telomeres resemble common fragile sites and that this feature of telomeres is partially repressed by TRF1.
S phase-dependent ATR signaling upon loss of TRF1
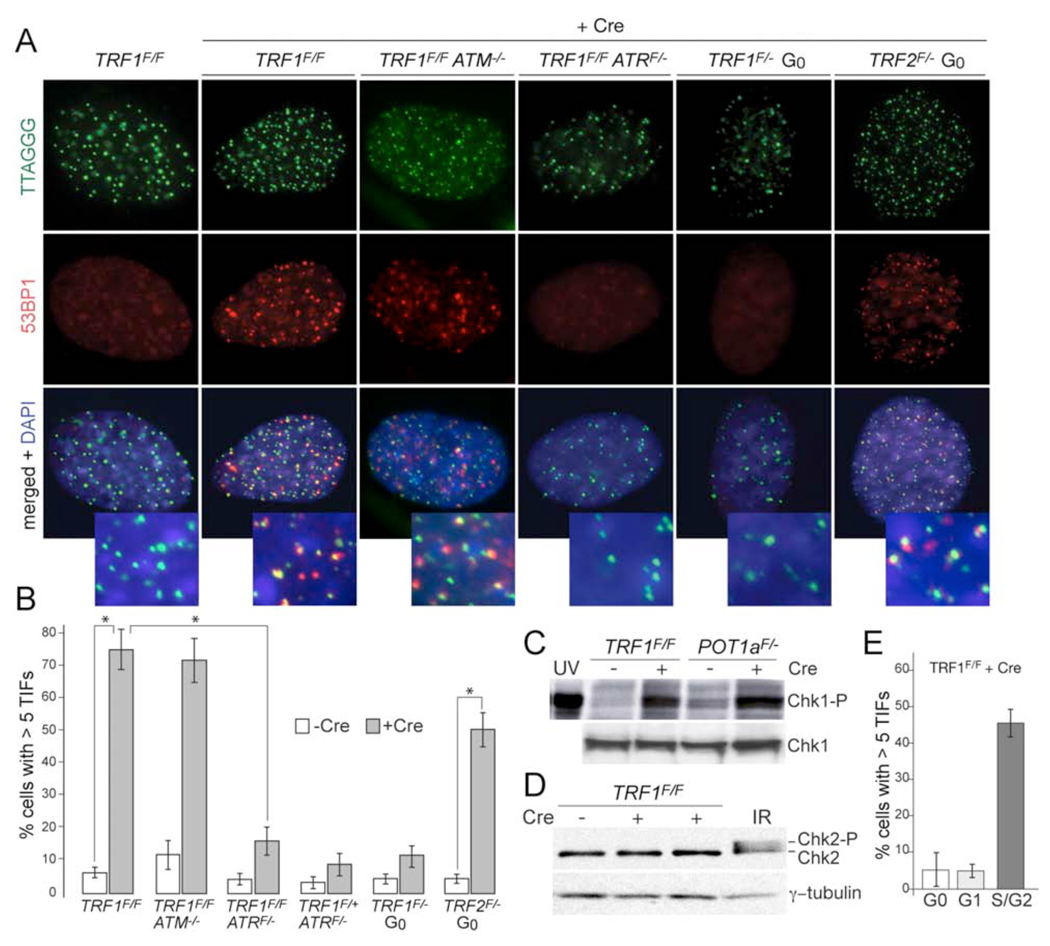
Consistent with a DNA replication defect, cells lacking TRF1 showed a strong telomere damage response phenotype, as evidenced by 53BP1 and γ-H2AX Telomere Dysfunction Induced Foci (TIFs) (Takai et al., 2003) (Fig. 3A, B, Suppl. Fig. 4). The TIF response was fully repressed by exogenous TRF1, establishing that it was due to TRF1 loss (Suppl. Fig. 1). Using MEFs with compound genotypes, we determined whether this DNA damage signal depended on the ATM, DNA-PKcs, or ATR kinase. Of these three kinases, only ATR was required for the TIF response (Fig. 3A and B; Suppl. Fig. 5). Consistent with ATR signaling, Chk1 became phosphorylated upon deletion of TRF1, whereas phosphorylation of the ATM target Chk2 was not detected (Fig. 3C and D).
Figure 3. Deletion of TRF1 results in an S phase dependent ATR kinase signal.
(A) ATR-dependent TIF formation upon deletion of TRF1 from cycling cells. Cells with the indicated genotypes were analyzed at day 4 post pWZL-Cre using FISH for telomeres (green), IF for 53BP1 (red), and DAPI DNA counterstain (blue). To circumvent the lethality associated with ATR deletion, f the TRF1F/F ATRF/F cells were arrested in G0 by contact inhibition and serum starvation, infected with Ad-Cre, released at day 3, and analyzed 1 day later. For the two right-hand panels, TRF1F/− and TRF2F/− cells were similarly arrested in G0 and infected with Ad-Cre, but analyzed at 4 day while in G0. Deletion of ATR, TRF1 and TRF2 was verified by immunoblotting (Suppl. Fig. 5).
(B) Quantification of the TIF response as shown in panel (A). Bargraphs represent mean values of three independent experiments and SDs. Asterisks: P < 0.01 based on a two-tailed Student's t-test.
(C) Immunoblot for Chk1 phosphorylation. Cells with the indicated genotypes were analyzed at day 6 post Cre. POT1a null MEFs and cells treated with UV (25 J/m2, 30 min recovery) serve as positive controls.
(D) Immunoblot for Chk2 phosphorylation. Cells were treated as in (C). MEFs treated with IR (2 Gy, 1 hr recovery) serve as a positive control.
(E) S phase dependent induction of TIFs. TRF1F/F cells were synchronized in G0 and infected with Ad-Cre and analyzed as in (A). For G1, cells were released into normal medium on day 3 post Cre and harvested 15 hrs post release. S/G2 cells were released into normal medium followed by an aphidicolin block and analyzed 7 hours after release from the G1/S block. Bargraphs represent mean values of three independent experiments and SDs. TRF1 was deleted in ∼50% of the cells (Suppl Fig. 5). FACS analysis of the G0, G1, and S/G2 cells is shown in Suppl Fig. 5.
We next asked whether progression through S phase was required for the activation of ATR at telomeres lacking TRF1. To test this, we deleted TRF1 from quiescent (G0) primary TRF1F/− cells. As a positive control, TRF2, which is known to be required for telomere protection in all stages of the cell cycle (Konishi and de Lange, 2008), was deleted from a parallel culture of quiescent primary TRF2F/− cells. While deletion of TRF2 resulted in the expected 53BP1 foci at telomeres, the TIF response was minimal in G0 cells lacking TRF1 (Fig. 3A and B; Suppl. Fig. 5). TIFs only became prominent when the cells were released from G0 and progressed through S phase (Fig. 3E; Suppl. Fig. 5). These results demonstrate that progression through S phase in absence of TRF1 induces an ATR-dependent DNA damage signal at telomeres. As most cells in an asynchronous population of immortalized TRF1 null cells showed had numerous TIFs, it is likely that much of the DNA damage generated at telomeres in S phase persists when TRF1 is absent.
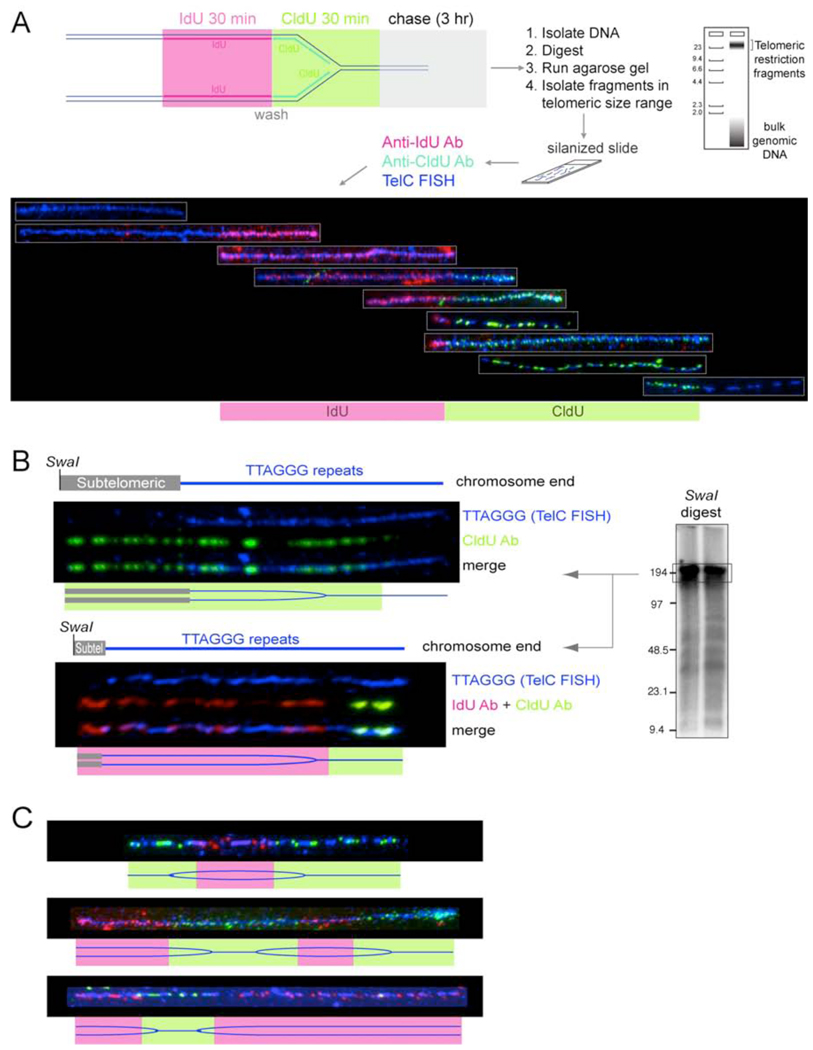
Analysis of telomere replication in wild type cells using SMARD
We used SMARD (Single molecule analysis of replicated DNA; (Norio and Schildkraut, 2001) to examine the progression of replication forks through telomeric DNA (Fig. 4A). SMARD relies on two sequential periods of in vivo labeling with different halogenated nucleotides (IdU and CldU) to mark replicating DNA molecules. Genomic DNA from the labeled cells was digested with frequently cutting restriction enzymes that cleave most of the genomic DNA into small fragments but do not cut in the long (>20 kb) TTAGGG repeat arrays, so that DNA fragments with a MW >25 kb isolated from an agarose gel were enriched for telomeric DNA. The incorporation of IdU and CldU was visualized with fluorescent antibodies in partially denatured DNA molecules stretched onto silanized glass slides. We identified the telomeric DNA fragments with a FISH-PNA probe (TelC) that anneals to the G-rich telomeric repeat strand. Although annealing of the TelC probe interferes with detection of the IdU and CldU in the TTAGGG repeats, substitutions in the CCCTAA repeat strand are detectable. Both the telomeric FISH signal and the IdU or CldU fluorescent signals appeared as strings of dots that were often interrupted due to the partial denaturation of the DNA (Fig. 4A). Nonetheless, long telomeric DNA molecules were readily identified among the mixture of DNA fragments. The optimized procedure used pulse-labelling periods of 30 min followed by a 3 hour chase. Since this procedure only labels the DNA in cells that are in S phase during the pulses, the protocol was further improved by repeating the pulse-chase six times. The total duration of the six rounds of pulse/chase was 21 hours, which is less than the cell doubling time. As replication forks progress at ∼2 kb/min (Anglana et al., 2003), even the longest telomeres (∼150 kb) should be fully replicated within one round of the double-pulse/chase procedure. As expected, the average length of the IdU and CldU segments was approximately equal and the two substitutions were observed in approximately equal fractions of the telomeric DNA molecules. Because the telomeric DNA fragments used for the analysis are of variable lengths, the rate of fork progression cannot be determined accurately in these experiments. However, given that we frequently observed telomeric fragments in the >25 kb size range that were completely labeled with either IdU or CldU in experiments using 30 min pulses, it is unlikely that the fork rate is lower than 1 kb/min.
Figure 4. SMARD analysis of telomere replication in wild type cells.
(A) Top: schematic depiction of the SMARD protocol to visualize the replication of single telomeric DNA molecules. See text for description. Bottom: Telomeric DNA molecules of variable lengths identified by telomeric FISH (TelC; blue) with incorporated IdU and CldU detected with fluorescent antibodies (red and green, respectively). The telomeric fragments are organized assuming that replication proceeds from a subtelomeric origin towards the chromosome end.
(B) Two examples of replication fork progression towards the chromosome end. SMARD on ∼180-kb telomeric DNA SwaI fragments containing subtelomeric DNA of variable lengths. Procedure as in (A) except that the DNA was digested with SwaI and the DNA was resolved on a pulse-field gel (see genomic blot inset). Duration of the IdU and CldU pulses was 1 hour each. The pattern is consistent with replication forks progressing from a subtelomeric origin towards the chromosome end as depicted in the cartoon below each SMARD image.
(C) Examples of three telomeric molecules with IdU/CldU incorporation patterns consistent with replication initiating in the TTAGGG sequence. Procedure as in (A).
The final protocol yielded telomeric DNA molecules with a pattern of IdU/CldU incorporation that could be consistent with replication proceeding from a subtelomeric origin towards the chromosome end (Fig. 4A). To determine the direction of the replication fork, we analyzed telomeric DNA molecules with an attached segment of subtelomeric DNA generated by digestion with SwaI (Fig. 4B). The length of the telomeric SwaI fragments is ∼180 kb as identified by genomic blotting (see inset in Fig. 4B). In this size fraction of SwaI-digested DNA, the telomeric fragments are not strongly enriched, limiting the number of telomeric molecules available for analysis. Nonetheless, we identified 90 telomeric SwaI restriction fragments, which had a labeled (IdU or CldU) segment that extended beyond the telomeric DNA labeled with FISH (Fig. 4B). The presence of IdU or CldU in the subtelomeric segment and the absence of substitution in the distal end of the molecules is consistent with progression of the replication fork from a subtelomeric site into the telomeric DNA. We also observed molecules with IdU in the subtelomeric segment that contained IdU at the proximal end and CldU at the distal end. Again, this configuration is consistent with replication from a subtelomeric origin.
Occasional replication initiation within telomeric repeats in wild type cells
We observed a small number of telomeric DNAs that suggested initiation of DNA replication within the telomeric repeats (Fig. 4C). These molecules contained an IdU segment flanked on both sides by segments of CldU, indicating that replication had started in the telomeric sequences during the IdU pulse and continued in both directions during the CldU pulse. In some cases, replication proceeded both from a subtelomeric origin and an origin within the telomere, leading to convergence of two forks within the telomeric repeats. The frequency of initiation events within the telomeric DNA was low; only ∼3% of telomeric molecules showed a pattern consistent this mode of replication. Occasional initiation of DNA replication in the telomeric repeat array is consistent with the relative lack of sequence specificity of mammalian ORC (Falaschi et al., 2007). In addition, the association of ORC components with shelterin (Deng et al., 2007; Tatsumi et al., 2008; Atanasiu et al., 2006) could contribute to formation of a pre-replication complex within the telomeric DNA.
Diminished telomeric replication upon deletion of TRF1
To determine whether TRF1 affected the efficiency of telomere replication, we measured the fraction of telomeric DNA molecules that contained either IdU or CldU (or both) in DNA obtained from TRF1-proficient and -deficient cells (Fig. 5). In three independent experiments, the deletion of TRF1 resulted in a ∼2 fold lower incorporation of halogenated nucleotides in telomeric DNA molecules regardless of their length. This effect was not due to a general reduction in DNA replication since the incorporation of BrdU was not altered at the time-point studied (Suppl. Fig. 2C). As a control, the well-studied replicating region derived from the Igh locus (Norio et al., 2005) was unaffected by deletion of TRF1 (Fig. 5). Thus, deletion of TRF1 had a specific effect on the replication of telomeric DNA. Furthermore, the efficiency of telomere replication was not altered when TRF2 was deleted from TRF2F/− Lig4−/− MEFs (Fig. 5) and deletion of TRF2 did not induce a fragile telomere phenotype (Celli and de Lange, 2005), indicating a specific role for TRF1 in facilitating telomere replication.
Figure 5. Deletion of TRF1 diminishes the replication efficiency of telomeric DNA.
SMARD assay results from three independent experiments in which TRF1 was deleted from TRF1F/F MEFs. Cells were labeled with IdU and CldU as indicated on the right at day 4 after infection with H&R Cre retrovirus (+Cre) or vector control (− Cre). In the upper panel (experiments 1 and 2), DNA was digested with frequently cutting enzymes and telomeric restriction fragments >25 kb were isolated (schematic on the left). Telomeric DNA molecules were identified by FISH and the % of molecules containing IdU and/or CldU was determined. In the middle panel, telomeric MboI/AluI fragments in the 130–180 kb range (see genomic blot inset) were isolated from a CHEF gel and the fraction of telomeric molecules that contained IdU and/or CldU was determined as above. SMARD assay was done in one experiment in which TRF2 was deleted from TRF2F/F DNA-Lig4−/− cells and the fraction IdU and/or CldU labeled telomeric molecules (130–180 kb range) was analyzed. In the lower panel, the DNA preparation of TRF1F/F MEFs used in the middle panel was digested with SwaI and a 180 kb restriction fragment from the Igh locus was isolated. DNA probes from that locus (see map below) were used to identify the Igh fragments on stretched DNA and the ratio of labeled vs. unlabeled fragments was determined.
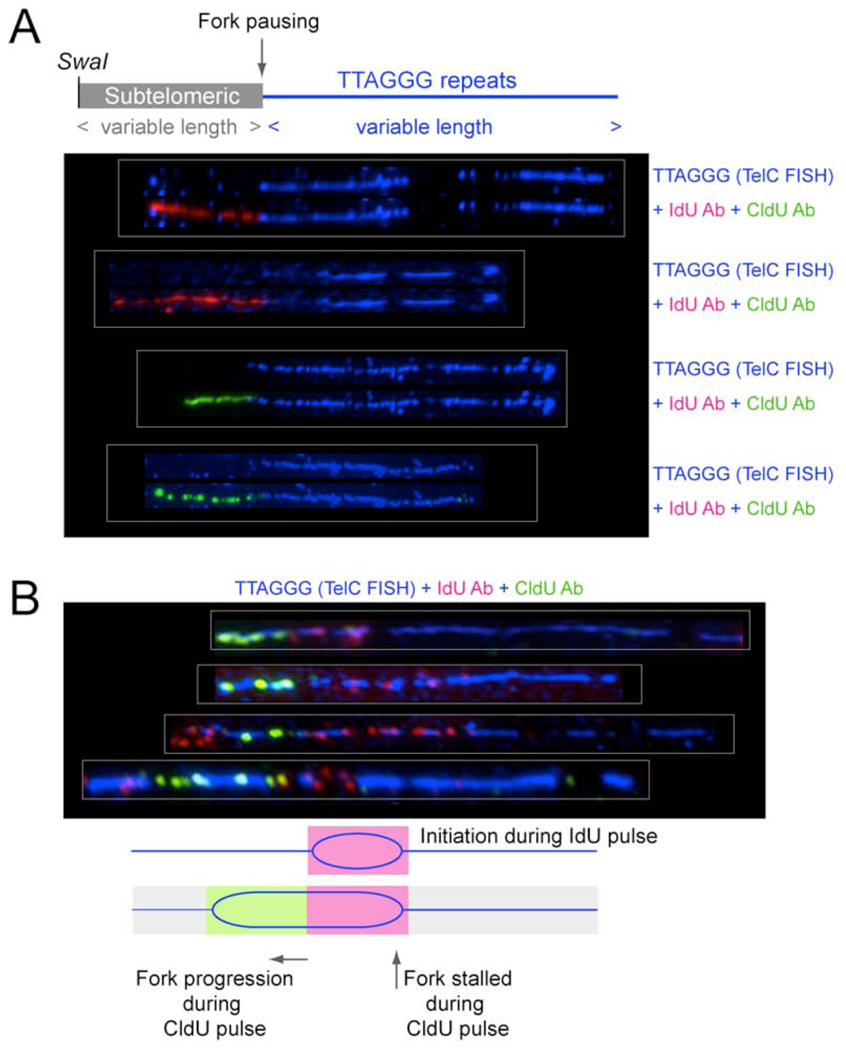
Evidence for replication fork stalling
Inspection of SwaI-digested telomeric DNA molecules, which carry a subtelomeric DNA segment, revealed several cases of IdU/CldU labeling patterns consistent with replication fork stalling in or before the telomeric DNA (Fig. 6A). Among 97 telomeric molecules from TRF1-deficient cells, 7 showed IdU or CldU incorporation in the subtelomeric DNA but no incorporation in the telomeric segment. These patterns would indicate that in the absence of TRF1, the fork has a greater tendency to stall when it encounters telomeric DNA. Such molecules were not identified among 78 telomeric SwaI fragments that were derived in a parallel experiment using TRF1-proficient cells.
Figure 6. Evidence for fork stalling at telomeres in cells lacking TRF1.
(A) Evidence for replication fork stalling at the subtelomeric/telomeric boundary. Shown are four SwaI DNA fragments containing telomeric repeats and subtelomeric DNA from cells lacking TRF1. For procedure see Fig. 4. Molecules shown represent incorporation patterns of IdU and CldU consistent with replication of the subtelomeric DNA (lacking TelC FISH signal) and fork stalling at the boundary of subtelomeric and telomeric DNA as shown in the schematic.
(B) Evidence for replication fork stalling after initiation of DNA replication within telomeric DNA in TRF1 deficient cells. DNA was cut with frequently cutting restriction enzymes and molecules >25 kb were isolated. Labeling was performed as in Fig. 5, top panel. The patterns of incorporation of IdU and CldU are consistent with initiation of replication within the telomeric DNA near the end of a IdU pulse followed by fork progression in only one direction during the CldU labeling period.
Additional evidence for fork stalling was obtained from the occasional molecules generated by replication initiation within the telomeric repeats. We observed telomeric DNA molecules with a non-terminal IdU segment that was short compared to other molecules in the same experiment, indicative of initiation of replication in the telomeric DNA at the end of the IdU pulse (Fig. 6B). In these molecules, the IdU segment is flanked on one side by CldU, indicating fork progression during the CldU labeling period. Importantly, a significant number of these molecules showed no CldU incorporation at the other side of the IdU segment, indicating that the fork on that side did not progress during the CldU labeling period. Although the number of this type of replication products was small (14 out of 250 IdU and/or CldU labeled molecules), they were never observed in DNA from TRF1-proficient cells processed in parallel (400 IdU and/or CldU labeled molecules examined), demonstrating again that absence of TRF1 impairs the normal progression of the replication fork.
Fragile telomeres in human cells
We next asked whether human telomeres also resemble fragile sites. Since it is difficult to fully inactivate human TRF1 with RNAi or dominant negative alleles, we determined whether treatment of human cells with aphidicolin induced the fragile telomere phenotype. Using the same low level of aphidicolin applied to mouse cells, we observed an increase in the frequency of fragile telomeres as compared to untreated cells (Suppl. Fig. 7A). Thus, it is likely that human and mouse telomeres are similar with regard to posing a challenge to the replication fork.
Effect on telomere maintenance
We considered the possibility that the fragile telomere phenotype might lead to loss of telomeric DNA. TRF1 null cells contained a small fraction of chromosome ends that appeared to lack telomeric signals and this phenotype was somewhat stronger when ATR was inhibited (Suppl. Fig. 2A). However, the length of the telomeric restriction fragments of TRF1 null cells was unaltered (Suppl. Fig. 2B). Because small telomere length changes are difficult to detect in mouse cells, we addressed the question of potential telomeric DNA loss in the human fibrosarcoma clone HTC75. We followed the effect of aphidicolin on telomere length in HTC75 cells over 50 population doublings (Suppl. Fig. 7C). As a control, parallel cultures were treated with a concentration of zeocin that induced approximately the same number of DNA damage foci per cell (Suppl. Fig. 7D). Neither zeocin nor aphidicolin induced loss of the telomeric DNA in HTC75 cells. Rather, aphidicolin, but not zeocin, resulted in moderate telomere elongation (Suppl. Fig. 7B). Thus, the replication problems in telomeres are not a major source of telomere loss in telomerase-positive cells and may even enhance the telomerase pathway. In budding yeast, partial inhibition of DNA replication also induces telomere elongation (Carson and Hartwell, 1985; Adams and Holm, 1996).
The mechanism by which TRF1 represses telomere fragility
As deletion of TRF1 but not TRF2 affected telomere replication, we asked whether a specific feature of TRF1 was responsible for its function. Although TRF1 is notably different from TRF2 in its N-terminal domain, which is acidic, this domain was not responsible for the repression of replication problems. TRF1ΔAc fully repressed the fragile telomere phenotype of TRF1 null cells, whereas TRF1ΔMyb, which lacks the ability to bind to telomeric DNA, was unable to complement the loss of the endogenous TRF1 (Fig. 7A, B). The repression of replication problems was also not due to a change in TERRA, a class of RNA polymerase II transcripts that contain UUAGGG repeats (Azzalin et al., 2007). Although TRF1 was shown to be in a complex with RNA polymerase II, and was suggested to contribute to TERRA metabolism (Schoeftner and Blasco, 2008), no change in the abundance of TERRA was observed in TRF1 null cells (Fig. 7B).
Figure 7. The mechanism by which TRF1 represses telomere fragility.
(A) Frequency of fragile telomeres in Cre-treated TRF1F/F MEFs complemented with TRF1ΔAc or TRF1ΔMyb (see Suppl. Fig. 6A for metaphase spreads and TRF1 immunoblots).
(B) TERRA detected by Northern blotting of RNA from cells with the indicated genotype at day 4 post Cre. Bottom: Ethidium bromide (EtBr) staining of rRNAs serves as loading control.
(C) Fragile telomere incidence in cells lacking Blm and/or Wrn.
(D) Representative metaphase spreads from TRF1F/F MEFs (+ or − Cre treatment) infected with Blm and Rtel1 shRNAs as indicated.
(E) Quantification of fragile telomeres in TRF1F/F MEFs (+ or − Cre treatment) infected with Blm and Rtel1 shRNAs as indicated. See Suppl. Fig. 6 for validation of the shRNAs.
(F) Schematic summarizing the proposed function of TRF1 in the context of shelterin. While TRF2 and POT1 repress the DNA damage response throughout the cell cycle, TRF1 is proposed to act in S phase to facilitate replication fork progression through the telomeric DNA. TRF1 is proposed to prevent formation of a fork barrier (most likely G4 DNA) in part by acting with BLM and RTEL1.
We next considered that TRF1 might repress replication problems by recruiting a class of helicases that can remove G4 DNA structures. G4 DNA can be formed by single-stranded TTAGGG repeats and might impede the replication fork. One candidate helicase is the BLM RecQ helicase, which contains the FxLxP TRF1 binding motif (FILMP at aa 311 of human BLM), binds TRF1 in vitro (Lillard-Wetherell et al., 2004), and binds and unwinds G4 DNA (Sun et al., 1998; Huber et al., 2002; Huber et al., 2006). Indeed, BLM-deficient mouse cells (Luo et al., 2000) showed a high frequency of spontaneous fragile telomeres (Fig. 7C), whereas cells lacking another RecQ helicase, WRN, did not show this phenotype. Furthermore, a BLM shRNA induced the fragile telomere phenotype and this effect appeared to be epistatic with TRF1 (Fig. 7D, E). A second candidate helicase that has been proposed to act on G-rich telomeric DNA is RTEL1, which affects telomere length setting in mouse species (Ding et al., 2004) and was recently shown to be functionally similar to Srs2, a budding yeast helicase that inhibit the Rad51 HDR pathway (Barber et al., 2008). Interestingly, the published metaphases of RTEL1 deficient ES cells show a mild fragile telomere phenotype although the frequency of this phenotype was not reported (Ding et al., 2004). RTEL1 shRNA induced fragile telomeres and, as was the case with BLM, this phenotype appeared epistatic with deletion of TRF1 (Fig. 7D, E). It will be necessary to derive Blm/TRF1 DKO and Rtel1/TRF1 DKO cells to further corroborate that TRF1 acts by recruiting/activating these helicases to telomeres.
DISCUSSION
Mammals employ TTAGGG repeats to mark the ends of their chromosomes. These repeats have been used to protect chromosome ends throughout eukaryotic evolution and remain the predominant telomeric repeat in most eukaryotic phyla. Despite the obvious utility of this sequence, there is a potential drawback of the TTAGGG repeat-based telomere protection strategy, which we report on here. Our data establish that the telomeric regions of mouse and human chromosomes challenge DNA replication, leading to a phenotype resembling common fragile sites.
Telomeres as fragile sites
Telomeres were not previously recognized as fragile sites, most likely because their terminal position prohibits the observation of the DNA distal to the gaps and breaks, unless the telomeric DNA is highlighted by FISH. Telomeric FISH showed that telomeres can attain a variety of aberrant structures, ranging from a simple gap to long strings of fragmented telomeric signals or even an extended strand of telomeric DNA. These cytological aspects of the fragile telomere phenotype are informative because they provide direct observation of the aberrant structure. In contrast, FISH probes that mark the center of instability of the very large common fragile sites often do not coincide with the actual breaks or gaps, which can occur at a distance (Becker et al., 2002). Since the structure of the fragile telomeres is highly varied, it is unlikely that the underlying lesion is a simple double-stranded DNA break. Our observations are more compatible with altered packaging and/or condensation of the chromatin perhaps due to extended areas of single-stranded DNA resulting from incomplete replication or processing of stalled forks.
The origin of the telomere replication problem
It will be important to determine what aspect of the telomeric DNA is causing problems during DNA replication. The fragile telomere phenotype is not a consequence of late replication as mammalian telomeres replicate throughout S phase (Ten Hagen et al., 1990; Wright et al., 1999). It is also unlikely that the terminal position of the TTAGGG repeats is responsible for the fragile nature of telomeres as fork arrest/pausing was observed many kbs from the actual telomere terminus. Furthermore, an aphidicolin-induced fragile site in Chinese hamster chromosomes is at or near interstitial telomeric DNA (Musio et al., 1996), suggesting that internally placed telomeric repeats can cause the same problems as terminal ones.
We favor the view that the telomeric DNA is a problematic substrate for the replication machinery, most likely due to formation of G-G Hoogsteen base-paired G4 DNA in the G-rich telomeric repeat strand. Our finding that BLM and RTEL1, two helicases that have been implicated in the removal of G4 DNA, repress the fragile telomere phenotype is consistent with the idea that the fork is primarily hindered by secondary structures formed by the G-rich telomeric DNA. If correct, this would predict that G4 ligands such as telomestatin and RHPS4 might induce a fragile telomere phenotype and that some of their effects on the growth of tumor cells (Salvati et al., 2007; Tahara et al., 2006; Gomez et al., 2006) may be due to their preferential inhibition of telomere replication.
Another possibility is that the telomeric DNA itself does not impair replication but becomes a challenge when bound to the telomeric protein complex. For instance, the single-stranded telomeric DNA binding protein, POT1, may compete with RPA, thereby hampering lagging strand DNA synthesis. However, TTAGGG repeats also impair DNA replication in budding yeast (R. Wellinger, pers. comm.). Since budding yeast lacks shelterin, this result argues that TTAGGG repeats provide an inherent problem to the replication fork, regardless of the proteins bound.
The function of TRF1
Within shelterin, TRF2 and POT1 proteins collaborate to repress all DNA damage response pathways that threaten chromosome ends: DNA damage signaling by the ATM and ATR kinases and NHEJ and HDR mediated DSB repair. These functions are needed throughout the cell cycle and loss of TRF2 or POT1 proteins in G0, G1, S, or G2 result in a DNA damage response (Celli and de Lange, 2005; Hockemeyer et al., 2006; Konishi and de Lange, 2008) (Y. Gong and TdL, unpubl.). In contrast, TRF1 has a specific function in S phase, facilitating the replication of telomeres, thereby preventing ATR activation and the formation of fragile telomeres in metaphase.
Our data suggest that TRF1 primarily acts through the recruitment of BLM and RTEL1, but other factors are not excluded. Neither RTEL1 nor BLM were observed in an exhaustive PICh-based analysis of proteins associated with HeLa cell telomeres, although BLM was found at ALT telomeres (Dejardin and Kingston, 2009). However, it is possible that the association of these helicases is transient and therefore escapes detection.
Deletion of the presumed fission yeast ortholog of TRF1 and TRF2, Taz1, results in a block in telomere replication (Miller et al., 2006). When Taz1 is absent, 2-D gels reveal an aberrant class telomeric fragments, referred to as the ‘plume’, speculated to represent replication problems. Deletion of Taz1 also resulted in fork stalling at a chromosome-internal segment of telomeric DNA. Furthermore, the telomeres of taz1− cells are rapidly lost and require constant re-synthesis by telomerase. These findings are consistent with a role for Taz1 in promoting replication through telomeric DNA and further underscore the similarity of fission yeast and mammalian telomeres (see also (Miyoshi et al., 2008)).
Implications
The finding that mammalian telomeres resemble fragile sites makes several predictions. Common fragile sites are prone to sister-chromatid exchanges, often undergo rearrangements, and are frequent targets of integration of exogenous DNA (reviewed in (Durkin and Glover, 2007)). With regard to the first hallmark of common fragile sites, the fragile telomeres in TRF1 null cells do not appear to undergo frequent telomere sister-chromatid exchanges (T-SCEs, Suppl. Fig. 2). However, T-SCEs are known to be repressed by TRF2 and Pot1a/b, which remain associated with telomeres in TRF1 null cells (Celli et al., 2006; Palm et al., 2009).
With regard to the second hallmark of common fragile sites, their propensity to undergo rearrangements, recent work on focal deletions in colon carcinomas has been revealing. A large fraction (16%) of focal deletions occur near telomeres (Scott Powers, pers. comm.), consistent with genome rearrangements due to the fragile nature of telomeres and providing a parallel with the tumor-like microdeletions at common fragile sites (Arlt et al., 2009; Durkin et al., 2008). In addition, human chromosome ends show frequent duplications in subtelomeric sequences and the rate of sister-chromatid exchange is high near the telomeres (reviewed by (Riethman, 2008)). Both phenomena may be related to the fragile telomere phenotype described here.
Finally, with regard to the integration of foreign DNA into common fragile sites, it is noteworthy that chromosome ends are often enriched for mobile elements. For instance, a human herpes virus (HHV-6) was recently show to preferentially integrate in telomeres (P.G. Medveczky, pers. comm.) and LINE-1 elements can transpose to telomeres in certain hamster cell lines (Morrish et al., 2007). An extreme version of telomere-tropic integration is found in the bdelloid rotifers which have chromosome ends littered with foreign DNA, including mobile elements and DNA derived from horizontal gene transfer (Gladyshev et al., 2008). Although the preferred telomeric invasion in the bdelloid rotifers and mammalian cells could be due to addition of DNA to the termini of deprotected telomeres followed by telomere healing by telomerase, it is also possible that integration is biased by frequent replication fork arrest within the telomeric repeat array. In the latter scenario, the invading element is less likely to compromise telomere function. One could speculate that frequent fork arrest in subtelomeric/telomeric regions could have adaptive value since it would provide a safe sink for mobile elements that might otherwise invade more precious parts of the genome. This could explain why throughout eukaryotic evolution, telomeres have not evolved away from the TTAGGG repeats that generate replication problems.
EXPERIMENTAL PROCEDURES
TRF1 gene targeting, isolation of MEFs, and cell culture procedures
The mouse TRF1 locus was modified using standard gene targeting techniques to generate the TRF1F and TRF− genotypes shown in Figure 1. The alleles were maintained in mice with a mixed background (129/C57Bl6). Compound genotypes were created by intercrosses of TRF1F/F with ATM+/−, ATRF/−, Lig4−/− and DNA-PKcs−/− mice (Barlow et al., 1996; Brown and Baltimore, 2003; Gao et al., 1998). MEFs were isolated from E13.5 embryos and immortalized with pBabeSV40-LT (a gift from Greg Hannon). SV40-LT immortalized Wrn−/−, Blm−/−, and Wrn−/− BIm−/− mouse ear fibroblasts were a gift from Brad Johnson. Cre recombinase was introduced using Hit&Run-Cre, Ad5 CMV Cre, or pWZL-Cre as described previously (Celli and de Lange, 2005). Aphidicolin treatments (0.2 µM) were for 16 hours. shRNAs for Blm (shBLM3c; GGACCTGCTGGAAGATTTA) and ATR (shATR3-1; (Lazzerini Denchi and de Lange, 2007)) were introduced using 4 infections at 6 hr intervals using pSuperior puromycin retrovirus. shRNA for Rtel1 (pLK0.1 from Open Biosystems) was introduced using 2 lentiviral infections at 12 hr intervals.
Analysis of telomeric DNA, IF, IF-FISH, immunoblotting, FACS, and SA-β-gal assays All techniques were as described previously (Celli and de Lange, 2005; Dimitrova et al., 2008).
SMARD assay
The SMARD assay was performed essentially as described previously (Norio and Schildkraut, 2001). Cells were sequentially labeled with 25 µM IdU (30 min or 1 hr) and 25 µM CldU (30 min or 1 hr) with 3 PBS washes in between followed by incubation with media without IdU/CldU for 3 hours. This process was repeated 6 times for 30 min pulses and 3 times for 1 hr pulses. DNA isolation and processing for SMARD was as described previously (Norio and Schildkraut, 2001). DNA was stretched on microscope slides coated with 3-aminopropyltriethoxysilane (Sigma). After stretching, the DNA was denatured in alkali-denaturing buffer (0.1 N NaOH in 70% ethanol and 0.1% b-mercaptothanoland) for 8, 12 or 15 min and fixed by adding 0.5% glutaraldehyde for 5 min. Telomeric DNA was identified by hybridizing with a Biotin-00-(CCCTAA)4 PNA probe and Alexa Fluor 350-conjugated NeutrAvidin antibody (Molecular Probes) followed by biotinylated anti-avidin antibody (Vector). Halogenated nucleotides were detected with a mouse anti-IdU monoclonal antibody (Becton-Dickinson) and a rat anti-CldU monoclonal antibody (Accurate). Alexa Fluor 568-conjugated goat anti-mouse (Molecular Probes) and Alexa Fluor 488-conjugated goat anti-rat were used as secondary antibodies (Molecular Probes).
ChIP analysis
ChIP was performed as previously described (Loayza and de Lange, 2003).
TERRA analysis
Total cellular RNA was prepared using RNeasy Mini Kit (Qiagen), according to the manufacturer instructions and Northern blot analysis was performed as previously described (Azzalin et al., 2007).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Devon White for expert mouse husbandry and Eros Lazzerini Denchi for assistance with protocols and helpful discussion. Megan van Overbeek is thanked for assistance with TRF1 antibody and members of the de Lange are thanked for comments on these experiments. We are grateful to Sara Buonomo for the BLM shRNA and to Brad Johnson for providing the Blm−/−, Wrn−/− and Blm−/− Wrn−/− cell lines. We thank Scott Powers, Peter Medveczky, and Mundy Wellinger for allowing us to cite unpublished data. AS is supported by a postdoctoral fellowship from Susan G. Komen for the Cure. This work was supported by grants from the NIH to TdL and CLS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams AK, Holm C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4614–4620. doi: 10.1128/mcb.16.9.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006;7:716–721. doi: 10.1038/sj.embor.7400730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, Mcllwraith MJ, O'Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, Tissenbaum H, West SC, Rose AM, Boulton SJ. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Becker NA, Thorland EC, Denison SR, Phillips LA, Smith DI. Evidence that instability within the FRA3B region extends four megabases. Oncogene. 2002;21:8713–8722. doi: 10.1038/sj.onc.1205950. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson MJ, Hartwell L. CDC17: an essential gene that prevents telomere elongation in yeast. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- Celli G, de Lange T. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Celli GB, Lazzerini Denchi E, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr Biol. 2007;17:1989–1995. doi: 10.1016/j.cub.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, Tarn PP, Nagy A, Lansdorp PM. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Ragland RL, Arlt MF, Mulle JG, Warren ST, Glover TW. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc Natl Acad Sci U S A. 2008;105:246–251. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L, Chapman L, Moss H, de Lange T, Rhodes D. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Molecular Cell. 2001;8:351–361. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- Falaschi A, Abdurashidova G, Sandoval O, Radulescu S, Biamonti G, Riva S. Molecular and structural transactions at human DNA replication origins. Cell Cycle. 2007;6:1705–1712. doi: 10.4161/cc.6.14.4495. [DOI] [PubMed] [Google Scholar]
- Feichtinger W, Schmid M. Increased frequencies of sister chromatid exchanges at common fragile sites (1)(q42) and (19)(q13) Hum Genet. 1989;83:145–147. doi: 10.1007/BF00286707. [DOI] [PubMed] [Google Scholar]
- Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver DT, Alt FW. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- Glover TW, Berger C, Coyle J, Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- Glover TW, Stein CK. Induction of sister chromatid exchanges at common fragile sites. Am J Hum Genet. 1987;41:882–890. [PMC free article] [PubMed] [Google Scholar]
- Gomez D, Wenner T, Brassart B, Douarre C, O'Donohue MF, El Khoury V, Shin-Ya K, Morjani H, Trentesaux C, Riou JF. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J Biol Chem. 2006;281:38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Huber MD, Lee DC, Maizels Np. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MD, Duquette ML, Shiels JC, Maizels N. A Conserved G4 DNA Binding Domain in RecQ Family Helicases. J Mol Biol. 2006;358:1071–1080. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J Biol Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, de Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol. 2003;23:6533–6541. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- LeBeau MM, Rowley JD. Heritable fragile sites in cancer. Nature. 1984;308:607–608. doi: 10.1038/308607a0. [DOI] [PubMed] [Google Scholar]
- Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- Musio A, Rainaldi G, Sbrana I. Spontaneous and aphidicolin-sensitive fragile site 3cen co-localizes with the (TTAGGG)n telomeric sequence in Chinese hamster cells. Cytogenet Cell Genet. 1996;75:159–163. doi: 10.1159/000134469. [DOI] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- Norio P, Schildkraut CL. Visualization of DNA replication on individual Epstein-Barr virus episomes. Science. 2001;294:2361–2364. doi: 10.1126/science.1064603. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Iwano T, Tachibana M, Shinkai Y. Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J Biol Chem. 2008;283:23981–23988. doi: 10.1074/jbc.M802395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Ann Rev Genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Philippe C, Coullin P, Bernheim A. Double telomeric signals on single chromatids revealed by FISH and PRINS. Ann Genet. 1999;42:202–209. [PubMed] [Google Scholar]
- Riethman H. Human telomere structure and biology. Annu Rev Genomics Hum Genet. 2008;9:1–19. doi: 10.1146/annurev.genom.8.021506.172017. [DOI] [PubMed] [Google Scholar]
- Salvati E, Leonetti C, Rizzo A, Scarsella M, Mottolese M, Galati R, Sperduti I, Stevens MF, D'lncalci M, Blasco M, Chiorino G, Bauwens S, Horard B, Gilson E, Stoppacciaro A, Zupi G, Biroccio A. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J Clin Invest. 2007;117:3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Karow JK, Hickson ID, Maizels Np. The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3' telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Ezura K, Yoshida K, Yugawa T, Narisawa-Saito M, Kiyono T, Ohta S, Obuse C, Fujita M. Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes Cells. 2008;13:1045–1059. doi: 10.1111/j.1365-2443.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- Ten Hagen KG, Gilbert DM, Willard HF, Cohen SN. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol Cell Biol. 1990;10:6348–6355. doi: 10.1128/mcb.10.12.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undarmaa B, Kodama S, Suzuki K, Niwa O, Watanabe M. X-ray-induced telomeric instability in Atm-deficient mouse cells. Biochem Biophys Res Commun. 2004;315:51–58. doi: 10.1016/j.bbrc.2004.01.014. [DOI] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Liao ML, Shay JW. Normal human telomeres are not late replicating. Exp Cell Res. 1999;251:492–499. doi: 10.1006/excr.1999.4602. [DOI] [PubMed] [Google Scholar]
- Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.