Abstract
Invasive disease following methicillin-resistant Staphylococcus aureus (MRSA) detection is common, regardless of whether initial detection involves colonization or infection. We assessed the genetic relatedness of isolates obtained ≥2 weeks apart representing either repeated infections or colonization-infection sets to determine if infections are likely to be caused by previously harbored strains. We found that MRSA infection following initial colonization or infection is caused by the same strain in most cases, suggesting that a single successful attempt at decolonization may prevent the majority of later infection.
Methicillin-resistant Staphylococcus aureus (MRSA) causes >278,000 hospitalizations and 56,000 septic episodes annually [1]. Hospitalized patients with newly identified MRSA colonization or infection have a 35% risk of subsequent invasive disease within 1 year [2, 3]. Strategies to prevent infection have included decolonization [–11], administration of anti-staphy-lococcal antibodies [12], and vaccination [13–14].
Decolonization has had variable success [4]. Its benefit may depend on whether subsequent MRSA infections are caused by the original infecting or colonizing strain, indicating persistent carriage or repeated acquisition from the local environment. Under these scenarios, a single successful decolonization of patients and potentially their immediate surroundings could prevent future infection. However, if subsequent infections are caused by different strains, decolonization would be of limited value unless sustained over long periods.
Colonizing strains are responsible for infections occurring soon after detected colonization [18]. Less is known about the association between colonizing strains and subsequent infections occurring in the intermediate or distant future, but 40%– 60% of carriers demonstrate prolonged carriage with the same strain [14–17]. We assessed the genetic relatedness of MRSA isolates obtained >2 weeks apart. We evaluated isolates representing both repeated infections and asymptomatic carriage followed by infection.
Methods
We conducted a multicenter cohort study in 4 academic hospitals identifying patients with MRSA invasive disease following prior MRSA infection or colonization (table 1). Hospitals selected a 1–2-year study period during which all patients with sterile site MRSA cultures >2 weeks apart were identified (the infection-infection group). Associated MRSA isolates were retrieved. Two institutions additionally collected isolates from patients with MRSA-positive nares surveillance cultures followed by invasive clinical cultures (the colonization-infection group). Patients could contribute to both groups. Decolonization was not routinely employed. This study was approved by each hospital’s institutional review board.
Table 1.
Participating institutions and cohorts.
| Institution | Study design | Collected specimens | Study period |
|---|---|---|---|
| 1 | Retrospective cohort, hospital-wide | Repeated bacteremia | 1 January 2002 to 31 December 2003 |
| 2 | Retrospective cohort, hospital-wide | Repeated invasive diseasea | 1 January 2002 to 31 December 2003 |
| 3 | Retrospective cohort, hospital-wide | Repeated bacteremia | 1 September 2003 to 31 August 2004 |
| 3 | Retrospective cohort, surgical intensive care unit | Nares-bacteremia | 1 September 2003 to 31 August 2004 |
| 4 | Prospective cohort, intensive care units (8)b | Repeated invasive diseasea,nares-invasive diseasea | 1 September 2003 to 30 June 2005 |
Defined by Centers for Disease Control criteria (see methods).
Intensive care units included cardiac (1), medical (1), surgical (1), burn/trauma (1), cardiac surgery (2), neurosurgery (1), and thoracic surgery (1).
Medical records were reviewed to confirm infection by Centers for Disease Control and Prevention (CDC) criteria [19]. Sequential infections were required to be discrete and unrelated, as determined through review by an infectious diseases physician. Sequential device-related bacteremia required device removal between events and complete resolution of the initial infection.
Data collected included age, sex, hospitalization dates, length of hospital stay(s), hospital day of cultures, pre-admission location(s), and postdischarge disposition(s). Risk factors for MRSA infection were collected, such as recent surgery, wounds, and comorbidities (hemodialysis, diabetes).
Isolates were sent to the Molecular Epidemiology and Fungus Testing Laboratory at the University of Iowa (Iowa City) for confirmation of methicillin resistance (mecA gene probe), antibiotic susceptibility testing, and SCCmec typing [20]. PCR assays for the Panton-Valentine leukocidin (PVL) gene were performed. Antibiotic susceptibility testing involved broth microdilution [21, 22] for oxacillin, erythromycin, clindamycin, tetracycline, rifampin, moxifloxacin, gentamicin, vancomycin, linezolid, and quinupristin-dalfopristin; mupirocin, tetracycline, and trimethoprim-sulfamethoxazole susceptibility were determined by Etest (AB Biodisk) [23]. D-tests for inducible clindamycin resistance were performed when isolates were resistant to erythromycin but susceptible to clindamycin.
Strain types based on PFGE [24] were classified as indistinguishable, similar, or different [25]. Identical banding patterns were considered to be indistinguishable, those differing by ≤3 bands were considered to be similar (includes indistinguishable strains), and those differing by >3 bands were considered to be different. PFGE patterns were compared to defined USA types [26] to distinguish common hospital and community-associated MRSA strains (BioNumerics). Total and hospital-specific dendrograms were created to display strain relatedness.
Patients’ characteristics were reported as proportions. Repeated infections were summarized by culture site, infection type by CDC criteria [14], duration between events, and location between events (inpatient, home, or skilled nursing facility or rehabilitation center [SNF/rehab]). Location between events was characterized by the highest acuity center.
Within the infection-infection group, strain relatedness was assessed by the proportion of the first 2 infection events per person that had indistinguishable, similar, or different strain types. Strain relatedness among the colonization-infection group was similarly assessed. Associations between strain relatedness and patient location or duration between events were assessed using Fisher’s exact test.
MRSA isolates were additionally described by SCCmec type and presence of PVL. We also assessed whether subsequent infecting strains exhibited increased antibiotic resistance, compared with original strains.
Results
Thirty-seven patients had ≥2 discrete MRSA infections, for a total of 83 infections. By study design, nearly all infections (79 [95%] of 83) involved bacteremia. Twenty-nine patients had sets of isolates representing colonization followed by infection, 27 (93%) of which involved bacteremia. Eight of the 29 had another subsequent MRSA infection and contributed to both groups. Patient characteristics and infection types are found in table 2 and table 3.
Table 2.
Characteristics of patients with methicillin-resistant Staphylococcus aureus infections.
| No. (%) of patients | ||
|---|---|---|
| Characteristic | Infection-infection (n = 37) |
Colonization-infection (n = 29) |
| Hospital | ||
| 1 | 10 (27) | 0 (0) |
| 2 | 2 (5) | 0 (0) |
| 3 | 14 (38) | 3 (10) |
| 4 | 11 (30) | 26 (90) |
| Male sex | 18 (49) | 17 (59) |
| Age group, years | ||
| 18–34 | 5 (14) | 4 (14) |
| 35–44 | 4 (11) | 1 (3) |
| 45–54 | 5 (14) | 4 (14) |
| 55–64 | 10 (27) | 3 (10) |
| 65–84 | 11 (30) | 17 (59) |
| ≥85 | 2 (5) | 0 (0) |
| Diabetes | 13 (35) | 8 (28) |
| Hemodialysis | 5 (14) | 3 (10) |
| Recent surgerya | 21 (57) | 26 (90) |
| Woundb | 17 (46) | 22 (76) |
Defined as surgery within 6 months of the initial isolate. The high proportion of surgery in the colonization-infection group reflects sampling in surgical intensive care units.
At time of initial isolate.
Table 3.
Characteristics of repeated methicillin-resistant Staphylococcus aureus events.
| Characteristic | Infection-infectiona (n = 37) |
Colonization-infectiona (n = 29) |
|---|---|---|
| Time between events, median days (range) | 74 (14–482) | 39 (15–250) |
| Source of first isolate | ||
| Primary bacteremia | ||
| Total | 28 (76) | 29 (100)b |
| Catheter-related | 23 (82) | … |
| Unknown source | 5 (18) | … |
| Secondary bacteremia | ||
| Total | 6 (16) | … |
| Surgical site infection | 3 (50) | … |
| Lung infection | 2 (33) | … |
| Bone/joint infection | 1 (17) | … |
| Other (nonbacteremic) | 3 (8)c | … |
| Source of second isolated | ||
| Primary bacteremia | ||
| Total | 24 (65) | 21 (72) |
| Catheter-related | 21 (88) | 11 (52) |
| Unknown source | 3 (12) | … |
| Secondary bacteremia | ||
| Total | 11 (30) | 6 (21) |
| Surgical site infection | 3 (27) | 0 (0) |
| Lung infection | 2 (18) | 3 (50) |
| Bone/joint infection | 3 (27) | 1 (17) |
| Vascular | 3 (27) | 1 (17) |
| Soft-tissue infection | 0 (0) | 1 (17) |
| Other (nonbacteremic) | 2 (5)e | 2 (7)f |
| Patient disposition between isolates | ||
| Not discharged | 13 (35) | 15 (52) |
| Homeg | 13 (35) | 4 (14) |
| Skilled nursing facility or rehabilitation center | 11 (30) | 10 (34) |
NOTE. All data are no. (%) of patients, unless otherwise indicated.
Description pertains to first 2 events per person.
All primary bacteremia events in the colonization-infection group were nares-bacteremias.
Urinary tract infection (1), lung infection (1), and gastrointestinal infection (1).
Ninety percent of patients experiencing serial bacteremia had an echocardiogram performed to exclude endocarditis as the source.
Urinary tract infection (1) and bone and joint infection (1).
Surgical site infection (1) and bone and joint infection (1).
Restricted to patients whose only disposition between events was to home.
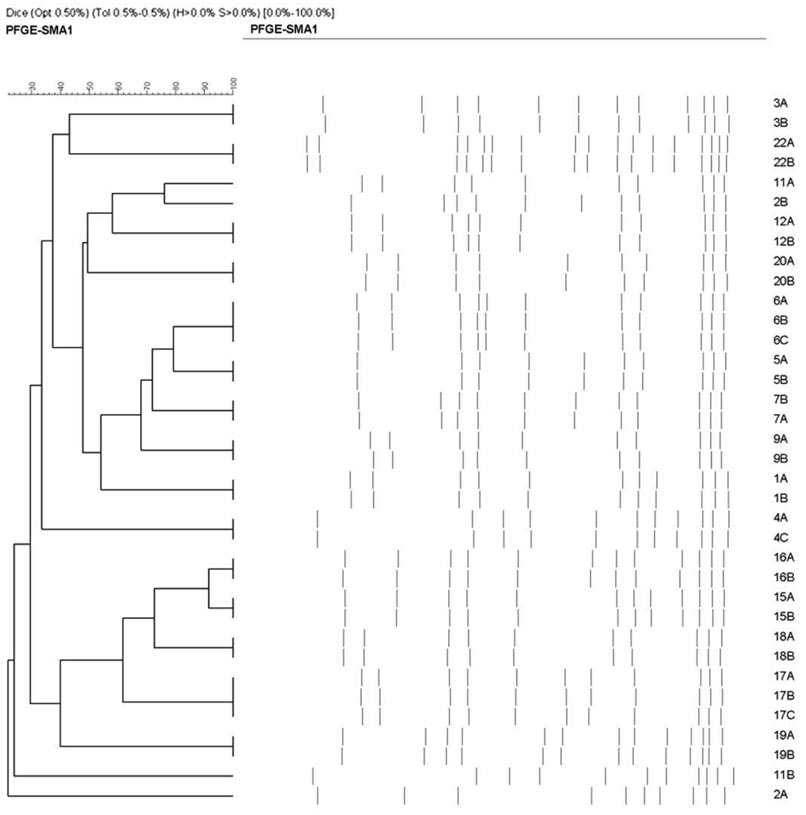
We found considerable strain diversity, with 97%–100% of patients having PFGE subtypes that were unique within each institution (example institution shown in figure 1). Nevertheless, despite population diversity, strains from a single patient representing either sequential infection or colonization-infection were highly related (table 4). Among the infection-infection group, only 4 (11%) of 37 patients had subsequent MRSA infections caused by a different strain. Patients with intervening SNF/rehab stays were more likely to have infections caused by different strains than were those who went home (P=.02) or remained in the hospital (P=.02). Longer durations between MRSA infections were not predictive of differing strain types (table 5).
Figure 1.
Dendrogram demonstrating the broad diversity of methicillin-resistant Staphylococcus aureus (MRSA) strains across patients at a single participating institution. The dendrogram is based on PFGE of MRSA isolates from discrete events. Seventeen patients are represented, each identified by a number on the right side of the figure. Fourteen patients had multiple discrete bacteremic events, and 3 patients had bacteremia following surveillance isolates. Letter designations within a given number refer to isolates from the same patient. In this example institution, no 2 patients have identical isolates based on PFGE. Identical strain types may be slightly offset in pattern as a result of small differences in lane alignment or gel characteristics when grouping strains for dendrogram analysis.
Table 4.
Genetic relatedness of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from individual patients.
| No. (%) of patients | |||||
|---|---|---|---|---|---|
| Source | Indistinguishable strain typea |
Similar strain typea,b |
Different strain typea |
PVL positivec | SCCmec IVd |
| Infection-infection | |||||
| Total (n = 37) | 28 (76) | 33 (89) | 4 (11) | 2 (5)d | 4 (11) |
| Bacteremia-bacteremiae(n = 34) | 26 (76) | 30 (88) | 4 (12) | 2 (6)d | 4 (12) |
| Colonization-infection | |||||
| Total (n = 29) | 21 (72) | 29 (100) | 0 (0) | 0 (0) | 2 (7) |
| Nares-bacteremiaf(n = 27) | 19(70) | 27 (100) | 0 (0) | 0 (0) | 2 (7) |
| Total (n = 66) | 49(74) | 62 (94) | 4 (6) | 2 (3)d | 6 (9) |
NOTE. PVL, Panton-Valentine leukocidin.
First 2 events per patient.
Includes indistinguishable and similar strains. Strains differ by ≤3 bands by PFGE.
No. of sets with any strain meeting this criterion.
One set was USA300, and one was USA400.
MRSA isolates from 2 discrete MRSA bloodstream events in a single patient.
MRSA isolate from a nares culture and a subsequent MRSA bloodstream event in a single patient.
Table 5.
Similarity of methicillin-resistant Staphylococcus aureus strain types, by interval days and location.
| No. (%) of patients | |||
|---|---|---|---|
| Characteristic | Indistinguishable strain type |
Similar strain typea |
Different strain type |
| Infection-infection setsb | |||
| Interval time, months | |||
| <2 (n = 14) | 11 (79) | 13 (93) | 1 (7) |
| 2–5 (n = 17) | 13 (76) | 15 (88) | 2 (12) |
| ≥6 (n = 6) | 4 (67) | 5 (83) | 1 (17) |
| Interval location | |||
| Not discharged (n = 13) | 11 (85)c | 13 (100)c | 0 (0)c |
| Home (only) (n = 14) | 14 (100)c | 14 (100)c | 0 (0)c |
| Skilled nursing facility or rehabilitation center (n = 10) | 3 (30) | 6 (60) | 4 (40) |
| Colonization-infection setsd | |||
| Interval time, months | |||
| <2 (n = 20) | 13 (65) | 20 (100) | 0 (0) |
| 2–5 (n = 7) | 6 (86) | 7 (100) | 0 (0) |
| ≥6 (n = 2) | 2 (100) | 2 (100) | 0 (0) |
| Interval location | |||
| Not discharged (n = 15) | 12 (80) | 15 (100) | 0 (0) |
| Home (only) (n = 7) | 5 (71) | 7 (100) | 0 (0) |
| Skilled nursing facility or rehabilitation center (n = 7) | 4 (57) | 7 (100) | 0 (0) |
Includes indistinguishable and similar strains. Strains differ by ≤3 bands by PFGE.
First 2 events per patient.
P < .05 when comparing proportions with those whose interval location included a skilled nursing facility or rehabilitation center. Since similar and different PFGE types compose the whole set and are mutually exclusive, the tests of proportions occurring in these groups are identical.
Infection reflects first invasive event after detection of colonization.
Six patients had >2 MRSA infections. These infections involved 20 bacteremic events spanning 30–302 days (median, 182.5 days). Five of 6 patients’ infections were repeatedly caused by indistinguishable strain types.
USA100 was the most common strain type in either group (54 of 66 sets), followed by USA500 (3 sets) and USA200 (2 sets). One patient each had repeated infection with USA300 and USA400, accounting for the 2 PVL positive, SCCmec IV isolates.
The percentage of initial invasive MRSA isolates that were susceptible to the following antibiotics is as follows: moxifloxacin, 6% of isolates; erythromycin, 6%; clindamycin, 33%; trimethoprim- sulfamethoxazole, 92%; tetracycline, 95%; gentamicin, 92%; rifampin, 100%; vancomycin, 100%; linezolid, 100%; quinupristin-dalfopristin, 100%; and mupirocin, 79%. Nine strains had inducible clindamycin resistance. Among the 26 repeated infections caused by identical strains, 4 strains (15%) showed increased antibiotic resistance (moxifloxacin, intermediate to resistant [1 strain]; gentamicin, susceptible to resistant [1 strain]; rifampin, susceptible to intermediate [1 strain]; and clindamycin, susceptible to resistant [1 strain]).
Discussion
With a 35% risk of infection in the year following detection [2, 3], there is a need to prevent disease among MRSA carriers. We show that most MRSA infections following initial colonization or infection are caused by identical strains. This likely represents ongoing risks of infection from persistently carried strains, because diverse strain types at participating institutions made repeated acquisition of an identical strain unlikely from any source other than a patient’s immediate surroundings. It also supports lack of strain-specific immunity. This finding suggests that single successful decolonization of patients and possibly their immediate surroundings may obviate the majority of later MRSA infections.
Patients who remained hospitalized and those discharged to home were significantly more likely to experience repeated infections due to identical strains, compared with patients who had intervening SNF/rehab stays. Given the low frequency (<1%) with which healthy people carry MRSA [27], this was not unexpected for MRSA carriers who returned home, where there was unlikely to be a preexisting reservoir of diverse strains among family members. This is in contrast to those with an intervening stay at an SNF/rehab, where MRSA prevalence is often high and where social networking is common and encouraged. Among hospitalized patients, contact precautions and the lack of patient interaction may explain the tendency for a single strain to cause repeated infections despite evidence that hospital strains are diverse. Thus, decolonization of patients in their home or hospital room may be more successful at preventing infection than similar attempts at an SNF/rehab, where repeated screening and decolonization may be needed to ensure lack of carriage.
In 24% of subsequent infections, strains were not identical, although most differed by <3 bands. These strains may represent new environmental acquisition. Another possibility is that the duration between infections was sufficiently long for minor genetic changes to produce highly related, but non-identical, strains.
We found that a patient’s location between infections was more predictive of strain relatedness than was time between infections. Nearly all isolates recovered from persons with repeated infection were of similar strain types, even when infections were separated by >6 months.
MRSA strains causing repeated infections generally retained the same susceptibility profile. Of the 15% of strains with a more resistant antibiotic profile at the time of the second infection, only 2 had a substantial change in their MIC, and 1 was confirmed to be related to inducible macrolide-lincosamide-streptogramin resistance.
There are several limitations to this study. First, the sample size is small, despite multiple participating centers. Second, all institutions were academic centers, and results may not be generalizable to other settings. In addition, these results are largely based on health care-associated MRSA strains. Further data are needed to assess the relatedness of strain types in recurrent community-associated MRSA infections. Third, because we did not perform environmental cultures, we do not know the contribution of a patient’s immediate surroundings in maintaining carriage with the same strain. Nevertheless, we recognize that several studies have shown that MRSA easily contaminates the environment [28–30].
Repeated MRSA infection and MRSA infection occurring after colonization are not uncommon. We show that the majority of these infections are due to strains identical to the initial strain, even when the events occur many months apart. This evidence of persistent carriage suggests that time-limited decolonization regimens for patients, possibly coupled with disinfection of their surroundings, may substantially reduce the risk of subsequent infection.
Acknowledgments
Financial support. The CDC Prevention Epicenters Program and the National Institutes of Health (K23AI64161-01).
Potential conflicts of interest. D.K.W. receives research support from Sage Products, and has served as a consultant to Enturia and 3M Healthcare. R.P. has received research grants from Sanofi-Aventis, GlaxoSmithKline, Pfizer, and TAP Pharmaceuticals in the past 2 years. All other authors: no conflicts.
References
- 1.Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang SS, Platt R. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin Infect Dis. 2003;36:281–285. doi: 10.1086/345955. [DOI] [PubMed] [Google Scholar]
- 3.Huang SS, Hinrichsen VL, Stulgis L, et al. Methicillin-resistant Staphylococcus aureus infection in the year following detection of carriage [abstract 157]. Programs and abstracts of the 16th Society of Healthcare Epidemiology of America Annual Meeting (Chicago); Arlington, VA. Society of Healthcare Epidemiology of America.2006. [Google Scholar]
- 4.Loeb M, Main C, Walker-Dilks C, Eady A. Antimicrobial drugs for treating methicillin-resistant Staphylococcus aureus colonization. Cochrane Database Syst Rev. 2003;4 doi: 10.1002/14651858.CD003340. CD003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simor AE, Phillips E, McGeer A, et al. Randomized controlled trial of chlorhexidine gluconate for washing, intranasal mupirocin, and rifampin and doxycycline versus no treatment for the eradication of methicillin-resistant Staphylococcus aureus colonization. Clin Infect Dis. 2007;44:178–185. doi: 10.1086/510392. [DOI] [PubMed] [Google Scholar]
- 6.Parras F, Guerrero C, Bouza E, et al. Comparative study of mupirocin and oral co-trimoxazole plus topical fusidic acid in eradication of nasal carriage of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:175–179. doi: 10.1128/aac.39.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura A, Mochizuko T, Nishizawa K, Mashiko K, Yamamoto Y, Otsuka T. Trimethoprim-sulfamthoxazole for the prevention of methi-cillin-resistant Staphylococcus aureus pneumonia in severely burned patients. J Trauma. 1998;45:383–387. doi: 10.1097/00005373-199808000-00032. [DOI] [PubMed] [Google Scholar]
- 8.Roccaforte JS, Bittner MJ, Stumpf CA, Preheim LC. Attempts to eradicate methicillin-resistant Staphylococcus aureus colonization with the use of trimethoprim-sulfamethoxazole, rifampin, and bacitracin. Am J Infect Control. 1988;16:141–146. doi: 10.1016/0196-6553(88)90024-7. [DOI] [PubMed] [Google Scholar]
- 9.Perl TM, Cullen JJ, Wenzel RP, et al. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N Engl J Med. 2002;346:1871–1877. doi: 10.1056/NEJMoa003069. [DOI] [PubMed] [Google Scholar]
- 10.Kauffman CA, Terpenning MS, He X, et al. Attempts to eradicate methicillin-resistant Staphylococcus aureus from a long-term-care facility with the use of mupirocin ointment. Am J Med. 1993;94:371–378. doi: 10.1016/0002-9343(93)90147-h. [DOI] [PubMed] [Google Scholar]
- 11.Harbarth S, Dharan S, Liassine N, Herrault P, Auckenthaler R, Pittet D. Randomized, placebo-controlled, double-blind trial to evaluate the efficacy of mupirocin for eradicating carriage of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1412–1416. doi: 10.1128/aac.43.6.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisman LE, Thackray HM, Gracia-Prats JA, et al. Phase I/II double blind, placebo controlled, dose escalation, safety and pharmacokinetics study in very low birth weight neoneates of BSYX-A110, an antistaphylococcal monoclonal antibody for the prevention of staphylococcal bloodstream infections [abstract LB14]. Programs and abstracts of the Pediatric Academic Societies Annual Meeting (San Francisco); Woodlands, TX. Pediatric Academic Societies.2004. [Google Scholar]
- 13.Shinefield H, Black S, Fattom A, et al. Use of a Staphylococcus aureus conjugate vaccine in patients receiving hemodialysis. N Engl J Med. 2002;346:491–496. doi: 10.1056/NEJMoa011297. [DOI] [PubMed] [Google Scholar]
- 14.Fattom A, Fuller S, Propst M, et al. Safety and immunogenicity of a booster dose of Staphylococcus aureus types 5 and 8 capsular polysaccharide conjugate vaccine (StaphVAX) in hemodialysis patients. Vaccine. 2004;23:656–663. doi: 10.1016/j.vaccine.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 15.Herwaldt LA, Pottinger JM, Coffman S, Tjaden J. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in a Veterans Administration medical center. Infect Control Hosp Epidemiol. 2002;23:502–505. doi: 10.1086/502096. [DOI] [PubMed] [Google Scholar]
- 16.Maslow JN, Brecher S, Gunn J, Durbin A, Barlow MA, Arbeit RD. Variation and persistence of methicillin-resistant Staphylococcus aureus strains among individual patients over extended periods of time. Eur J Clin Microbiol Infect Dis. 1995;14:282–290. doi: 10.1007/BF02116520. [DOI] [PubMed] [Google Scholar]
- 17.Lim MSC, Marshall CL, Spelman D. Carriage of multiple subtypes of methicillin-resistant Staphylococcus aureus by intensive care unit patients. Infect Control Hosp Epidemiol. 2006;27:1063–1067. doi: 10.1086/507959. [DOI] [PubMed] [Google Scholar]
- 18.Von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. New Engl J Med. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 19.Horan TC, Gaynes RP. Surveillance of nosocomial infections. In: Mayhall CG, editor. Hospital epidemiology and infection control. 3rd ed. Philadelphia: Lippincott Williams & Wilkins; 2004. pp. 1659–1702. [Google Scholar]
- 20.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards [NCCLS]) Approved standard M7-A5. 5th ed. Wayne, PA: NCCLS; 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. CLSI document M100-S16. Wayne, PA: CLSI; 2004. Performance standards for antimicrobial susceptibility testing: 16th informational supplement. [Google Scholar]
- 23.Palepou MF, Johnson AP, Cookson BD, et al. Evaluation of disc diffusion and Etest for determining the susceptibility of Staphylococcus aureus to mupirocin. J Antimicrob Chemother. 1998;42:577–583. doi: 10.1093/jac/42.5.577. [DOI] [PubMed] [Google Scholar]
- 24.Diekema DJ, Pfaller MA, Turnidge J, et al. Genetic relatedness of multidrug- resistant, methicillin (oxacillin)-resistant Staphylococcus aureus bloodstream isolates from SENTRY antimicrobial resistance surveillance centers worldwide, 1998. Microb Drug Resist. 2000;6:213–221. doi: 10.1089/mdr.2000.6.213. [DOI] [PubMed] [Google Scholar]
- 25.Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists: molecular typing working group of the society for healthcare epidemiology of america. Infect Control Hosp Epidemiol. 1997;18:426–439. doi: 10.1086/647644. [DOI] [PubMed] [Google Scholar]
- 26.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuehnert MJ, Kruszon-Moran D, Hill HA, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States 2001–2002. J Infect Dis. 2006;193:172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 28.Boyce JM, Potter-Bynoe G, Chenevert C, King T. Environmental contamination due to methicillin-resistant Staphylococcus aureus: possible infection control implications. Infect Control Hosp Epidemiol. 1997;18:622–627. [PubMed] [Google Scholar]
- 29.Sexton T, Clarke P, O’Neill E, et al. Environmental reservoirs of methicillin-resistant Staphylococcus aureus in isolation rooms: correlation with patient isolates and implications for hospital hygiene. J Hosp Infect. 2006;62:187–194. doi: 10.1016/j.jhin.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 30.French GL, Otter JA, Shannon KP, et al. Tackling contamination of the hospital environment by methicillin-resistant Staphylococcus aureus (MRSA): a comparison between conventional terminal cleaning and hydrogen peroxide vapour decontamination. J Hosp Infect. 2004;57:31–37. doi: 10.1016/j.jhin.2004.03.006. [DOI] [PubMed] [Google Scholar]