Abstract
In an effort to extend the peptide aptamer approach, we have developed a scaffold protein that allows small molecule ligand control over the presentation of a peptide aptamer. This scaffold, a fusion of three protein domains, FKBP12, FRB, and GST, presents a peptide linker region for target protein binding only in the absence of the small molecule Rapamycin or other non-immunosuppressive Rapamycin derivatives. Here we describe the characterization of ligand-regulated peptides aptamers that interact with and inhibit the 5′-AMP-Activated Protein Kinase (AMPK). AMPK, a central regulator of cellular energy homeostasis, responds to high cellular AMP/ATP ratios by promoting energy producing pathways and inhibiting energy consuming biosynthetic pathways. We have characterized 15 LiRPs of similar, poly-basic sequence and have determined that they interact with the substrate peptide binding region of both AMPK α1 and α2. These proteins, some of which serve as poor substrates of AMPK, inhibit the kinase as pseudosubstrates in a Rapamycin regulated fashion in vitro, an effect that is largely competitive with substrate peptide and mediated by an increase in the kinase's apparent Km for substrate peptide. This pseudosubstrate inhibition of AMPK by LiRP proteins reduced the AMP stimulation of AMPK in vitro and caused the inhibited state of the kinase to kinetically resemble the basal, unstimulated state of AMPK, providing potential insight into the molecular mechanisms of AMP stimulation of AMPK.
Keywords: AMPK, peptide aptamer, Ligand-Regulated peptide aptamers, pseudosubstrate, substrate inhibition
Introduction
The genomic era has provided much to the biological sciences, not least of which has been the identification of the coding sequence for the ∼25,000 known and hypothetical proteins that make up the human proteome1. Bridging the divide between the primary sequence of these proteins and their cellular function now stands as one of the primary tasks in the biological sciences. Methods to do this are varied, and range from traditional knockout genetics2 or the use of RNA interference3 technologies to the use of small molecules that block specific actions of a target protein4. Each particular method has its own set of advantages and disadvantages. Genetic removal techniques can offer high or absolute specificity for a target protein and can be used to remove a protein in a spatially controlled fashion using tissue specific promoter technologies, but these methods can often be plagued by compensation, complicating identification of protein function. Chemical agents can provide exquisite temporal control of protein function and allow for the inhibition of individual domains of multidomain proteins, yet these molecules often interact with evolutionarily conserved binding sites and have questionable specificity for target proteins. Recently, techniques that combine chemical and genetic approaches have been developed that can conceptually combine the advantages of small molecule inhibitors (temporal control and inhibition of single domains) and genetic agents (high specificity, tissue specific expression, and subcellular localization), creating hybrid chemical-genetic agents that expand the biologist's tool kit and allow for more controlled techniques to inhibit protein function, such as chemical inducers of dimerization5; 6; 7; 8; 9; 10, engineered receptor-ligand pairs11; 12; 13; 14; 15, and ligand-regulated macromolecules16; 17; 18; 19; 20; 21.
With protein-protein interaction networks continuing to emerge as a fundamental organizing characteristic of the cell22; 23; 24, it is no surprise that the protein-protein interactions that a given protein makes contribute greatly to the overall function of that protein. One promising approach for the rapid generation of agents for protein-protein interaction disruption is the use of selected peptide aptamers25. In this method, combinatorial libraries of peptide sequences, typically displayed from a surface loop of a scaffold protein, are used to select individual members that interact with a specific target molecule, often in the yeast two-hybrid system. This method has been employed in the selection of aptamers targeting multiple proteins (Cdk225; 26, E2F27, Ras28, the HPV protein E629, and others), displayed from various scaffold proteins (E.coli TrxA25, GFP 30, Staphylococcus nuclease31, SteA32, and others33), and selected or screened in multiple high-throughput systems (yeast two-hybrid 25 and retroviral delivery to mammalian cells 34).
We have worked on extending the peptide aptamer approach by developing a scaffold that will control the presentation of the randomized peptide aptamer region with a cell permeable, small molecule ligand16, merging the benefits of small molecules (inhibition of single domains, exquisite temporal control, disability) with the powers of the genetically-encoded, selection based peptide aptamer approach (tissue specific expression, rapid generation of protein targeting agents). Towards this goal, we have engineered a novel Ligand-Regulated Peptide (LiRP) scaffold protein16 (figure 1), based on the trimeric complex of FKBP-12, Rapamycin, and the FRB domain of mTOR35. By fusing these small protein domains with a randomized peptide linker, the addition of Rapamycin imparts a conformational constraint on the peptide aptamer. The scaffold is completed with the presence of a steric occlusion domain, the protein GST, which is fused to the c-terminus of FRB and blocks access to the randomized linker region when it is constrained in the Rapamycin-bound state. Using the LiRP scaffold we were able to impart ligand regulation (using both Rapamycin and it non-immunosuppressive analogue AP23102) on the interaction of two small peptides with their cellular protein partners, the c-Src SH3 domain and the Retinoblastoma protein. In addition, we demonstrated the utility of our LiRP scaffold by selecting ligand-regulated peptide aptamers from a combinatorial library that bound conditionally to the Retinoblastoma protein, as well as the 5′AMP-Activated Protein Kinase (AMPK) α1 subunit16.
Figure 1. LiRP Protein Scaffold and Displayed Sequences Selected for AMPK α1 Binding.
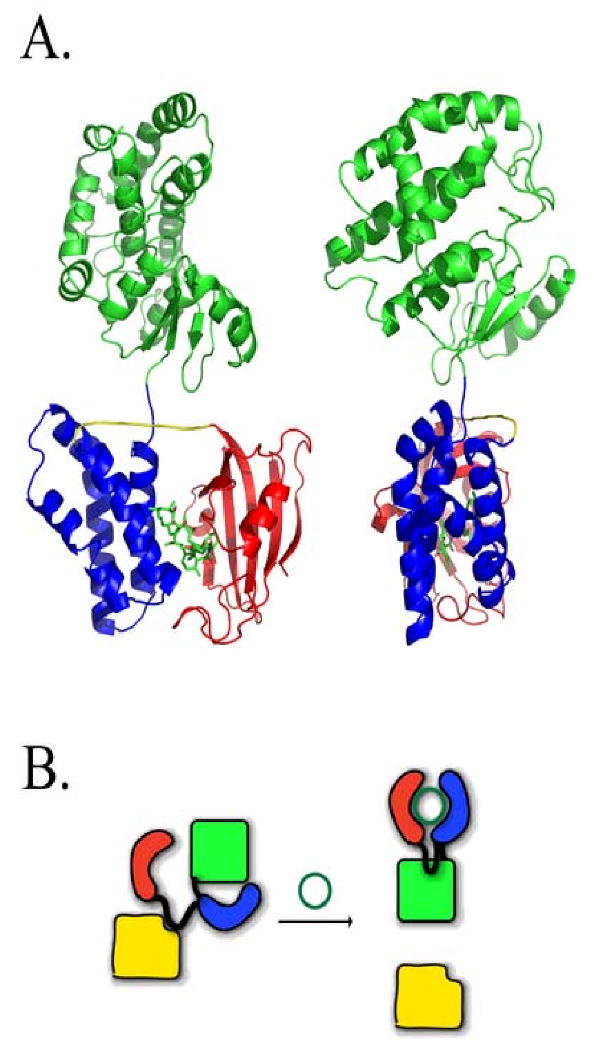
A. A model, displayed in two views differing by rotation on the vertical axis by 90°, exhibiting the steric occlusion of the randomizable peptide region to target proteins of the LiRP protein scaffold, created in SYBYL (trypos corporation) and rendered in PyMol (Delano Scientific). In the model, FKBP is colored red, FRB is colored Blue, GST is colored green, and Rapamycin, shown in stick form, is colored by atom type.
The 5′AMP-Activated Protein Kinase (AMPK) is a heterotrimeric protein kinase that is stimulated by increased AMP concentrations as well as other organismal signals such as the adipocyte hormones adiponectin and leptin36. AMPK responds to these activating stimuli by phosphorylating target proteins, inactivating key biosynthetic enzymes in energy-consuming, anabolic pathways (Acetyl-CoA Carboxylase37, HMG-CoA Reductase38, Glycogen Synthase39, mTOR40, enzymes in fatty acid, cholesterol, glycogen, and protein synthesis, respectively) or promoting energy-producing, catabolic cellular pathways (glycolysis41 and glucose transport42). Active AMPK exists as a stable heterotrimeric protein complex, consisting of an α catalytic subunit that contains a protein kinase domain, a regulatory γ subunit that binds AMP43 and is thought to mediate its stimulatory effects, and a β scaffolding subunit which interacts with both the α and γ subunits44. AMP stimulates AMPK in two ways, by direct allosteric activation of the kinase activity and by causing an increase in the steady state phosphorylation level of AMPK's activation loop. In the presence of AMP, AMPK exhibits a 2-6 fold increase in in vitro kinase activity, mediated largely by an increase in the maximal velocity without a major decrease in the Km of the kinase for its peptide substrate38; 45. While the exact molecular mechanism by which AMP stimulates AMPK is unknown, it is thought that either binding of AMP to the γ subunit either confers a positive, stimulatory signal to the α subunit43; 46, or it relieves an inhibitory communication between γ and α subunits, possibly mediated by γ-bound ATP47. The uncertainty of the precise molecular mechanisms for AMPK stimulation, as well as the general utility that modulators of AMPK may have in aiding the study of its cellular function, led us to target the AMPK α subunit with the peptide aptamer approach.
Previously we described the selection of 15 LiRPs from a peptide aptamer library that interacted with AMPK α1 in a Rapamycin dependent fashion16. Upon identification of these LiRPs, it became clear that they shared a large amount of sequence similarity (table 1). All of the sequences were poly-basic in the first 4 of the 7 amino acid randomized peptide aptamer linker, and many had hydrophobic residues in the c-terminal portion of the aptamer region. In the present study we have characterized in more detail the interaction of these LiRPs with AMPK, and demonstrate that they inhibit AMPK's kinase activity in a rapamycin-regulated fashion. We provide multiple pieces of evidence to suggest that these LiRP proteins interact with the AMPK substrate binding pocket and inhibit the kinase as pseudosubstrates. In addition to simple inhibition of AMPK, the LiRP proteins decrease the AMP stimulation of the kinase. The reduction of AMP stimulation, as well as the similarity between the basal- and LiRP-inhibited states of the kinase, suggests the possibility that the AMP stimulation of the kinase may be due to relief of a pseudosubstrate inhibited heterotrimeric complex.
Table 1. 15 LiRPs Identified as Interactors With AMPK α1.
Primary sequence of 15 Ligand-Regulated Peptides identified from a selection experiment targeting AMPK α1.
| LiRP Protein | Aptamer Sequence |
|---|---|
| LiRP A1-1 | YRQRDKF |
| LiRP A1-2 | RRQRFMFGSCIGLTYS |
| LiRP A1-3 | RRMWPWS |
| LiRP A1-4 | RRMRPCR |
| LiRP A1-5 | PPQCRHL |
| LiRP A1-6 | RRQSHWS |
| LiRP A1-7 | KRMRTWK |
| LiRP A1-8 | KRQRWCY |
| LiRP A1-9 | KRQRDFT |
| LiRP A1-10 | KRQTLWW |
| LiRP A1-11 | RRCRERT |
| LiRP A1-12 | QRMRDRS |
| LiRP A1-13 | RHQSWWH |
| LiRP A1-14 | KRMRSSV |
| LiRP A1-15 | RMMRSRT |
Results
YTH Characterization of AMPK α1 Interacting LiRPs
After the initial selection of the 15 LiRPs that interacted with AMPK α1, we sought a more detailed description of their interaction with the target protein, beginning with an assessment of differences in the various LiRPs ability to interact with AMPK and ability of Rapamycin to disrupt that interaction. Relying on a qualitative assessment of strain growth to determine differences between the LiRPs, we found that there was indeed heterogeneity in the strength of interactions and that there was an inverse relationship between strain growth and the ease with which rapamycin disrupted the interaction, with strong interactors requiring higher concentrations of ligand to completely prevent strain growth. Specifically, one LiRP exhibited a more rapid growth rate (LiRP A1-10) and three LiRPs had a slower growth rate (LiRPs A1-9, A1-11, and A1-15) (figure 2, panels b and c). The ability of Rapamycin to disrupt the interactions was assessed at 3 Rapamycin concentrations, and showed that weakest interactors (LiRP A1-9, A1-11, and A1-15) were unable to interact with AMPK α1 at lower doses of Rapamycin than the stronger interactors (LiRP A1-2 and A1-10) (figure 2, panels d, e, and f). Importantly, these clear differences in the various LiRP-AMPK α1 interactions, as assessed by cell growth, were not caused by corresponding differences in LiRP expression level or cellular stability, as all LiRPs exhibited similar protein expression levels in western blots (data not shown).
Figure 2. Characterization of Growth Rate and Switching Phenotype of LiRP Proteins in the Yeast Two-Hybrid.
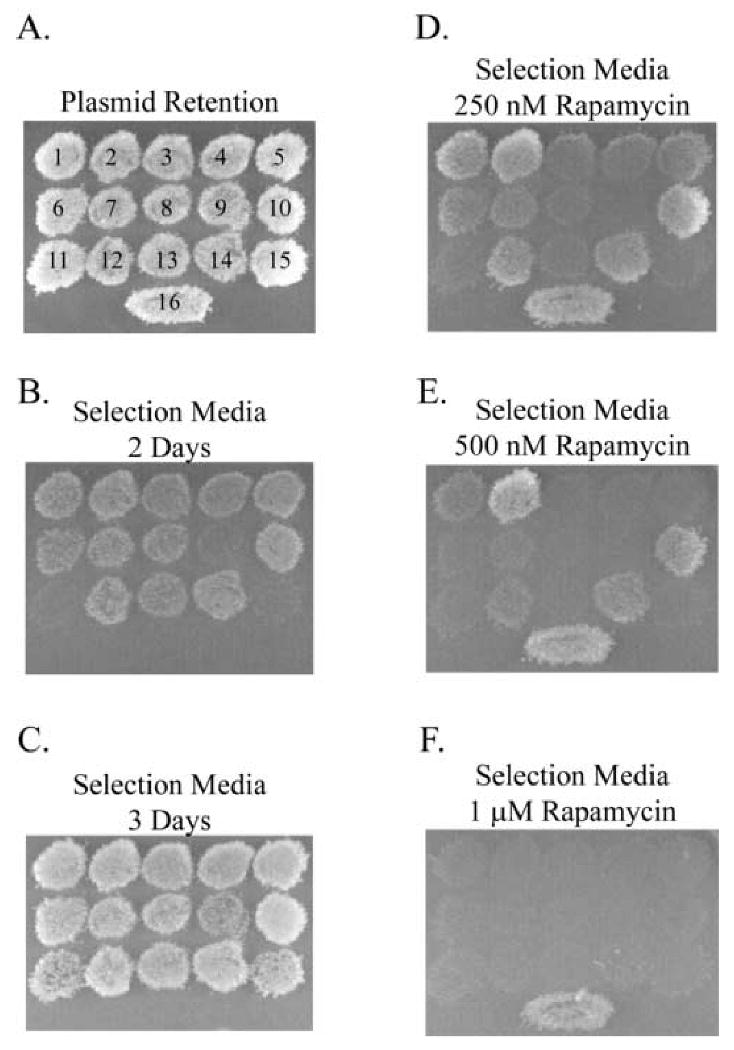
Shown is one representative experiment depicting the variation in LiRP-AMPK interaction strength and the ability of Rapamycin to disrupt the interactions. Various yeast strains were patched onto plasmid retention media and replica plated onto experimental plates with selection media. A. Plasmid retention plate showing growth of strains containing DB-AMPKα1 and all 15 LiRP proteins (labeled 1-15) and the control strain for Rapamycin containing DB-FKBP12 and AD-FRB (labeled 16). B. and C. Images depicting colony growth on selection media at 2 days growth (B.) and 3 days growth (C.). D.-F. Images of selection media plates after 3 days growth with either 250 nM Rapamycin (D.), 500 nM Rapamycin (E.), or 1 μM Rapamycin (F.).
To determine what regions of AMPK were involved in the LiRP interaction and whether the interaction was specific for the AMPK α1 isoform, we created a series of truncations of both the AMPK α1 isoform and the similar, but functionally distinct, AMPK α2 isoform (amino acids 1-312 and 313-548 for α1 and 1-312 and 313-552 for α2). We found that all 15 LiRPs interacted with the N-terminal 312 amino acids of AMPK α1 and α2 (table 1), a region of the AMPK α protein that contains the catalytic kinase domain.
Identification of Amino Acid Residues on AMPK α1 Required For LiRP Interaction
We set up a selection experiment to identify point mutations in the AMPK α subunit that disrupted the LiRP-AMPK interaction to more finely identify the area of AMPK α1 required for the interaction. To do this, we screened an AMPKα1 mutant library mutated by error prone PCR between amino acids 1-364. We identified 2 mutant AMPK clones that did not interact with LiRP A1-2, maintained an interaction with AMPK β1, and expressed a full length Gal4DB-AMPK α1 protein. In addition to these 2 full length clones, 2 clones that contained truncated DB-AMPK α1 proteins expected to be greater than the pDB-AMPKα1(1-312) bait protein sufficient for a LiRP-AMPK α1 interaction were collected and sequenced. These 4 clones contained in total 23 missense mutations from which we selected 10 to retest individually. Three of the ten mutations (E183K, E100A, and E143A) prevented the LiRP-A1-12–AMPK interaction (figure 3, panel a), while the remaining mutations had no effect on the AMPK-A1-12–LiRP interaction. While all three mutations disrupted the weaker LiRP-A1-12–AMPK α1 interaction, only the E100A mutation fully disrupted the LiRP-A1-2–AMPK α1 interaction. No tested mutations had any effect on the AMPKα1-AMPKβ1 interaction.
Figure 3. Identification of AMPK α1 Residues Required For LiRP-AMPK Interaction.
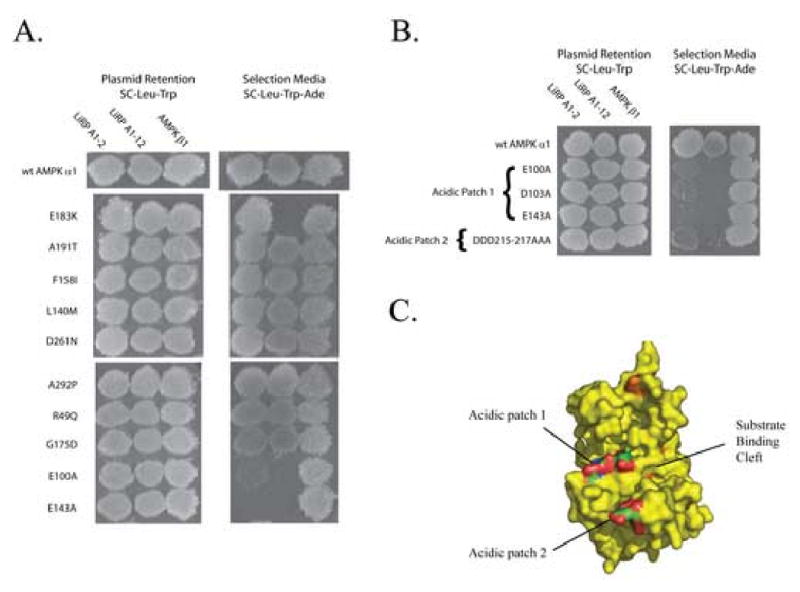
A. A yeast two-hybrid experiment showing the interaction of various individual DB-AMPK α1 point mutants identified in the AMPKα1mut selection and their interactions with LiRP A1-2, LiRP A1-12, or AMPK β1 activation domain fusions. B. Mutations of residues in two acidic patches in the AMPK substrate binding pocket prevent the interaction of LiRP A1-2 and LiRP A1-12 with AMPK, but do not effect the interaction with AMPK β1. C. Surface depiction of Snf1 crystal structure showing the location of two acidic patches required for AMPK-substrate and AMPK-LiRP interactions. Acidic patches are colored by atom type, mutations identified in the AMPKα1mut screen that do not modulate the LiRP A1-2–AMPK interaction from panel A. are colored orange.
When the interaction disrupting mutations were mapped to the Snf1 crystal structure (the yeast orthologue of AMPK α1)48, it became clear that two of the mutations were located in a highly acidic region of AMPKs substrate binding pocket, consisting of amino acids E100, D103, E143, D215, D216, and D217). We targeted the remaining acidic residues with alanine mutagenesis (D103A and the DDD215-217AAA triple mutant) and found that their mutation prevented or decreased the LiRP-AMPK interaction (figure 3, panel b and c), suggesting that these acidic residues participate in the LiRP interaction.
Inhibition of In Vitro AMPK Kinase Activity by Bacterially Expressed LiRPs
We used bacterially expressed and purified LiRPs in in vitro AMPK kinase assays to determine if the LiRPs modulated AMPK activity. For these in vitro tests we chose five LiRPs (A1-1, A1-2, A1-4, A1-11 and A1-12) with a spectrum of yeast two-hybrid interaction characteristics to express in E. coli and purify. In addition to these AMPK α1 interacting LiRPs, we also expressed and purified a LiRP selected for its ability to interact with the Retinoblastoma protein, LiRP Rb-1-3, to ensure that the LiRP scaffold alone did not modulate AMPK activity. We found that all 5 AMPK-targeted LiRPs tested inhibited AMPK activity, with A1-4 exhibiting the greatest inhibitory effect at the concentration tested (figure 4a). We also found that the inhibition caused by all AMPK-targeted LiRPs was completely relieved upon addition of Rapamycin. LiRP Rb-1-3, however, was unable to modulate AMPK activity, as was the presence of Rapamycin alone.
Figure 4. LiRP Proteins Inhibit AMPK and Reduce the AMP Stimulation of the Kinase.
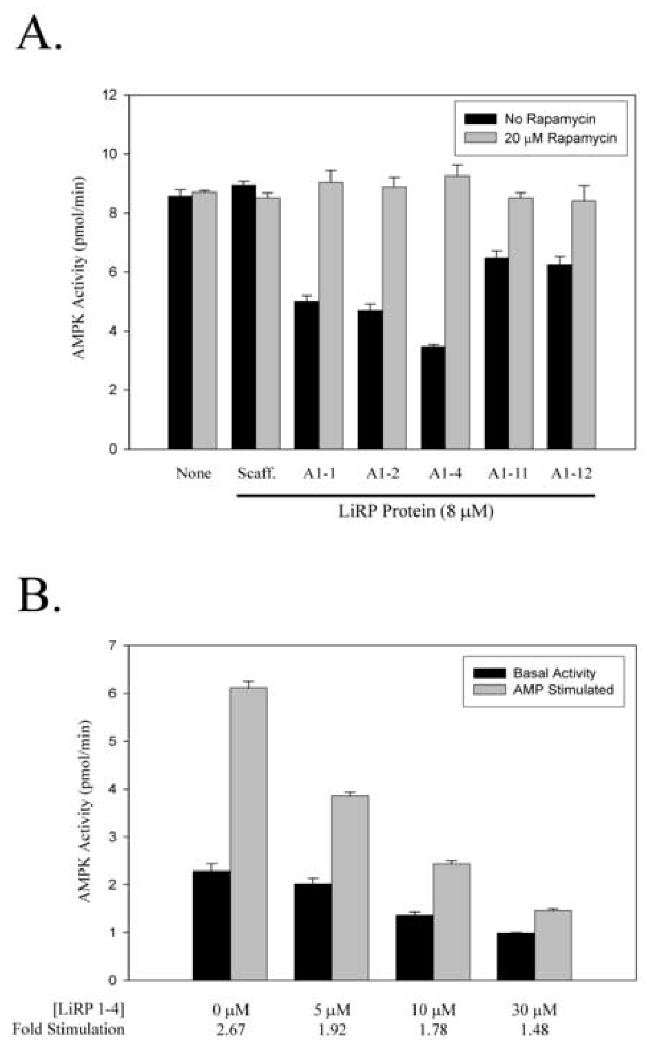
A. Graph showing the in vitro kinase activity of AMPK under varied conditions. Dark bars indicate the kinase activity in the absence of Rapamycin while light bars are the activity in the presence of 20 μM Rapamycin. Kinase activities were determined in the absence or presence of various LiRP proteins (A1-1, A1-2, A1-4, A1-11, A1-12), or the LiRP scaffold protein with a sequence not selected for binding to AMPK α1 (Scaff insert sequence is LYCYE). All LiRP protein concentrations are 8 μM, and 10 mU of purified AMPK (Upstate Biotech) was used (10 mU at 759U/mg protein in 25 μL reactions equals less than 3 nM AMPK heterotrimeric complex). B. AMPK kinase assays depicting the LiRP A1-4 inhibition of AMPK activity in the presence or absence of 200 μM AMP. Below the graph is the calculated AMP stimulation values (AMP stimulated/Basal) at the various concentrations of LiRP A1-4.
In the presence of AMP, purified rat liver AMPK exhibits a ∼3 fold increase in kinase activity in vitro (figure 4b). To determine if the LiRP proteins had any modulatory effect on this AMP stimulation we tested LiRP A1-4, the most potent of the 5 LiRPs tested, for inhibition of both basal and AMP stimulated AMPK. The presence of the LiRP A1-4 protein inhibited both the basal and AMP stimulated kinase activity of AMPK, inhibiting the AMP stimulated state to a greater extent (figure 4b). This effect led to a decrease in the AMP stimulation of the kinase, from a 2.7 fold increase in activity to a less than 1.5 fold increase in activity after AMP stimulation. This effect was also seen with LiRP A1-2 inhibition of AMPK (data not shown).
LiRP Inhibition of AMPK at Varied Substrate Concentrations
To examine the inhibition of AMPK by LiRP A1-4 further, we chose to study the kinetics of inhibition at various concentrations of AMPK substrate peptide. These studies were complicated by the fact that AMPK appears to be substrate inhibited by the SAMS peptide substrate at high concentrations (above 200 μM SAMS peptide), and this substrate inhibition was relieved by LiRP A1-4 (figure 5a). However, utilizing substrate concentrations that did not cause substrate inhibition, we found that LiRP A1-4 did indeed exhibit a dose dependent inhibition of AMPK, and that a substantial component of this inhibition was due to a significant increase in the apparent Km of the enzyme for substrate peptide (figure 6a), and that there appeared to be only a modest decrease in the Vmax of the enzyme in the presence of the largest concentrations of the LiRP protein. In addition, we were able to create a plot of the apparent Km values versus the LiRP A1-4 inhibitor concentrations to determine an approximate Ki value of 3.9 μM (figure 6b).
Figure 5. Kinase Activity of Basal, AMP Stimulated, and LiRP Inhibited AMPK.
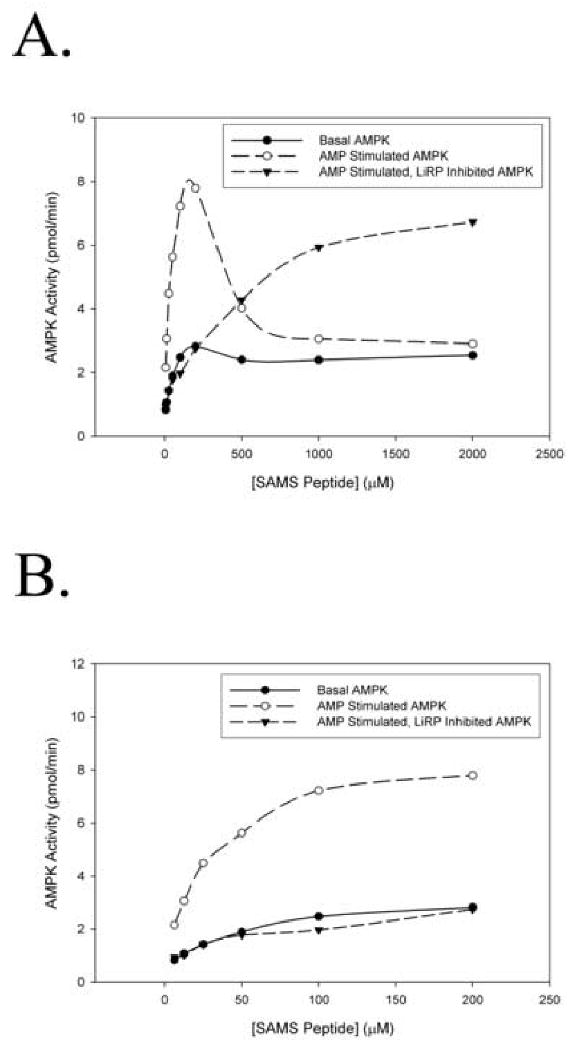
A. AMPK activity at various concentrations of SAMS peptide substrate, exhibiting the substrate inhibition at high concentrations of SAMS peptide, and relief of substrate inhibition in the presence of 30 μM LiRP A1-4. B. AMPK activity at various concentrations of SAMS peptide, only observing the data points below the SAMS peptide concentration that starts to show substrate inhibition of the basal state of AMPK, AMP stimulated AMPK, and AMP stimulated, LiRP A1-4 (30 μM) inhibited AMPK.
Figure 6. LiRP Inhibition of AMPK In Vitro Kinase Activity at Varied SAMS Peptide Substrate Concentrations.
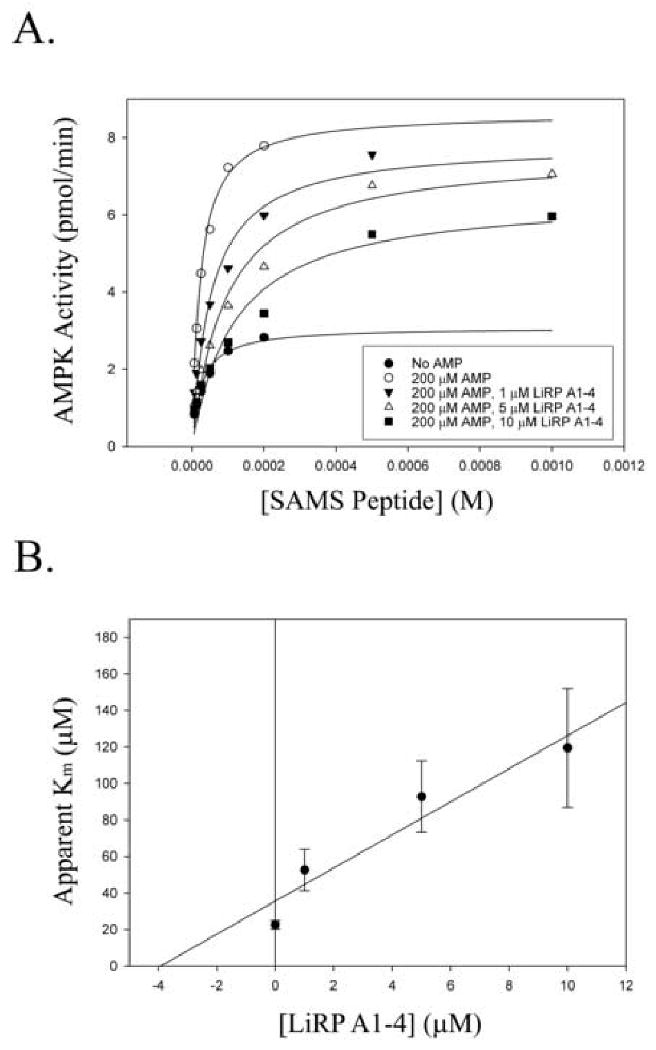
A. In vitro AMPK kinase assays (using 10 mU AMPK per data point) were performed at varied substrate peptide concentrations in the presence of 200 μM AMP and increasing concentrations of LiRP A1-4 inhibitor. Kinetic parameters were determined using non-linear regression analysis in SigmaPlot 9.0 by fitting data points to Michalis-Menten equation. Also included is uninhibited AMPK in the absence of AMP. Kinetic parameters (apparent Km:Vmax +/- standard errors) for the various states are: for uninhibited, basal AMPK (●) (25.4 +/- 4.9 μM : 3.08 +/- .18 pmol/min); AMP stimulated AMPK (○) (22.6 +/- 2.3 μM : 8.63 +/- .27 pmol/min); AMP stimulated AMPK with 1 μM LiRP A1-4 (▼) (52.7 +/- 11.3 μM : 7.86 +/- .53 pmol/min); AMPK with 5 μM LiRP A1-4 (Δ) (92.8 +/- 19.5 μM : 7.61 +/- .48 pmol/min); AMPK with 10 μM LiRP A1-4 (□) (119.5 +/- 32.5 μM : 6.51 +/- .56 pmol/min). B. Plot of Apparent Km values (+/- standard error), versus the LiRP A1-4 inhibitor concentrations, yielding the apparent Ki value of 3.9 μM.
Some LiRP Proteins serve as Rapamycin-Regulated, Partial Substrates of AMPK
After determining that the LiRP-AMPK interaction required key AMPK residues involved in the kinase-subsrate interaction and that the LiRPs could inhibit AMPK activity, we wanted to test the ability of a set of LiRP proteins to serve as AMPK substrates. We tested 5 LiRP proteins (A1-1, A1-2, A1-4, A1-11, and A1-12) for radiolabel incorporation in radioactive kinase assays and found that 3 served as partial substrates for AMPK, and did so in a Rapamycin-regulated fashion (A1-2, A1-11, and A1-12) (figure 7a). When quantified, the phosphorylation of these proteins was between 20 and 100 fold lower than the classical AMPK substrate, the SAMS peptide, clearly showing that some LiRPs could serve as substrates, albeit at a fraction of the efficiency of typical peptide substrates (figure 7b).
Figure 7. Some LiRP Proteins Can Be Phosphorylated By AMPK in a Rapamycin Regulated Fashion.
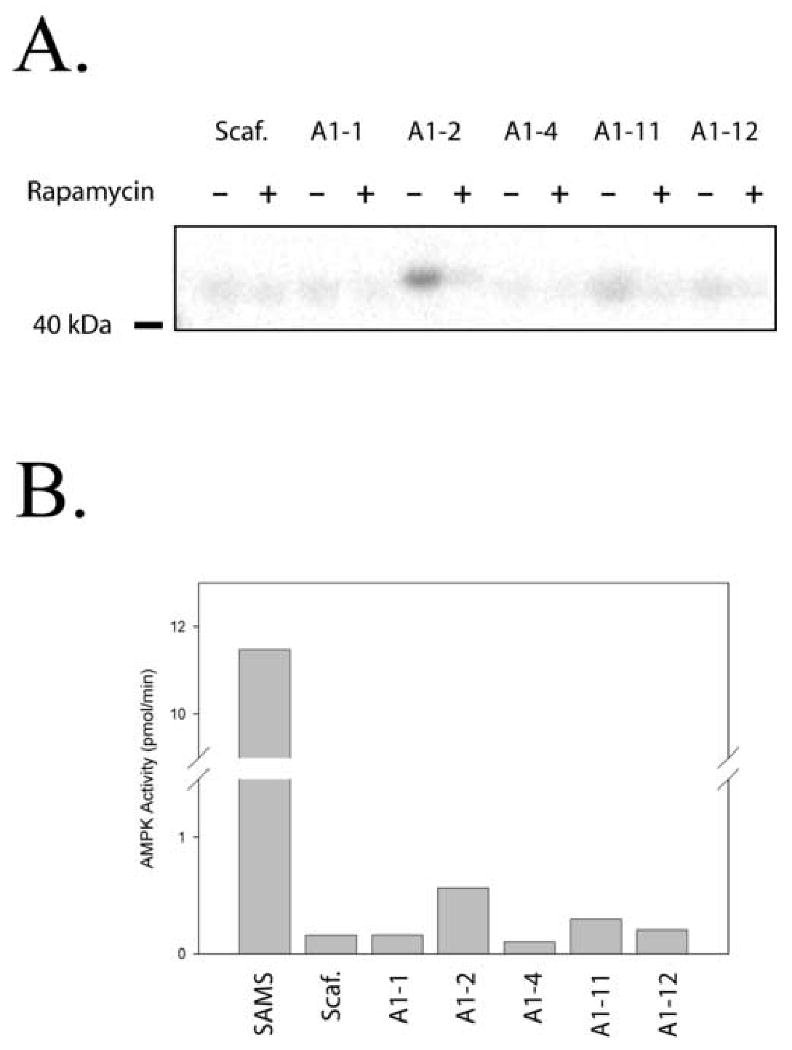
A. Phosphoimager detection of LiRP proteins, phosphorylated by AMPK, resolved on SDS-PAGE gels show a Rapamycin regulated phosphorylation of 3 of the 5 LiRP proteins tested (A1-2, A1-11, and A1-12). AMPK phosphorylation of the remaining LiRP proteins (A1-1 and A1-4) was not above the phosphorylation levels of the LiRP protein scaffold (Scaff. insert sequence is LYCYE). B. Quantification of AMPK phosphorylation of LiRP proteins and SAMS peptide reveals that LiRP proteins serve as poor AMPK substrates (between 20 and 200 fold poorer than SAMS peptide).
Discussion
In this work we report the characterization of 15 ligand-regulated peptide aptamers that inhibit AMPK as pseudosubstrates. In doing this, we have shown that our ligand-regulated peptide scaffold can display functional peptides in a conditional fashion in vitro. We have also demonstrated that, in the yeast two-hybrid system, there appears to be an inverse relationship between interaction strength and switchability, suggesting the LiRP scaffold exhibits mutually exclusive binding of either a target protein or Rapamycin, with stronger interactions requiring larger concentrations of Rapamycin to completely abolish LiRP-target binding. These data fit well with our proposed steric-occlusion model for the conditional display of peptides from our LiRP scaffold, whereby the binding of rapamycin to the scaffold conformationally constrains the three domain protein, blocking access to the randomized peptide with the GST domain. In subsequent studies it may be possible to exploit this effect, creating a gradient of binding ability where the Rapamycin concentration controls the amount of displayed, accessible peptide that is present. One negative aspect of this phenomenon is that peptide aptamers that bind their targets with high affinity may prove more difficult to conditionally display, requiring larger concentrations of Rapamycin or its non-immunosuppressive analogues to prevent aptamer-target interactions. However, the high affinity of rapamycin for FKBP and FRB49, an affinity that is likely to be even higher when FKBP and FRB are unimolecular, should compensate for high affinity LiRP-target interactions.
We have also demonstrated that 5 representative LiRPs, selected for their ability to interact with the AMPK α1 subunit, inhibit AMPK activity in vitro. Multiple pieces of evidence suggest that this inhibition occurs by competitively blocking access of the substrate peptide to the active site. First, we demonstrated that the LiRPs interact specifically with the N-terminal 312 amino acids of AMPK α1, a region containing the kinase domain, and that residues in the two acidic patch regions of the substrate binding pocket48, known to be involved in recognition of basic residues in the P-3 to P-6 regions of AMPK substrates50; 51; 52, are vital for the LiRP-AMPK interaction. In addition, we found that some LiRP proteins were rapamycin regulated substrates of AMPK. Finally, and perhaps most convincingly, we demonstrate that 5 LiRP proteins inhibit AMPK activity and that when the kinetics of LiRP A1-4 inhibition were studied it was evident that this inhibition is relieved at saturating concentrations of substrate peptide, a situation that strongly suggests competitive binding for the same site and a pseudosubstrate mode of inhibition.
It is interesting to note that in earlier work on the peptide aptamer approach26, Brent and coworkers selected aptamers that competitively inhibited the kinase Cdk2, showing that this was likely due to competition for the substrate binding region of the kinase. The similarity between their proposed mechanism of inhibition and ours is striking, and suggests that the application of the peptide aptamer approach could be successful in producing competitive inhibitors of substrate binding for more protein kinases. Broadly, this evidence also supports the hypothesis that the peptide aptamer approach is well suited for the identification and disruption of protein-protein interactions, and also suggests that peptide aptamers will often interact with a target protein at functional, protein-binding regions. Our LiRP scaffold, unlike the thioredoxin A scaffold, is expected to display extended peptides and does not provide the large entropic constraint that a surface loop insertion scaffold exhibits. Because of this, the LiRP scaffold has distinct disadvantages, of which one is the decreased interaction strength when compared to entropically constrained surface loop inserted peptide aptamers. However, the LiRP scaffold, because of its extended peptide display may be well suited for the identification and disruption of protein-peptide interactions, such as kinase-substrate or SH3-polyproline helix interactions, that require of the peptide component an extended, linear structure. These intermediate-strength, transient protein-peptide interactions are often some of the most interesting functional interactions a protein can make, and are especially prevalent in signal transduction pathways where these reversible interactions help mediate the coordinated responses of various inputs.
While all the evidence suggests that these LiRP proteins inhibit AMPK as pseudosubstrates, it is noteworthy that few are good matches with the traditional AMPK phosphorylation motif, Φ-(β,X)-X-X-S/T-X-X-X-Φ (Φ = hydrophobic residue and β = basic residues). Indeed, the most dominant feature contained in all of LiRPs is their poly-basic, and often poly-arginine, composition. When observing some of the known AMPK substrates it becomes clear that substrates are quite often poly basic in the P-3 to P-6 region (figure 8), a fact that has been clearly established in previous explorations of the AMPK-substrate interaction52. Some of the LiRPs, however, do have a well positioned phospho-accepting serine or threonine (LiRP A1-11 and A1-12) and were phosphorylated by AMPK in a Rapamycin regulated fashion. Indeed, all of the LiRP proteins have a glycine-serine linker immediately c-terminal to the aptamer region, a product of the cloning strategy for peptide aptamer library creation, and are therefore all potentially phosphorylatable. Interestingly, the three LiRPs that had the weakest interaction in the yeast two-hybrid (LiRPs A1-9, A1-11, and A1-14) were amongst those LiRPs whose sequence most closely resembled an AMPK substrate, raising the possibility that their weaker interactions were due to their ability to be partially phosphorylated, decreasing their ability to interact with the AMPK kinase domain. The LiRP protein that was phosphorylated most efficiently, LiRP A1-2, was also the only LiRP that had an extended insert sequence of 16 amino acids (an aberrant clone which resulted from two random inserts being cloned during library creation), suggesting that sequence c-terminal from the potential phosphoaccepting residue may cause the other LiRPs to be poor substrates. In addition, the fact that LiRP A1-2 served as the most effective AMPK substrate of the LiRPs tested suggests that the use of peptide aptamer libraries with longer aptamer sequences may be useful in the generation of higher affinity inhibitors, as the 7 amino acid, single-insert library members may be too small to take advantage of the full length of the AMPK substrate binding pocket, which can accommodate an ∼20 amino acid peptide substrate52.
Figure 8. AMPK Target Proteins Are Often Poly-basic in the P-3 to P-6 Region.
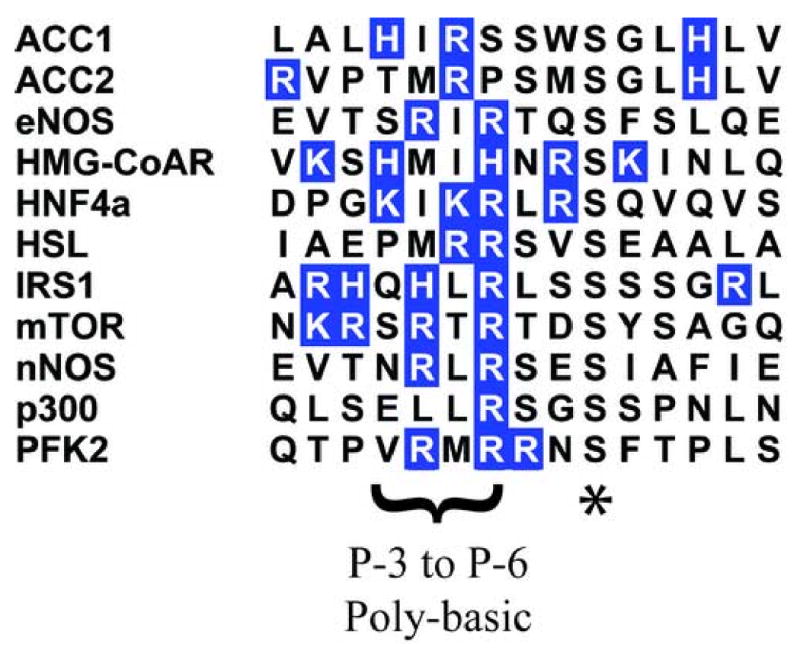
Pileup of some AMPK target proteins and displaying basic residues in blue exhibits the prevalence of poly-basic stretches in the P-3 to P-6 region of AMPK substrates (previously reported and described by Hardie and coworkers50; 52; 59) and shows the resemblance to the sequences found in LiRP proteins. Proteins included were collected from PhosphoELM database (phspho.elm.eu.org) for AMPK substrates.
For kinetic analysis and determination of the apparent Km and Vmax parameters of the various states of AMPK it was important to utilize data points in which the kinase was not substrate inhibited, a phenomenon described here and noted in earlier work53. It is unclear whether this is a general phenomenon or one unique to the SAMS peptide substrate. Some of the previously reported studies of AMPK activity at varied substrate concentrations were lacking kinetic analyses at high peptide substrate concentrations, possibly due to these same complicating factor38; 45; 53. In our system, it is clear that the ability to view the LiRP-inhibited, AMP stimulated state of the kinase at high substrate concentrations is vital for recognizing that these substrate concentrations relieve the LiRP inhibition of AMPK (figure 5a). If one were to only examine the kinetic analyses of LiRP inhibition at substrate peptide concentrations below 200 μM the inhibition could easily be mistaken for non-competitive inhibition that is not relieved by substrate peptide (figure 5b). It was interesting that while the LiRP proteins clearly inhibited AMPK, they also relieved the substrate inhibition caused by SAMS peptide. The mechanism for how these conflicting actions occur is unclear, but we speculate that one potential explanation for the observed substrate inhibition is an ordered two-substrate kinetic mechanism of AMPK activity (the effects of which are described theoretically and experimentally for Phosphofructokinase by Ferdinand54), with ATP binding first followed by the peptide substrate. If this is indeed the case then the LiRP proteins may relieve this substrate inhibition by interacting with only a portion of the substrate binding pocket, preventing the substrate peptide from binding out of order, yet still allowing ATP binding, thus relieving the inhibition observed when large concentrations of SAMS peptide prevent ATP binding.
Upon determining that the LiRPs inhibited AMPK activity we were immediately intrigued by two aspects of LiRP inhibited-AMPK that seemed to resemble the basal, unstimulated state of the kinase. First, it was clear that LiRP inhibition of AMPK caused a reduction in the AMP stimulation of AMPK, an effect that could not be relieved with saturating concentrations of AMP (figure 4b). Second, at peptide concentrations where substrate inhibition was not occurring, AMP-stimulated, LiRP A1-4–inhibited AMPK very closely resembled the basal state of uninhibited AMPK (figure 5b). These two apparent similarities between LiRP-inhibited, AMP stimulated AMPK and the basal state of the kinase suggest the possibility that the basal state of the kinase could be due to a pseudosubstrate inhibition as well, and that the pseudosubstrate LiRP inhibitors mimic this state of the kinase. This is only one potential explanation of the similarities between the LiRP-inhibited and basal states of the kinase, as the LiRP protein could also block a putative stimulatory interaction of the kinase in the AMP bound state, or the similarities could simply be coincidental. However, because a precise molecular explanation of the basal and AMP stimulated states of AMPK have not been definitively described, we feel a more detailed kinetic description of these two states of AMPK may provide insight into this enzymes mechanism of regulation. Previous studies, and indeed the work presented here, concluded that the basal state of the kinase is the result of a lower maximal activity with little change in Km when compared to AMP stimulated AMPK (effectively a non-competitively inhibited state). However, if high substrate peptide concentrations were used these conclusions may not hold and we may instead find that the AMPK basal state is the result of a pseudosubstrate inhibited form of the kinase. These are fundamental issues of the AMP stimulation of AMPK, and get at the nature of the intramolecular interactions that hold AMPK in a basal, inhibited state, and would therefore be of general utility in understanding the regulation of this kinase as well as aiding the rational design of therapeutic modulators of AMPK activity.
In this report we show the characterization of a class of peptide inhibitors of AMPK that can be displayed conditionally from our Ligand-Regulated Peptide scaffold. These inhibitors interact with the substrate binding region of AMPK and act as pseudosubstrate inhibitors of the kinase and behave as competitive inhibitors with respect to substrate peptide. The presence of Rapamycin prevents this interaction and inhibition, both in vitro and in the yeast two-hybrid system. We feel this represents a significant step towards the ultimate goal of using these agents for the spatial and temporal control of protein function in vivo. The aptamers selected here exhibit significant inhibition at concentrations ranging from 5 to 20 μM, and although it is difficult to use this information to estimate binding constants due to the competitive nature of their binding with SAMS peptide, we suspect that they interact with AMPK with a low μM Kd. In the presented in vitro kinase assays, LiRP proteins inhibited AMPK only when present in vast excess (in figure 4 LiRP protein is at 8 μM while the AMPK concentration is below 3 nM). While the inhibitory effect in vivo may be greater due to a much lower substrate concentration than the 200 μM SAMS peptide used in the above in vitro studies, it may still be necessary to identify new, higher affinity LiRPs targeting AMPK or mature the described LiRPs using random mutagenesis. In addition to the characterization of these inhibitory LiRPs, we also describe here the apparent similarity between the pseudosubstrate inhibited kinase and the basal state of AMPK, suggesting that more detailed kinetic studies of AMPK in its basal and AMP stimulated states may provide insight into the molecular mechanisms of AMP stimulation of AMPK.
Experimental Section
Plasmids and Reagents
All reagents, unless otherwise noted, were obtained from Sigma. The yeast two-hybrid strain MaV203 (Invitrogen) is commercially available, and strain PJ69-4A (tor2-1 Δfpr1) has been previously reported16. Lyticase enzyme was purchased from Promega. SAMS peptide was custom synthesized by UW-Biotech Peptide Synthesis Facility. PKA for control phosphorylation studies was purchased from NEB. pPrey-LiRP plasmids and pBait-AMPKα1 plasmids were described elsewhere. TEV protease expression plasmid, pTEV-his, was the generous gift of Prof. Brian Fox. For the MBP-fusion expression vector pET-MBPtev, MBP coding sequence was PCR amplified using primers 1 and 2 (primer 2 incorporated the tev protease cleavage site) from plasmid pAIDL1455. This PCR product was inserted as an NdeI/BamHI fragment into the pET-37b plasmid, creating the MBP-fusion-6his expression plasmid pET-MBPtev. The multiple MBP-LiRP expression vectors, designated pMBP-LiRP-x, were created by PCR amplifying LiRP coding sequence using primers 3 and 4. The resulting HindIII/SalI fragment was cloned into HindIII/XhoI cut pET-MBPtev. pDB-AMPKα2 was made by PCR amplifying α2 sequence from the plasmid pGEM-AMPKα2 (generous gift of Prof. Alan Attie) using primers 5 and 6 and cloning the BamHI/PstI fragment into similarly cut pGBKT7. Plasmids pBait-AMPKα1-1-312, pBait-AMPKα1-312-552, pBait-AMPKα2-1-312, and pBait-AMPKα2-312-554 were constructed by PCR amplifying the corresponding portion of AMPKα1 or AMPKα2 using primer pairs 7/8, 9/10, 11/12, and 13/14, respectively. The resulting fragments (NcoI/SalI, NdeI/XhoI, NcoI/EcoRI, NdeI/EcoRI respectively) were cloned into similarly cut pGBKT7 vector. Mutations that were observed in the AMPKa1mut screen were inserted individually into the pBait-AMPKα1 plasmid by quick change mutagenesis. The mutations E183K, A191T, F158I, L140M, D261N, A292P, R49Q, F175D, E100A, E143A, D103A, and DDD(215-217)AAA were installed with mutagenic primer pairs 15/16, 17/18, 19/20, 21/22, 23/24, 25/26, 27/28, 29/30, 31/32, 33/34, 35/36, and 37/38, respectively. Parental pBait-AMPKα1 DNA was removed with DpnI, and PCR products were transformed into bacteria. Individual clones were amplified and DNA was sequenced to confirm incorporation of the mutation.
YTH Characterization
Bait plasmids containing the TRP1 selection marker and prey plasmids containing the LEU2 selection marker were transformed into strain PJ69-4A (tor2-1 Δfpr1) and plated onto SC-Leu-Trp media. Single colonies containing both bait and prey plasmids were patched onto SC-Leu-Trp plates and replica plated onto SC-Leu-Trp-Adenine selection plates +/- various concentration of Rapamycin. Replica plates were allowed to incubate 2-3 days prior to image acquisition.
AMPKα1mut Library Synthesis
A randomly mutagenized AMPK α1 bait plasmid library, pLib-AMPKα1mut, was created by using error-prone pcr as described previously56. Briefly, AMPKα1 was amplified using primer pair 7/10 under the following mutagenic conditions (10× Polymerase Buffer, 2.5 mM MgCl2, 0.5 mM MnCl2, 100 μM dCTP, 100 μM dATP, 1 mM dGTP, 1 mM dTTP, 10 ng pDB-AMPKα1 template DNA, 100 pmol of each primer, and 20 U KOD-Hot Start Polymerase). The resulting mutated AMPKα1 PCR product was cut with NcoI and ApaI (ApaI cuts the wt AMPKα1 sequence at amino acid 374). This library of mutated fragments was cloned into similarly cut pDB-AMPKα1 vector and transformed into chemically competent DH5α E. coli. Transformed cells were allowed to recover for 1 hour in LB media, serially diluted and plated onto LB-Amp plates to obtain a measure of the library complexity. Pooled transformations were then incubated overnight in 500 mL LB media + 50 μg/ml Ampicilin followed by large scale DNA purification (Promega megaprep kit). Overall, 4.45 × 105 independent transformants were amplified to produce 0.7 mg of pLib-AMPKα1mut DNA. 9 single colonies were picked from titer plates for sequencing and analysis of mutation rate. From 8100 bp of random sequence read, 54 mutations were found, giving the library an error rate of 0.6% in the randomized region. Of the 54 nucleotide mutations, 37 accounted for missense amino acid substitutions and one was a frame shifting deletion.
AMPKα1mut Selection Experiment
Large-scale transformations of YTH strain containing pPrey-LiRP-A1-2 MaV-203 followed published protocols57. Serial dilutions of the original transformation were plated to determine the overall number of yeast transformants. The remaining amount of the yeast transformation was plated onto SC-Leu-Trp + 0.1% 5-FOA (negative selection media). After 4 days, colonies were picked from transformation plates, grown in patches on SC-Leu-Trp plates, and replica plated onto SC-Leu-Trp-His + 10 mM 3-AT, SC-Leu-Trp-Ura, or SC-Leu-Trp + 0.1% 5-FOA plates to confirm the selected phenotype. To isolate the plasmid DNA from positive strains, yeast were grown in 2 mL of liquid SC-Leu-Trp media overnight, yeast were pelleted and resuspended in lysis buffer (50 mM Tris-Cl, 50 mM EDTA, 10 mM β-mercaptoethanol, 1.2 M sorbitol, and 200 U lyticase) and incubated in a rotary shaker at 37° C overnight. Cells were fully lysed and the plasmid DNA purified using the Promega Miniprep kit. This DNA was then transformed into chemically competent DH5-α E. coli for amplification and then purifed with Promega Miniprep kits. Isolated DNA was sequenced and retransformed into AD-LiRP-A1-2 expressing PJ69-4A (tor2-1 Δfpr1) yeast to reconfirm phenotype. These yeast strains were also used in Western Blot analysis to look for full length DB-AMPKα1 via the c-Myc tag. Isolated clones were also transformed with pAD-AMPKβ1 to determine if they retained the AMPKα-AMPKβ interaction.
Protein Expression and Purification
For expression of Tev protease, the plasmid pTEV-his was transformed into BL21-DE3 cells, grown overnight to saturation, pitched into 1 liter of LB media, grown to an OD600 of 0.6, and induced with 0.2 mM IPTG at 16° for 36 hours. Bacteria were pelleted and resuspended in buffer A (50 mM Phosphate pH 7.4, 120 mM NaCl). Cells were lysed by sonication and the insoluble materials pelleted by centrifugation at 40,000 × g for 30 min. The soluble material was applied to a nickel-NTA column (equilibrated with buffer A). The column was washed with buffer B (60 mM Imidazole, 500 mM NaCl, 20 mM Tric-Cl pH 7.9) and TEV protease was eluted with buffer E (1 M Imidazole, 500 mM NaCl, 20 mM Tric-Cl pH 7.9). Protein was concentrated, exchanged into buffer A, supplemented with 1 mM DTT and 30% v/v glycerol, and stored at -20° at a final concentration of 1 mg/mL.
For expression of the LiRP proteins, pET-MBP-LiRP expression plasmids were transformed into BL21-DE3 RIL cells, single colonies were grown overnight, and saturated cultures were pitched into 1L of LB media at 37°. At OD600 of 0.6, cultures were chilled to 16°, and induced with 100 mM IPTG for 48 hours. Bacteria were peleted, resuspended in PBS + 1 mM PMSF, and lysed by sonication. The insoluble materials were pelleted by centrifugation at 40,000 × g for 30 min. The soluble material was applied to an amylase-agarose resin (NEB), washed with PBS, and eluted with 10 mM maltose. Eluted protein was cleaved with Tev protease at 4° C until completion, to liberate the LiRP protein from its MBP fusion. LiRP proteins were then bound to Glutathion Uniflow resin (clontech) and eluted with 10 mM Glutathione. LiRP protein was exchanged into pH 7.5 PBS, concentrated, supplemented with 1 mM DTT and 25% v/v glycerol, and stored at -20° C.
In vitro Kinase Assays
Kinase assays were performed as described elsewhere58. Briefly, 10 mU active rat liver AMPK (Upstate) was added to 25 μL assay buffer (200 μM ATP, +/- 200 μM AMP, 100 μM SAMS peptide (or varied [SAMS peptide] for kinetic experiments) and ATPγP32) containing various concentrations of LiRP protein. Reactions were incubated for 10 minutes at 30° C and stopped by spotting 15 μL of the reaction mixture on P81 phosphocellulose filter paper discs (Whatman) to specifically immobilize the SAMS peptide. Unincorporated radioactive ATP was then removed by washing in 1% phosphoric acid twice. Filter discs were then rinsed in water and acetone, allowed to dry, and radioactivity was counted using a scintillation counter. For kinetic analyses data points were fit to the Michalis-Menten equation, in SigmaPlot 9.0, to obtain kinetic parameters.
Protein Phosphorylation Assays
Active AMPK (20 mU) was incubated with various LiRP proteins, at 20 μM concentrations, in kinase assay buffer (200 μM ATP, ATPγP32, and 200 μM AMP in Hepes-Brij Buffer) for 30 minutes at 30° C. Reactions were stopped by adding 2× SDS-PAGE loading buffer. Proteins were run on 12% SDS-PAGE gels, and radioactivity was detected using a phosphor-imager cassette. For quantification of phosphate incorporation, exhaustive phosphorylation of a known PKA tag containing protein (GST-PKA-tag-FKBP12 expressed from pGEX-2TK-FKBP12) by active PKA (NEB) was loaded on the gel for comparison and quantification. The ImageJ program was used for quantification of phosphorylation.
Table 2. LiRP Interaction with AMPK α Truncations.
Results from YTH experiments showing selectivity of 15 LiRPs for the amino acids 1-312 of both AMPK α1 and α2.
| LiRP | Alpha 1 (1-548) |
Alpha 1 (1-312) |
Alpha 1 (313-548) |
Alpha 2 (1-552) |
Alpha 2 (1-312) |
Alpha 2 (313-552) |
|---|---|---|---|---|---|---|
| A1-1 | + | + | - | + | + | - |
| A1-2 | + | + | - | + | + | - |
| A1-3 | + | + | - | + | + | - |
| A1-4 | + | + | - | + | + | - |
| A1-5 | + | + | - | + | + | - |
| A1-6 | + | + | - | + | + | - |
| A1-7 | + | + | - | + | + | - |
| A1-8 | + | + | - | + | + | - |
| A1-9 | + | + | - | + | + | - |
| A1-10 | + | + | - | + | + | - |
| A1-11 | + | + | - | + | + | - |
| A1-12 | + | + | - | + | + | - |
| A1-13 | + | + | - | + | + | - |
| A1-14 | + | + | - | + | + | - |
| A1-15 | + | + | - | + | + | - |
Acknowledgments
We would like to thank A. Attie for AMPK related plasmids. We also thank the Research Corporation, W.M. Keck Foundation, NIH (GM065406), and Novartis (fellowship to B.F.B) for funding.
Abreviations
- AMP
5′-Adenosine monophosphate
- ATP
5′-Adenosine triphosphate
- AMPK
AMP-Activated protein kinase
- LiRP
Ligand-Regulated Peptide
- 5-FOA
5-fluoroorotic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu Rev Genet. 2002;36:153–73. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- 3.Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber SL. The small-molecule approach to biology. Chem Eng News. 2003;81:51–61. [Google Scholar]
- 5.Clackson T. Controlling mammalian gene expression with small molecules. Curr Opin Chem Biol. 1997;1:210–8. doi: 10.1016/s1367-5931(97)80012-9. [DOI] [PubMed] [Google Scholar]
- 6.Liberles SD, Diver ST, Austin DJ, Schreiber SL. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc Nat Acad Sci U S A. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mootz HD, Blum ES, Tyszkiewicz AB, Muir TW. Conditional protein splicing: A new tool to control protein structure and function in vitro and in vivo. Journal of the American Chemical Society. 2003;125:10561–10569. doi: 10.1021/ja0362813. [DOI] [PubMed] [Google Scholar]
- 8.Rivera VM, Wang XR, Wardwell S, Courage NL, Volchuk A, Keenan T, Holt DA, Gilman M, Orci L, Cerasoli F, Rothman JE, Clackson T. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science. 2000;287:826–830. doi: 10.1126/science.287.5454.826. [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–24. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 11.Belshaw PJ, Schoepfer JG, Liu KQ, Morrison KL, Schreiber SL. Rational Design of Orthogonal Receptor-Ligand Combinations. Angewandte Chemie-International Edition in English. 1995;34:2129–2132. [Google Scholar]
- 12.Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–72. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 13.Koh JT. Engineering selectivity and discrimination into ligand-receptor interfaces. Chem Biol. 2002;9:17–23. doi: 10.1016/s1074-5521(02)00087-x. [DOI] [PubMed] [Google Scholar]
- 14.Maly DJ, Allen JA, Shokat KM. A mechanism-based cross-linker for the identification of kinase-substrate pairs. J Am Chem Soc. 2004;126:9160–9161. doi: 10.1021/ja048659i. [DOI] [PubMed] [Google Scholar]
- 15.Shi YH, Koh JT. Selective regulation of gene expression by an orthogonal estrogen receptor-ligand pair created by polar-group exchange. Chemistry & Biology. 2001;8:501–510. doi: 10.1016/s1074-5521(01)00028-x. [DOI] [PubMed] [Google Scholar]
- 16.Binkowski BF, Miller RA, Belshaw PJ. Ligand-regulated peptides: a general approach for modulating protein-peptide interactions with small molecules. Chem Biol. 2005;12:847–55. doi: 10.1016/j.chembiol.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 17.Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem Biol. 2004;11:1157–63. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Buskirk AR, Liu DR. Creating small-molecule-dependent switches to modulate biological functions. Chem Biol. 2005;12:151–61. doi: 10.1016/j.chembiol.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 20.Vuyisich M, Beal PA. Controlling protein activity with ligand-regulated RNA aptamers. Chem Biol. 2002;9:907–13. doi: 10.1016/s1074-5521(02)00185-0. [DOI] [PubMed] [Google Scholar]
- 21.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 22.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M, Burgess S, McDaniel L, Stimpson E, Spriggs F, Williams J, Neurath K, Ioime N, Agee M, Voss E, Furtak K, Renzulli R, Aanensen N, Carrolla S, Bickelhaupt E, Lazovatsky Y, DaSilva A, Zhong J, Stanyon CA, Finley RL, Jr, White KP, Braverman M, Jarvie T, Gold S, Leach M, Knight J, Shimkets RA, McKenna MP, Chant J, Rothberg JM. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 25.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380:548–50. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 26.Cohen BA, Colas P, Brent R. An artificial cell-cycle inhibitor isolated from a combinatorial library. Proc Natl Acad Sci U S A. 1998;95:14272–7. doi: 10.1073/pnas.95.24.14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabbrizio E, Le Cam L, Polanowska J, Kaczorek M, Lamb N, Brent R, Sardet C. Inhibition of mammalian cell proliferation by genetically selected peptide aptamers that functionally antagonize E2F activity. Oncogene. 1999;18:4357–63. doi: 10.1038/sj.onc.1202825. [DOI] [PubMed] [Google Scholar]
- 28.Kurtz SE, Esposito K, Tang W, Menzel R. Inhibition of an activated Ras protein with genetically selected peptide aptamers. Biotechnol Bioeng. 2003;82:38–46. doi: 10.1002/bit.10538. [DOI] [PubMed] [Google Scholar]
- 29.Butz K, Denk C, Ullmann A, Scheffner M, Hoppe-Seyler F. Induction of apoptosis in human papillomaviruspositive cancer cells by peptide aptamers targeting the viral E6 oncoprotein. Proc Natl Acad Sci U S A. 2000;97:6693–7. doi: 10.1073/pnas.110538897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abedi MR, Caponigro G, Kamb A. Green fluorescent protein as a scaffold for intracellular presentation of peptides. Nucleic Acids Res. 1998;26:623–30. doi: 10.1093/nar/26.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norman TC, Smith DL, Sorger PK, Drees BL, O'Rourke SM, Hughes TR, Roberts CJ, Friend SH, Fields S, Murray AW. Genetic selection of peptide inhibitors of biological pathways. Science. 1999;285:591–5. doi: 10.1126/science.285.5427.591. [DOI] [PubMed] [Google Scholar]
- 32.Woodman R, Yeh JT, Laurenson S, Ko Ferrigno P. Design and validation of a neutral protein scaffold for the presentation of peptide aptamers. J Mol Biol. 2005;352:1118–33. doi: 10.1016/j.jmb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–68. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 34.Hitoshi Y, Gururaja T, Pearsall DM, Lang W, Sharma P, Huang B, Catalano SM, McLaughlin J, Pali E, Peelle B, Vialard J, Janicot M, Wouters W, Luyten W, Bennett MK, Anderson DC, Payan DG, Lorens JB, Bogenberger J, Demo S. Cellular localization and antiproliferative effect of peptides discovered from a functional screen of a retrovirally delivered random peptide library. Chem Biol. 2003;10:975–87. doi: 10.1016/j.chembiol.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Choi JW, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 36.Hardie DG, Hawley SA. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays. 2001;23:1112–9. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- 37.Sim AT, Hardie DG. The low activity of acetyl-CoA carboxylase in basal and glucagon-stimulated hepatocytes is due to phosphorylation by the AMP-activated protein kinase and not cyclic AMP-dependent protein kinase. FEBS Lett. 1988;233:294–8. doi: 10.1016/0014-5793(88)80445-9. [DOI] [PubMed] [Google Scholar]
- 38.Carling D, Clarke PR, Zammit VA, Hardie DG. Purification and characterization of the AMP-activated protein kinase. Copurification of acetyl-CoA carboxylase kinase and 3-hydroxy-3-methylglutaryl-CoA reductase kinase activities. Eur J Biochem. 1989;186:129–36. doi: 10.1111/j.1432-1033.1989.tb15186.x. [DOI] [PubMed] [Google Scholar]
- 39.Jorgensen SB, Nielsen JN, Birk JB, Olsen GS, Viollet B, Andreelli F, Schjerling P, Vaulont S, Hardie DG, Hansen BF, Richter EA, Wojtaszewski JF. The alpha2-5′AMP-activated protein kinase is a site 2 glycogen synthase kinase in skeletal muscle and is responsive to glucose loading. Diabetes. 2004;53:3074–81. doi: 10.2337/diabetes.53.12.3074. [DOI] [PubMed] [Google Scholar]
- 40.Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277:23977–80. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- 41.Marsin AS, Bouzin C, Bertrand L, Hue L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J Biol Chem. 2002;277:30778–83. doi: 10.1074/jbc.M205213200. [DOI] [PubMed] [Google Scholar]
- 42.Kurth-Kraczek EJ, Hirshman MF, Goodyear LJ, Winder WW. 5′ AMP-activated protein kinase activation causes GLUT4 translocation in skeletal muscle. Diabetes. 1999;48:1667–71. doi: 10.2337/diabetes.48.8.1667. [DOI] [PubMed] [Google Scholar]
- 43.Cheung PC, Salt IP, Davies SP, Hardie DG, Carling D. Characterization of AMP-activated protein kinase gamma-subunit isoforms and their role in AMP binding. Biochem J. 2000;346(Pt 3):659–69. [PMC free article] [PubMed] [Google Scholar]
- 44.Iseli TJ, Walter M, van Denderen BJ, Katsis F, Witters LA, Kemp BE, Michell BJ, Stapleton D. AMP-activated protein kinase beta subunit tethers alpha and gamma subunits via its C-terminal sequence (186-270) J Biol Chem. 2005;280:13395–400. doi: 10.1074/jbc.M412993200. [DOI] [PubMed] [Google Scholar]
- 45.Davies SP, Carling D, Hardie DG. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur J Biochem. 1989;186:123–8. doi: 10.1111/j.1432-1033.1989.tb15185.x. [DOI] [PubMed] [Google Scholar]
- 46.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of the eukaryotic cell? Annual Review of Biochemistry. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 47.Adams J, Chen ZP, Van Denderen BJ, Morton CJ, Parker MW, Witters LA, Stapleton D, Kemp BE. Intrasteric control of AMPK via the gamma1 subunit AMP allosteric regulatory site. Protein Sci. 2004;13:155–65. doi: 10.1110/ps.03340004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudolph MJ, Amodeo GA, Bai Y, Tong L. Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1. Biochem Biophys Res Commun. 2005;337:1224–8. doi: 10.1016/j.bbrc.2005.09.181. [DOI] [PubMed] [Google Scholar]
- 49.Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–21. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- 50.Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–5. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 51.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annual Review of Biochemistry. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 52.Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol. 2002;317:309–23. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- 53.Henin N, Vincent MF, Van den Berghe G. Stimulation of rat liver AMP-activated protein kinase by AMP analogues. Biochim Biophys Acta. 1996;1290:197–203. doi: 10.1016/0304-4165(96)00021-9. [DOI] [PubMed] [Google Scholar]
- 54.Ferdinand W. The interpretation of non-hyperbolic rate curves for two-substrate enzymes. A possible mechanism for phosphofructokinase. Biochem J. 1966;98:278–83. doi: 10.1042/bj0980278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCafferty DG, Lessard IA, Walsh CT. Mutational analysis of potential zinc-binding residues in the active site of the enterococcal D-Ala-D-Ala dipeptidase VanX. Biochemistry. 1997;36:10498–505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 56.Vartanian JP, Henry M, Wain-Hobson S. Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 1996;24:2627–31. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woods RA, Gietz RD. High-efficiency transformation of plasmid DNA into yeast. Methods Mol Biol. 2001;177:85–97. doi: 10.1385/1-59259-210-4:085. [DOI] [PubMed] [Google Scholar]
- 58.Hardie DG, Salt IP, Davies SP. Analysis of the role of the AMP-activated protein kinase in the response to cellular stress. Methods Mol Biol. 2000;99:63–74. doi: 10.1385/1-59259-054-3:63. [DOI] [PubMed] [Google Scholar]
- 59.Weekes J, Ball KL, Caudwell FB, Hardie DG. Specificity determinants for the AMP-activated protein kinase and its plant homologue analysed using synthetic peptides. FEBS Lett. 1993;334:335–9. doi: 10.1016/0014-5793(93)80706-z. [DOI] [PubMed] [Google Scholar]


