Summary
The p16INK4a tumor suppressor gene is a frequent target of epigenetic inactivation in human cancers, which is considered to be an early event in breast carcinogenesis. Here we describe the existence of a chromatin boundary upstream of the p16 gene that is lost when this gene is aberrantly silenced. We show that the multifunctional protein CTCF associates in the vicinity of this boundary and that absence of CTCF binding strongly coincides with p16 silencing in multiple types of cancer cells. CTCF binding also correlates with activation of the RASSF1A and CDH1 genes and this interaction is absent when these genes are methylated and silenced. Interestingly, defective poly(ADP-ribosyl)ation of CTCF and dissociation from the molecular chaperone Nucleolin occurs in p16-silenced cells, abrogating its proper function. Thus, destabilization of specific chromosomal boundaries through aberrant crosstalk between CTCF, poly(ADP-ribosyl)ation, and DNA methylation may be a general mechanism to inactivate tumor suppressor genes and initiate tumorigenesis in numerous forms of human cancers.
Introduction
Aberrant transcriptional silencing of tumor suppressor genes by epigenetic deregulation is a common occurrence in human malignancies. This is characterized by altered patterns of DNA hypermethylation in specific promoter regions and acquisition of histone modifications that are characteristic of repressed chromatin (Feinberg, 2008; Jones and Baylin, 2007). Because of its importance in cell proliferation, the human INK4 gene locus is a frequent target of inactivation by deletion or aberrant DNA methylation in a wide variety of human cancers (Kim and Sharpless, 2006). This locus encompasses approximately 42 kb on chromosome 9 and encodes three distinct tumor suppressor proteins, p15INK4b, p14ARF and p16INK4a (referred to hereafter as p15, p14 and p16). p16 is a key regulator of G1 phase cell cycle arrest and senescence, which it achieves primarily through inhibiting the cyclin-dependent kinases CDK4 and CDK6. In fact, inactivation of the p16 gene by promoter methylation or genetic change is one of the earliest losses of tumor suppressor function in numerous types of human cancers, such as breast, lung, colorectal cancers and multiple myeloma (Belinsky et al., 1998; Foster et al., 1998; Ng et al., 1997). Notably, p16 promoter methylation and transcriptional silencing have been shown to exist in histologically normal mammary tissue of cancer-free women. This suggests that these aberrant epigenetic changes may represent a cancerous pre-condition and an early event in promoting genomic instability that leads to tumorigenesis (Holst et al., 2003).
Although the precise mechanisms underlying epigenetic loss-of-function of the p16 gene remain unresolved, an examination of proteins important for its regulation may provide insight into the cause of aberrant silencing. One study revealed that transcription of all three INK4/ARF genes is controlled by a common CDC6-binding regulatory element (Gonzalez et al., 2006). While this is intriguing, RNA expression and DNA methylation profiles in a variety of tumors and cancer cell lines show no obvious coupling of p16 silencing with that of the p15 and p14 genes (Paz et al., 2003). p16 silencing could also result from gain-of-function or aberrant targeting of repressor proteins that modulate epigenetic processes. For example, ID1 regulates p16 expression during senescence through exchange of ID for ETS activators (Ohtani et al., 2001). However, it is unclear if this contributes to p16 deregulation during tumorigenesis. Another repressor of p16, the polycomb protein BMI1, has oncogenic activity and controls cell proliferation and senescence through the INK4a locus (Jacobs et al., 1999). In primary breast tumors, however, no correlation between BMI1 and p16 expression is observed (Silva et al., 2006). Other polycomb members such as EZH2 and SUZ12 also interact with p16 in proliferating fibroblasts (Bracken et al., 2007; Kotake et al., 2007). Although data linking EZH2 and p16 silencing is lacking, recent evidence indicates that EZH2 can recruit DNA methyltransferases and maintain methylation patterns at silenced genes in cancer cells (Vire et al., 2006).
To further understand the mechanism(s) by which the p16 gene becomes aberrantly silenced in human cancers, we examined epigenetic regulation at the level of the INK4/ARF chromosomal locus rather than solely at the p16 promoter. Here we present evidence that a chromosomal boundary exists at approximately 2 kb upstream of the p16 transcriptional start site. This boundary separates the p16 gene locus into discrete domains characterized by the presence or absence of repressive epigenetic marks and the histone variant H2A.Z, a functionally diverse protein recently shown to confer memory of transcriptional status and facilitate re-activation of target promoters (Raisner and Madhani, 2006). By contrast, in breast cancer cells containing aberrantly silenced p16 genes, the epigenetically defined domain at -2 kb disappears and regions 3′ of this boundary acquire characteristics of heterochromatin which is accompanied by loss of histone H2A.Z. Upon further examination, we noticed the presence of a recognition sequence for the zinc finger protein CTCF 3′ of the boundary. CTCF is a multi-functional transcription factor known to have a critical role in regulating chromosomal boundaries/insulators (Filippova, 2008; Wallace and Felsenfeld, 2007). Unexpectedly, we observed CTCF association with this region in numerous p16-expressing cell lines but complete absence in p16 non-expressing breast cancer cells, even though CTCF binds to another target gene, c-Myc, in all cases. Moreover, ablation of CTCF protein from p16-expressing cells by shRNA results in epigenetic changes to the p16 promoter and loss of transcription. In addition to breast cancer, aberrant p16 gene silencing is widely documented in a variety of human malignancies. We examined multiple myeloma cell lines and found that inactivation of the p16 gene is also correlated with absence of CTCF binding. Thus, our studies indicate that p16 gene repression can result from destabilization of a chromosomal boundary through dissociation of CTCF.
A similar examination of other well characterized epigenetically silenced genes in human cancers, RASSF1A and CDH1 (E-cadherin), also revealed a strong correlation between transcriptional inactivation and a loss of CTCF binding. The insulator function of CTCF has been shown to require its post-translational modification by poly(ADP-ribosyl)ation (PARlation) (Yu et al., 2004) and crosstalk between PARP-1 and CTCF strongly affects DNA methylation (Guastafierro et al., 2008). Strikingly, we found a defect in the poly(ADP-ribosylation) pathway in p16-silenced cells resulting in the absence of CTCF PARlation and dissociation from a new coregulator, Nucleolin. Furthermore, we demonstrated that chemical inhibition of PARlation or knockdown of PARP-1 directly impacts p16 and RASSF1A expression. We propose that destabilization of specific chromosomal boundaries is caused by aberrant interactions between CTCF and the poly(ADP-ribosyl)ation enzymatic machinery and can be a general mechanism to initiate potentially reversible genomic instability and tumorigenesis in human cancers.
Results
Loss of a Chromosomal Boundary at the p16 Gene Locus in Epigenetically Silenced Breast Cancer Cells
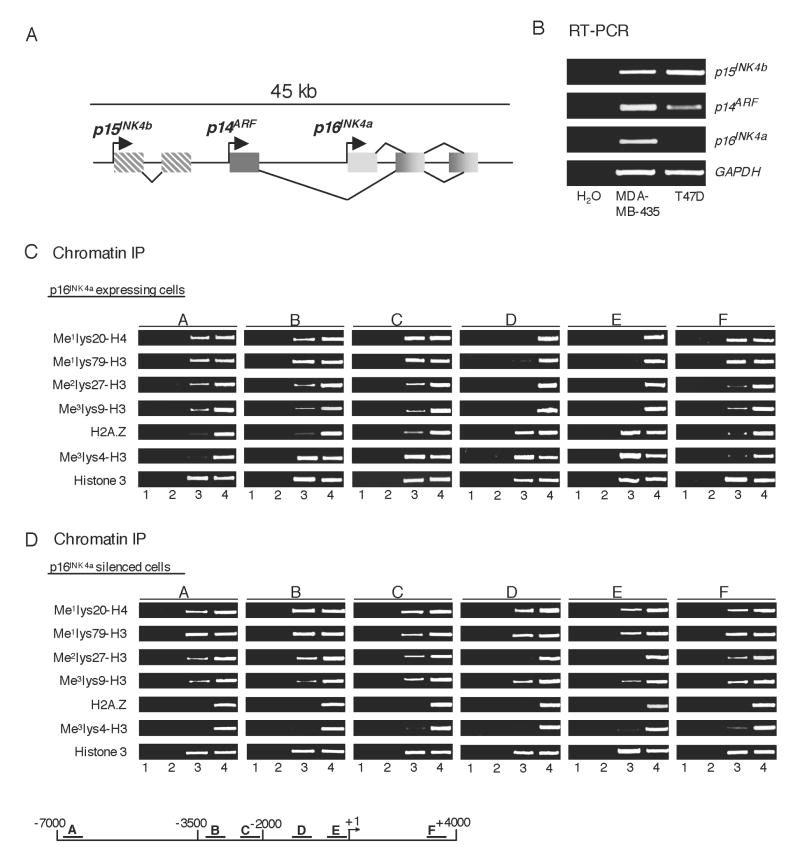
Aberrant transcriptional silencing of tumor suppressor genes is accompanied by dynamic changes in chromatin structure as revealed by the acquisition of histone modifications that are characteristic of repressed chromatin. To gain insight into chromatin structural alterations that may accompany p16 gene inactivation, we analyzed histone modifications surrounding the gene in p16-expressing (MDA-MB-435) and non-expressing (T47D) human breast cancer cell lines (Figure 1). Initially, we examined the transcriptional status of the three genes within the INK4/ARF locus, p15, p14, and p16 (diagrammed in Figure 1A). We found that each gene is active in MDA-MB-435 cells whereas p16 alone is silenced in T47D cells (Figure 1B). This indicates that event(s) leading to p16 deregulation in these cells specifically impacts this gene without affecting the entire INK4/ARF locus.
Figure 1.
Analyses of Histone Modifications at the p16 Gene
(A) Diagram of gene organization at the INK4/ARF chromosomal locus.
(B) RT-PCR analysis of gene expression at the INK4 locus in p16-expressing (MDA-MB-435) and non-expressing (T47D) breast cancer cells.
(C) ChIP analyses of histone modifications surrounding the p16 gene in p16-expressing cells. Lanes: 1, H2O; 2, no antibody; 3, antibodies to various histone modifications; 4, 1.6% total input DNA. ChIP-enriched DNA was PCR-amplified using specific amplicons (A-F) distributed throughout the p16 gene locus.
(D) A similar ChIP analysis in p16 non-expressing cells.
We next performed chromatin immunoprecipitations (ChIPs) to analyze a variety of histone modifications within the vicinity of the active p16 gene in MDA-MB-435 cells. These modifications include those that are typically associated with repressed chromatin, like Me1H4K20, Me2H3K27, and Me3H3K9, as well as marks that correlate with mammalian gene activation, such as Me3H3K4 and the histone variant H2A.Z. We also examined the presence of Me1H3K79 which is generally correlated with transcriptionally active genes in mammalian cells (Klose and Zhang, 2007). Localization of bulk histone 3 was measured to control for any large changes in nucleosomal placement and density. Surprisingly, in p16-expressing cells we found an enrichment of marks that are normally associated with silenced genes, as well as Me1H3K79, between 2-7 kb upstream of the proximal promoter (Figure 1C; amplicons A-C, Table S1). This chromatin structural organization is lost in the vicinity of the p16 proximal promoter between -2 kb and +1 (amplicons D-E). As expected in this region of an expressed gene, Me3H3K4 is enriched and H2A.Z is distributed in a similar pattern. The region of “active” chromatin between -2 kb and +1 is reversed downstream of the p16 gene at +4 kb where chromatin again becomes repressed (amplicon F). These data indicate that the 11 kb region encompassing the p16 gene is arranged into clearly demarcated domains of repressive versus active chromatin structures. Moreover, the data are consistent with the presence of a distinct chromosomal boundary/insulator within 2 kb upstream of the p16 transcriptional start site.
A similar ChIP analysis was conducted in T47D breast cancer cells in which the p16 gene is silenced and methylated (Figure S1) (Di Vinci et al., 2005). In these cells the chromatin structure of the aberrantly inactivated p16 gene is quite different from that found in p16-expressing MDA-MB-435 cells and, most strikingly, the chromosomal domain organization is lost (Figure 1D). This is apparent from the spread of repressive histone marks, Me1H4K20 and Me3H3K9 as well as Me1H3K79 from the upstream 2-7 kb domain through the -2 kb demarcation to encompass the entire 11 kb p16 gene locus. A dramatic loss of H2A.Z and Me3H3K4 from approximately -3 kb to +1 (amplicons B-E) is also evident. However, no significant change in the pattern of Me2H3K27 is observed indicating that the enzymatic activity associated with this mark may function in an independent manner. Overall, these data substantiate the existence of a chromatin boundary upstream of the p16 initiation site that functions to maintain the promoter in an active configuration by preventing the spread of repressive nucleosomal modifications from a neighboring domain. Interestingly, the disappearance of this boundary is correlated with aberrant epigenetic silencing of the p16 gene in certain breast cancer cell lines.
The Boundary/Insulator Protein CTCF Associates with the Transcriptionally Active but not Silenced p16 Gene
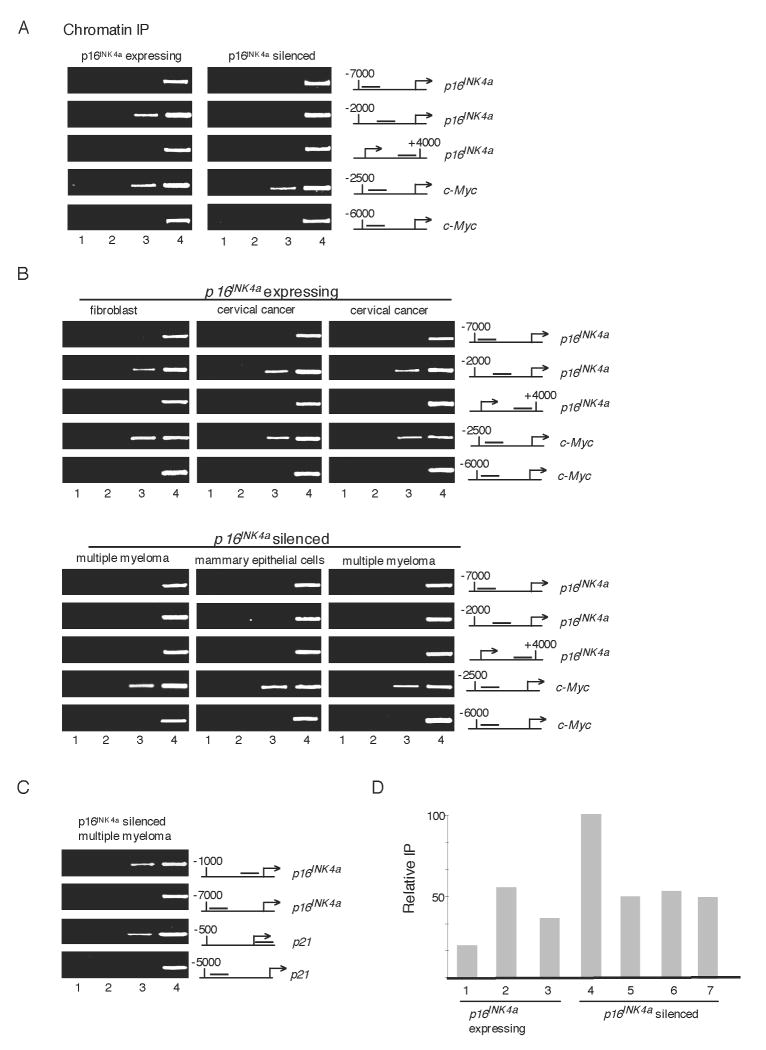
CTCF is a ubiquitous, multifunctional protein that has a critical role in organizing distinct chromosomal domains through boundary/insulator formation and repressing or activating transcription. Because we saw a pronounced change in chromatin structure upstream of the p16 gene when active or silenced (Figure 1), we explored the possibility that CTCF may associate within this region. To address this issue, a ChIP analysis was performed to identify sites of CTCF interaction within -7 to +4 kb of the p16 gene locus in expressing and non-expressing cells. Our data revealed that in p16-expressing cells, CTCF clearly binds downstream (amplicon D) of the region enriched for marks of heterochromatin within -2 kb and +1 of the active p16 gene (Figures 2A, S2A). However, no CTCF binding was observed at other distal regions in the locus near -7 kb (amplicon A) or +4 kb (amplicon F). Surprisingly, when we examined cells containing a silenced p16 gene, CTCF interaction at the upstream promoter site was not apparent. Yet, we detected CTCF binding at a well-characterized target gene, c-Myc, in both p16-expressing and non-expressing cell types. This indicates that loss of CTCF binding from the p16 gene in T47D cells is not due to a general defect in the ability of the endogenous protein to associate with its chromosomal targets. Moreover, CTCF dissociation from the p16 gene is not mechanistically linked to the stability of other CTCF interactions that we examined in the INK4/ARF locus (Figure S2B). No significant reduction in bulk CTCF protein levels or in cellular localization was observed between T47D and MDA-MB-435 cells (Figure S2C). Importantly, loss of CTCF interaction is not a consequence of cessation of p16 transcription since CTCF binding remains stable upon p16 gene inactivation by pharmacological inhibitors (Figure S3).
Figure 2.
CTCF Associates with the Active but not Silent p16 Gene
(A) ChIP analysis of CTCF binding in p16-expressing and non-expressing breast cancer cells. CTCF binding was also measured at the c-Myc gene in both cell lines. Lanes: 1, H2O; 2, no antibody; 3, anti-CTCF antibody; 4, 1.6% input DNA.
(B) CTCF Binding Correlates with p16 Expression in Multiple Types of Human Cancer Cells. ChIP analyses showing binding to the p16 gene in human primary fibroblasts (IMR90) and human cervical cancer cell lines (HeLa, C33A), but reduced association in p16 non-expressing cells: human multiple myeloma (KMS12, U266) and a primary breast epithelial-derived cell line (vHMEC). Lanes 1-4 are described as in (A).
(C) ChIP analyses of Sp1 binding to the p16 promoter in multiple myeloma (U266) cells. p21 is a positive control. Lane order is as described in (A).
(D) qPCR ChIP analyses of Sp1 binding reveal no clear correlation between Sp1 interaction and p16 expression. Lanes correspond to Sp1 IPs from the following cells: 1, cervical cancer (HeLa); 2, human primary fibroblasts (IMR90); 3, breast cancer (MDA-MB-435); 4, multiple myeloma (U266); 5, multiple myeloma (KMS12); 6, breast cancer (T47D); 7, primary breast epithelial-derived (vHMEC).
p16 Gene Expression Correlates with CTCF Binding Near its Chromosomal Boundary in Multiple Types of Human Cancer Cells
Having established a strong correlation between CTCF interaction with the p16 upstream promoter and p16 expression in breast cancer cell lines, we asked whether our observations could be extended to other types of human cancer cells. For example, the p16 gene is a frequent target of epigenetic inactivation in primary multiple myeloma cells (Ng et al., 1997). As shown in the ChIP analysis in Figure 2B, CTCF binding is highly correlated with p16 expression in diverse cell types such as non-transformed fibroblasts (IMR90) and cervical cancer cell lines (HeLa, C33A). Conversely, in two multiple myeloma cell lines (U266, KMS12) and a primary breast epithelial-derived cell line (vHMEC), each of which harbors a silenced p16 gene, CTCF binding was lost from the upstream promoter. Consistent with our findings in the MDA-MB-435 and TD47 breast cancer cell lines (Figure 2A), CTCF interaction at the c-Myc locus was constant in all cell types examined (Figure 2B). Thus, loss of CTCF binding from the p16 upstream promoter near its chromosomal boundary is correlated with transcriptional silencing in both human breast cancer and multiple myeloma cell lines even though CTCF interaction with c-Myc remains unaffected. The loss of CTCF binding at p16 could not be attributed to aberrant expression and recruitment of BORIS to replace CTCF, as we saw no correlation between p16 silencing and BORIS expression (Figure S4A and data not shown).
Next we asked whether the striking relationship between CTCF binding and p16 gene transcription could be extended to other factors that may regulate p16 expression. To address this, we examined association of the ubiquitous nuclear factor Sp1 with the p16 promoter in both p16-expressing and non-expressing cell lines because this protein has been implicated in p16 gene transactivation (Wu et al., 2007). Unexpectedly, we found strong Sp1 binding to the p16 promoter in multiple myeloma cells (U266) where the gene is silent (Figure 2C). The extent of Sp1 interaction with p16 was comparable to its association with the p21 gene. To extend this analysis, Sp1 binding to the p16 promoter was examined using real-time PCR. Quantitative analysis of Sp1 binding revealed that, unlike CTCF, there is no clear correlation between Sp1 interaction with the p16 promoter and its transcriptional activity (Figure 2D). From these data we conclude that transcriptional silencing of the p16 promoter is not due to occlusion of binding to regulatory factors in general. Instead, we hypothesize that silencing more likely results from the spread of heterochromatin caused by loss of the upstream chromatin boundary that is maintained by CTCF binding. Dissociation of another promoter-bound activator, Sp1, does not have this effect.
CTCF Epigenetically Regulates the p16 Promoter and Gene Expression
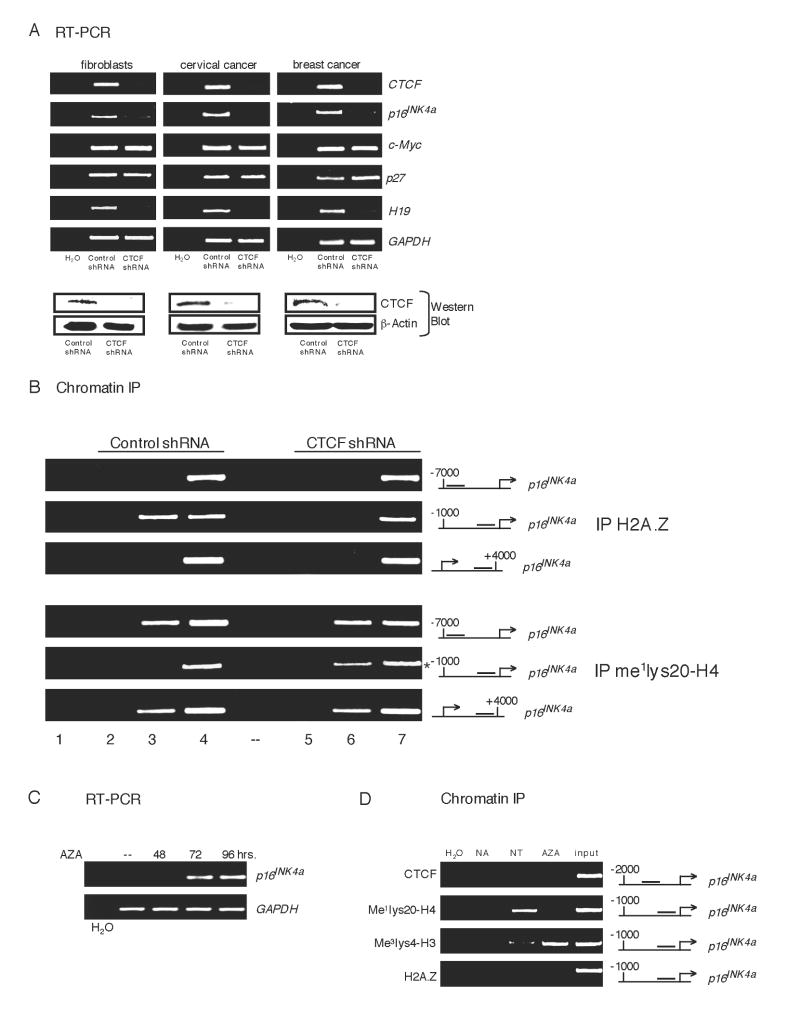
To investigate the functional role of CTCF at the p16 upstream region, we used shRNA to decrease expression of CTCF in several cell lines that contain an active p16 gene. Reduction of CTCF in each cell line was confirmed at the level of mRNA by RT-PCR (Figure 3A, upper panels) and protein by Western analysis (lower panels). Near complete ablation of cellular CTCF resulted in considerably reduced p16 mRNA levels in fibroblasts (IMR90), cervical cancer cells (HeLa), and breast cancer cells (MDA-MB-435) whereas no effect on p16 expression was observed in cells infected with a scrambled shRNA (Figures 3A, S4B). In addition, mRNA abundance of the H19 gene was significantly decreased in each CTCF knockdown cell line, consistent with the demonstrated involvement of CTCF in H19 expression (Szabo et al., 2004). Expression of the GAPDH gene, which served as a control for total mRNA abundance, was also unchanged by the absence of CTCF. In contrast to a previous report (Qi et al., 2003), we observed no change of the cell cycle inhibitor p27 transcript levels upon CTCF knockdown, which may reflect tissue-specific consequences of CTCF depletion. Unexpectedly, the CTCF target gene c-Myc remained impervious to loss of CTCF. This suggests that CTCF may have distinct functional roles at the p16, H19, and c-Myc genes with different requirements for continuous, compared to transient, binding.
Figure 3.
CTCF Knockdown Results in Transcriptional Silencing of the p16 Gene and Acquisition of Repressive Chromatin Modifications
(A) RT-PCR analysis of mRNA from p16-expressing cells infected with either control (scrambled) or CTCF-specific shRNA.
Lower panel: Western blot of CTCF protein expression in these shRNA-infected cells.
(B) ChIP analysis of the p16 gene after CTCF knockdown in p16-expressing cells (MDA-MB-435) using antibodies to the histone variant H2A.Z and Me1H4K20. Reactions from cells infected with scrambled shRNA (lanes 2-4) and CTCF-specific shRNA (lanes 5-7) are designated.
Lanes: 1, H2O; 2, no antibody; 3, anti-H2A.Z antibody; 4, 1.6% input DNA; 5, no antibody; 6, anti-Me1H4K20 antibody; 7, 1.6% input DNA; * denotes 0.25% input DNA in lanes 4 and 7 for this panel (p16 proximal promoter) only.
(C) RT-PCR analysis showing time course of AZA induction of p16 mRNA in breast cancer cells (T47D) containing a silenced p16 gene.
(D) ChIP analysis of the p16 promoter in T47D cells treated with AZA for 96 hours using the indicated antibodies. NA = no antibody, NT = no AZA treatment, AZA = AZA treatment, input = 1.6% of total DNA.
To explore the possibility that CTCF may influence chromatin organization at the p16 locus, we analyzed histone modifications at the p16 promoter in breast cancer cells (MDA-MB-435) whose CTCF levels were ablated by shRNA treatment (Figure 3B). Most strikingly, we observed a significant reduction of the histone variant H2A.Z at the p16 promoter upon loss of cellular CTCF (Figure 3B, upper panels) and an increase in Me1H4K20 in the same region (lower panels). The loss of H2A.Z and 3′ shift of the repressive histone mark to the region downstream of the -2 kb boundary corresponds to the epigenetic characteristics of the silenced p16 gene (Figure 1D) which no longer interacts with CTCF (Figure 2A) and apparently undergoes heterochromatin “spreading” from upstream regions. Thus, our results are consistent with the idea that CTCF binding is required to maintain a chromosomal boundary near -2 kb which preserves the p16 gene in a transcriptionally active chromatin domain.
Pharmacological Treatment of Cancer Cells Restores Temporary p16 Gene Transcription but not CTCF Binding
The p16 tumor suppressor gene is commonly silenced in numerous types of human cancers and remains a relevant therapeutic target of wide interest. One method that is extensively employed to restore p16 expression, both clinically and in vitro, is treatment of cancer cells with hypomethylating-nucleoside analogues such as 5′-AZA-2′-deoxycytidine (AZA) (Otterson et al., 1995), which reverses DNA methylation. We reasoned that treatment of cells with AZA might also restore CTCF binding at the p16 upstream promoter through one of two mechanisms. First, CTCF is known to bind DNA in a methylation-sensitive fashion (Hark et al., 2000) thus, demethylation of the p16 locus might allow CTCF to reassociate. Second, demethylation of target promoters by AZA can change the surrounding chromatin structure (Fahrner et al., 2002) which may facilitate rebinding of regulatory proteins, as observed for Sp1 (Zhang et al., 2005). We conducted a time course of p16 mRNA induction after AZA treatment of T47D breast cancer cells, which contain a methylated and silenced p16 gene. Significant reactivation of p16 expression occurred by 72 hours (Figures 3C, S4C). No synergistic p16 gene reactivation was observed in cells treated with both AZA and the HDAC inhibitor Trichostatin A (Figure S4C).
To examine whether any changes in histone modifications or potential reassociation of CTCF had occurred after reversal of p16 transcriptional silencing, a ChIP analysis was performed at 96 hours post-AZA treatment. As shown in Figure 3D, several alterations in nucleosome modification at the p16 promoter were apparent. Notably, Me1H4K20 and Me3H3K4 were reversed in accordance with gene activity. However, AZA treatment did not result in recruitment of CTCF or H2A.Z to the reactivated p16 gene, or Sp1 (data not shown). While AZA can reactivate p16 transcription, it does not entirely reverse alterations that occur during gene silencing. This is consistent with a recent study showing only partial reversal of the histone code to an active state at the MLH1 promoter after AZA treatment (McGarvey et al., 2006). In fact, the general inability to sustain long-term p16 gene expression after reversal of epigenetic silencing by AZA (Egger et al., 2007) may, in part, be explained by failure to reestablish the upstream chromatin domain boundary by CTCF.
Absence of CTCF PARlation in p16-Silenced Cells
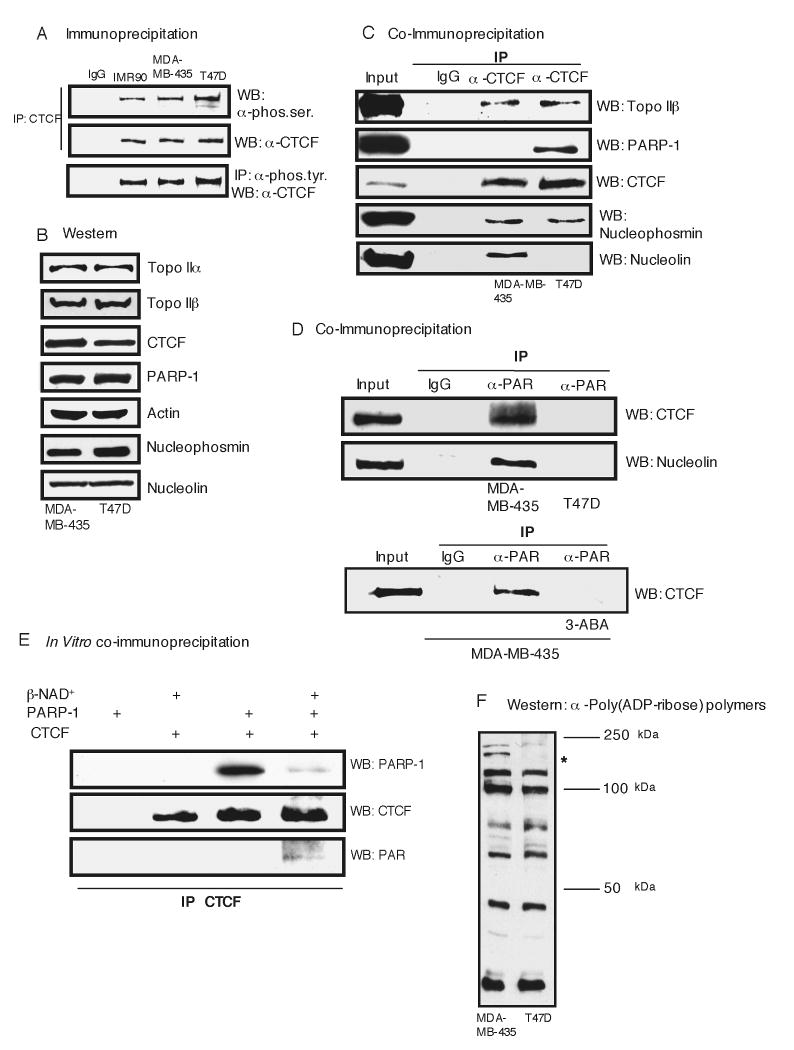
As described above, CTCF may be important for maintaining an active p16 gene. To further investigate possible CTCF defects that may impact its function in p16-silenced T47D breast cancer cells, we examined its post-translational modifications and association with several known protein interaction partners. CTCF is post-translationally modified by phosphorylation and poly(ADP-ribosyl)ation (PARlation) and interacts with multiple proteins, such as Topoisomerase IIβ, Nucleophosmin and PARP-1 (Yusufzai et al., 2004). As shown in Figure 4A, similar extents of CTCF phosphorylation at both serine and tyrosine residues were observed in normal fibroblasts, p16-expressing MDA-MB-435 and non-expressing TD47 breast cancer cells as determined by immunoprecipitation. In addition, cellular levels of CTCF and several known interaction partners, Topo IIα, Topo IIβ, Nucleophosmin and PARP-1, were comparable in MDA-MB-435 and TD47 cells as well as a new interactor, Nucleolin (Figure 4B). Co-immunoprecipitation of CTCF complexes indicated that CTCF interacts with Topo IIβ and Nucleophosmin similarly in both cell types but has opposite interaction characteristics with PARP-1 which, surprisingly, only associates with CTCF in p16 silenced cells (Figure 4C). Moreover, PARP-1 and Nucleolin appear to associate with CTCF in a mutually exclusive manner, with Nucleolin being present in CTCF complexes only in p16 expressing cells. To explore this further, we examined the PARlation status of CTCF in each cell type using an antibody that recognizes ADP-ribose polymers (PAR). Unexpectedly, we found PARlation associated with both CTCF and Nucleolin in p16-expressing MDA-MB-435 cells but not in p16-silenced TD47 cells (Figure 4D, upper panel). This PARlation is inhibited upon addition of the PARlation inhibitor 3-Aminobenzamide (3-ABA) demonstrating the specificity of this reaction (Figure 4D, lower panel). Initially, it appeared counterintuitive that CTCF could associate with PARP-1 but be unPARlated in TD47 cells whereas in MDA-MB-435 cells the opposite was true, CTCF was PARlated and dissociated from PARP-1. We surmised that differences in CTCF-PARP-1 interaction dynamics might reflect defects in the poly(ADP)ribosylation enzymatic pathway in TD47 cells. To substantiate this, we performed an in vitro binding assay using recombinant PARP-1 and CTCF proteins with or without the obligate poly(ADP)ribosylation substrate, β-NAD+, under enzymatic reaction conditions. Interestingly, we found that in the absence of β-NAD+ a stable complex formed between CTCF and PARP-1 when co-precipitated. Yet when β-NAD+ was present and CTCF PARlated, the CTCF/PARP-1 complex dissociated (Figure 4E). This supports the notion that upon CTCF PARlation, it dissociates from the PARP-1 enzyme as is observed in MDA-MB-435 cells. If the enzymatic reaction is not productive and CTCF remains unPARlated, the enzyme-substrate complex fails to release as seen in T47D cells (Figures 4C, 4D). An examination of total cellular proteins that are PARlated in either MDA-MB-435 or T47D extracts revealed a very similar pattern with the primary exceptions being two proteins in the size range of CTCF and larger whose modification is clearly impaired in T47D cells (Figure 4F). This indicates that while there are no apparent gross defects in the poly(ADP)ribosylation enzymatic machinery, its ability to react with specific protein substrates, such as CTCF, is deregulated. Further supporting this hypothesis is our finding that reintroduction of exogenous CTCF into T47D cells does not reestablish CTCF PARlation (Figure S5), possibly reflecting altered dynamics between PARP-1 Nucleolin, and Nucleophosmin.
Figure 4.
CTCF is Differentially Poly(ADP)ribosylated in p16 Expressing and Non-Expressing Breast Cancer Cells.
(A) Immunoprecipitations show similar levels of CTCF phosphorylation on serine (left panel) and tyrosine (right panel) residues in multiple cell lines.
(B) Western blot showing protein levels of CTCF and putative interacting partners in MDA-MB-435 and T47D cells.
(C) Co-IP of CTCF with Topo IIβ, PARP-1, Nucleophosmin and Nucleolin.
(D) Anti-PAR antibody co-IPs CTCF and Nucleolin in MDA-MB-435 cells. Bottom panel shows similar IP using material treated with 3-ABA for 24 hours. Inputs are equal to 2.5% starting material.
(E) In vitro binding of recombinant CTCF and PARP-1. IP of CTCF from the reactions following protein binding assays.
(F) Western blot of poly(ADP)ribosylated proteins in MDA-MB-435 and T47D cells. Asterisk denotes protein with similar molecular weight as CTCF that is differentially PARlated in MDA-MB-435 and T47D cells.
Differential Patterns of PARlation and CTCF Partner Binding at the p16 Gene
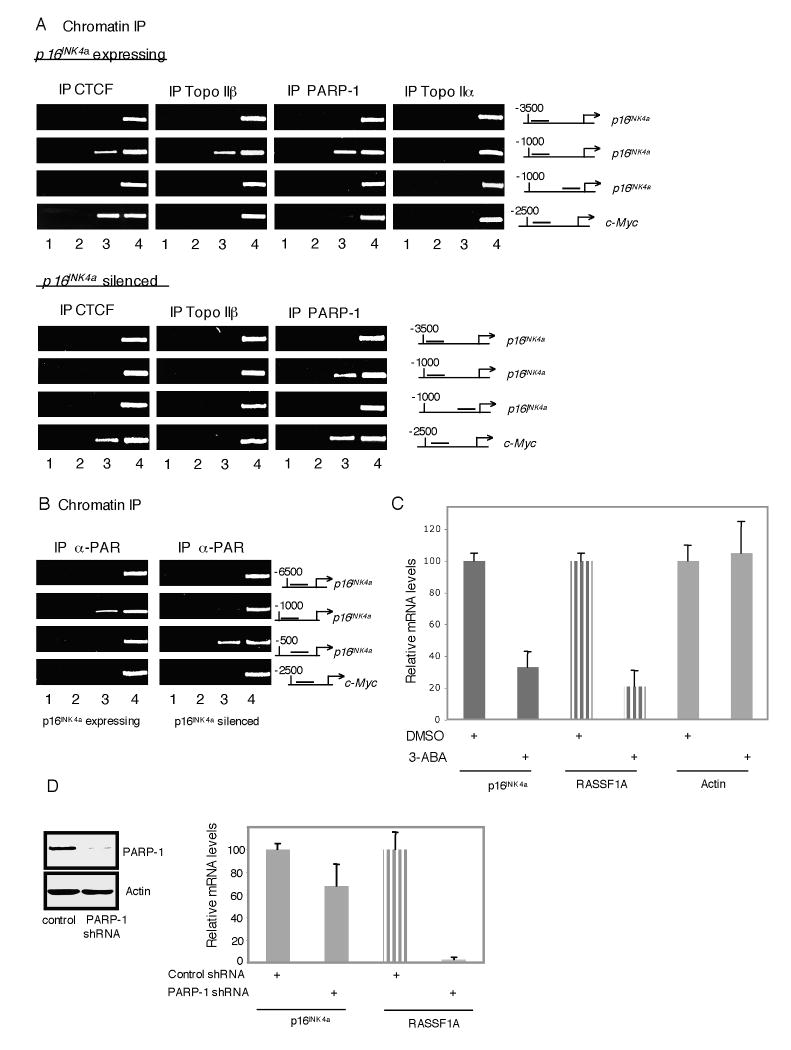
To determine whether known interacting partners of CTCF associate with the p16 gene when active or epigenetically silenced, we performed ChIP analyses on MDA-MB-435 or T47D extracts using antibodies to Topo IIα, Topo IIβ, and PARP-1. As shown in Figure 5A, in p16-positive cells CTCF, Topo IIβ, and PARP-1 each bind to the p16 gene in the region around -1 kb whereas no Topo IIα was detected in the distal or proximal promoter. At the CTCF binding site upstream of the c-Myc promoter, weak PARP-1 binding was observed but no association of Topo IIα or Topo IIβ. By contrast, in p16-negative cells not only is CTCF lost from the silent p16 gene but Topo IIβ is also dissociated. Interestingly, PARP-1 remains bound to the inactive p16 gene, apparently interacting independently of CTCF. PARP-1 binding to the CTCF site proximal to the c-Myc gene is also highly enriched in T47D cells. The presence of PARP-1 at the p16 and c-Myc genes led us to examine the distribution of chromatin-bound PARlated proteins using an anti-PAR antibody. As shown in Figure 5B, PARlation is enriched at the -1 kb region of the expressed p16 gene, (possibly indicating the presence of a PARlated CTCF), with low level PARlation within the proximal promoter. However when the p16 gene is silenced upon loss of CTCF, the pattern of PARlation shifts from -1 kb to being highly enriched at the proximal promoter even though PARP-1 is still bound near -1 kb. This redistribution may reflect PARlation of heterochromatin components that are enriched after CTCF dissociates, such as histone H1. Notably, modification of these components is unaffected by the aberration in the pathway that prevents poly(ADP)ribosylation of CTCF, underscoring the specific nature of this defect. In contrast to the p16 locus, no PARlation was observed at the c-Myc insulator site even in the vicinity of bound CTCF and PARP-1. These results indicate that separate CTCF binding sites are distinct from one another in terms of cofactor interactions and PARlation, potentially allowing CTCF to exert specialized regulatory functions on different target genes.
Figure 5.
Pattern of PARlation at the p16 Promoter Region Changes in p16 Silenced Cells.
(A) ChIP analyses of CTCF and putative interacting partners at the p16 and c-Myc genes in p16 positive MDA-MB-435 and p16 silenced T47D cells. Lanes: 1, H2O; 2, no antibody; 3, anti-H2A.Z antibody; 4, 1.6% input DNA;
(B) ChIP analyses of PARlation pattern at the p16 and c-Myc genes in the same cells as described in (A). Lane order is as in (A) but showing amplification of 0.25% input material.
(C-D) qPCR expression analysis of the CTCF target genes p16 and RASSF1A upon inhibition of PARP activity. Results are normalized to c-Myc levels. Error bars represent ± STDEV (C) Amplification of mRNA from MDA-MB-435 cells treated for 24 hours with 5 mM 3-ABA. (D) Amplification of mRNA from MDA-MB-435 cells infected with shRNA directed towards PARP-1.
Next, we examined whether PARlation of target proteins impacts the expression of CTCF-regulated tumor suppressor genes. To achieve this, we perturbed cellular PARlation activity in p16-expressing cells by two approaches: first, incubation with the broad-spectrum PARP inhibitor 3-ABA and second, shRNA-mediated ablation of PARP-1. In both cases, we observed a significant reduction of p16 mRNA levels as well as a dramatic decrease of the new CTCF target gene RASSF1A (Figures 5C,D, 6A). Collectively, these data show that normal PARP activity is required for full activation of these CTCF target genes and a disruption of this pathway may play a role in the long-term silencing of these tumor suppressors.
Figure 6.
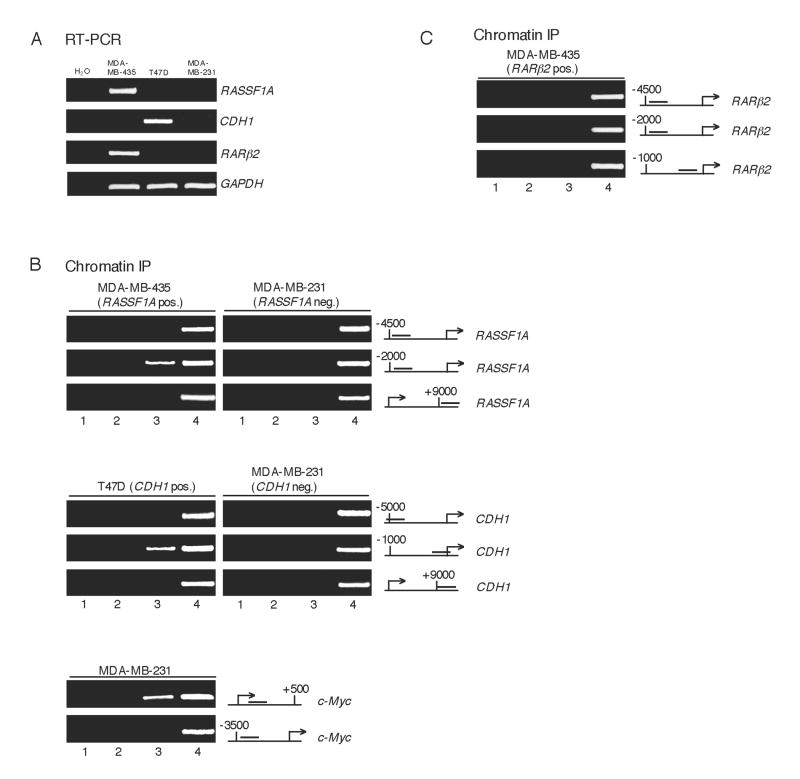
Loss of CTCF Binding from Genes that are Commonly Silenced in Human Cancers
(A) RT-PCR of CDH1, RASSF1A and RARβ2 expression in the indicated breast cancer cells.
(B) ChIP analyses of CTCF binding to the RASSF1A and CDH1 genes in expressing cells but not in cells where the genes are silenced. Lanes: 1, H2O; 2, no antibody; 3, anti-CTCF antibody; 4, 1.6% input DNA.
(C) CTCF does not bind to all genes commonly hypermethylated in cancer such as RARβ2. Lane order is as described in (B).
Epigenetic Silencing of the Tumor Suppressor Genes RASSF1A and CDH1 also Correlates with Loss of CTCF Binding
Having established that CTCF interaction upstream of the p16 promoter is abolished in several different types of human cancer cell lines in which the gene is hypermethylated and silenced, we speculated that CTCF binding sites might be present at other genes commonly inactivated in cancer. To this end, we identified potential CTCF recognition sequences in the promoters of the RASSF1A, CDH1 and RARβ32 genes and analyzed these regions for expression (Figure 6A) and CTCF binding in several breast cancer cell lines. Similar to p16, the RASSF1A protein is a tumor suppressor and aberrant methylation of its gene may represent an early event in breast tumorigenesis (Strunnikova et al., 2005). As shown in Figure 6B (upper panels), a ChIP analysis of RASSF1A-positive cells clearly demonstrates recruitment of CTCF to a region upstream (∼1.8 kb) of the promoter. However, in breast cancer cells in which RASSF1A is silenced and methylated no such binding of CTCF was detected.
Hypermethylation of CDH1 in breast cancer results in a loss of E-cadherin expression (Graff et al., 1995) and is highly associated with an invasive and infiltrating phenotype (Shinozaki et al., 2005). CDH1-positive breast cancer cells were examined to determine whether CTCF was bound to the CDH1 promoter when transcriptionally active. Again, we found CTCF binding at the immediate upstream (∼200 bp) region of a gene commonly methylated in cancer (Figure 6B, middle panels). As with the p16 and RASSF1A genes, CTCF binding was not detectable in cells where CDH1 is hypermethylated (Figure 6B, middle panels). Interestingly, although CTCF is absent from the RASSF1A and CDH1 promoters in MDA-MB-231 cells, it is still bound to its c-Myc site (Figure 6B, lower panels). This is consistent with our observations in other cell lines and indicates that CTCF in these cells is still functional to bind a subset of its target promoters even if it can no longer interact with specific tumor suppressor genes.
The RARβ32 gene is another common target of hypermethylation in breast cancer (Bovenzi et al., 1999) and, along with RASSF1A, may be a useful marker of increased breast cancer risk. However, upon examination of RARβ2-positive cells by ChIP we were unable to find CTCF association within the RARβ2 promoter or regions upstream (Figure 6C). From these data we conclude that loss of CTCF binding from critical sites is a common feature of several genes that are frequently silenced in human cancers, however this correlation does not apparently exist for all targets of aberrant hypermethylation.
Discussion
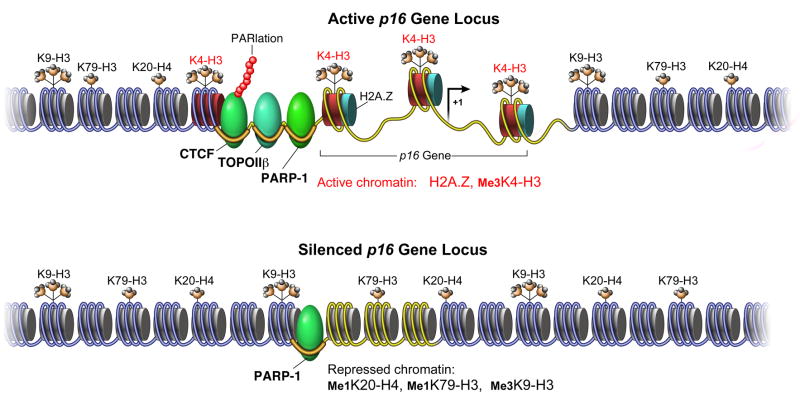
Our studies reveal an epigenetic mechanism of p16 transcriptional control that is deregulated when the gene is aberrantly silenced in human cancer cells (Figure 7). We observed that the chromatin structure surrounding the active p16 gene is highly organized with a discrete partition of histone modifications at approximately 2 kb upstream of the transcription start site. We were surprised to find highly enriched marks of repressed chromatin so close to a transcriptionally active gene. In p16-silenced breast cancer cells, partitioning of the p16 upstream region into distinct chromatin domains is lost and accompanied by disappearance of H2A.Z and Me3H3K4 within 2 kb of the inactivated promoter. This demonstrates that deregulation of epigenetic processes at silenced genes is not limited to DNA methylation and histone modification but can include placement of variant histones like H2A.Z. Upon loss of the -2 kb boundary, repressive histone marks spread throughout the entire p16 promoter region. Overall, our analyses indicate that a chromatin boundary exists upstream of the p16 gene which is destabilized in certain human cancer cells leading to aberrant transcriptional inactivation.
Figure 7.
Model of CTCF Function in Aberrant Tumor Suppressor Gene Silencing in Human Cancers. See text for discussion.
CTCF is a multifunctional protein that has previously been associated with establishing transitions between distinct chromatin domains and acting as a shield against the spread of heterochromatin (Bell et al., 1999). These studies, coupled with the detection of a chromatin boundary in the p16 upstream region led us to look for CTCF binding in the p16 promoter. Our analyses demonstrated that CTCF interaction with this region is strongly correlated with p16 transcription in a variety of human cell types. shRNA-knockdown of CTCF revealed that it plays an active role in maintaining p16 gene expression when associated near the upstream boundary, perhaps through stabilization of chromatin in this region. A dramatic loss of H2A.Z and gain of Me1H4K20 upon depletion of CTCF emphasizes an integral epigenetic organizational function for CTCF at this locus. It also suggests that CTCF facilitates the stabilization or deposition of this histone variant. Considering that both CTCF and H2A.Z may play important structural roles, we speculate that these two proteins may act cooperatively to organize nuclear chromatin in a spatial manner.
Intriguingly, we observed that CTCF is absent from the p16 upstream region in multiple types of human cancer cells where the p16 gene is silenced and methylated. We extended this finding to two other genes that are commonly silenced in cancer, RASSF1A and CDH1 (E-cadherin). Together, this indicates that loss of CTCF binding to critical regions may be a common event in epigenetic silencing of cancer-related genes. Indeed, CTCF has been suggested to regulate other tumor suppressor genes such as BRCA1 (Butcher et al., 2004) and RB (De La Rosa-Velazquez et al., 2007). Because CTCF is involved in blocking the spread of heterochromatin and directing interactions between chromosomes (Ling et al., 2006), its dissociation from these tumor suppressors may have multiple consequences that are detrimental to transcription and localized genomic stability. Furthermore, silencing of the p16 gene is an early step in breast carcinogenesis (Foster et al., 1998), which can lead to subsequent genomic instability (McDermott et al., 2006) and downstream methylation events (Reynolds et al., 2006). Thereby making CTCF a potentially critical target in tumor progression.
Surprisingly, we found that the probable cause of impaired CTCF binding to the p16 upstream region is defective poly(ADP-ribosyl)ation, resulting in the absence of CTCF PARlation in p16-silenced cells. Poly(ADP-ribosyl)ation has been shown to regulate multiple biological processes including DNA methylation, DNA repair, genotoxic stress, and epigenetic programming by post-translational modification of critical regulatory proteins and chromatin components (Kraus, 2008). In fact, inhibition of CTCF PARlation is correlated with failure to maintain IGF2 gene imprinting and insulator function in general (Yu et al., 2004).
We find that in p16-expressing cells, PARlated CTCF dissociates from PARP-1 and is complexed with cofactors Topo IIβ, Nucleophosmin, and a new interactor, Nucleolin. Nucleolin is a multifunctional protein with roles in cell membrane signaling, ribosomal RNA processing within the nucleolus, chromatin remodeling and transcription (Mongelard and Bouvet, 2007). The functional connection between PARP-1, Nucleolin and Nucleophosmin is very intriguing. These proteins have been isolated as a complex, and PARP-1 and Nucleolin have been shown to organize genomic DNA into topologically distinct domains through interaction with matrix/scaffold attachment regions that anchor chromatin onto the nuclear matrix (Galande, 2002). Strikingly, unPARlated CTCF fails to release from PARP-1 and loses its association with Nucleolin, but not Topo IIβ or Nucleophosmin. Such a complex is apparently insufficient to generate the p16 gene boundary even in the presence of chromatin-bound PARP-1. This is supported by previous work that underscored the importance of PARlation for proper CTCF insulator function (Yu et al., 2004). Our data also reveal that functionally distinct CTCF complexes associate with the p16 and c-Myc genes that differ in the requirement for specific cofactors. These include PARP-1 and Topo IIβ, which can coregulate transcription of some genes through transient DNA breakage and repair mechanisms (Ju et al., 2006). Thus, the absence of CTCF binding to p16, or other epigenetically silenced genes may result from defects in specific post-translational modifications, such as PARlation, or cofactor interactions without affecting the majority of CTCF genomic functions.
As expected, AZA treatment reactivates p16 transcription in non-expressing breast cancer cells. This was associated with an increase in Me3H3K4 and a decrease of Me3H3K9 but neither CTCF binding nor H2A.Z deposition was restored. These data may have clinical applications as AZA or AZA and HDAC inhibitors are incapable of completely restoring the normal histone code (McGarvey et al., 2006) or post-translational modifications of CTCF or other proteins to reestablish long-term expression of tumor suppressors, thus limiting their usefulness as therapeutic agents.
Overall, our results substantiate the critical role of CTCF in establishing and maintaining p16 and other tumor suppressor genes in higher-order chromosomal domains through appropriate boundary formation. These data raise the possibility that dissociation of CTCF from p16 during early tumorigenesis is not due to DNA methylation alone but may result from loss of PARlated CTCF that impairs the ability of CTCF to act as a functional component of a boundary or insulator element. This would result in secondary changes in chromatin structure that are incompatible with CTCF binding to DNA. When this integrity is breached by destabilized CTCF binding and loss of long-range epigenetic organization, aberrant gene silencing can ensue. Thus, the ability to restore CTCF interactions at vulnerable gene loci may have important therapeutic implications. Current efforts are now focused on targeted pharmacological intervention to restore CTCF PARlation and potentially reverse silencing of p16 and other tumor suppressor genes in human cancer cells.
Experimental Procedures
Cell Culture
All cell lines were maintained as described in Supplemental Materials.
Western Blotting and RT-PCR
Nuclear extracts were separated on 10% SDS-PAGE, transferred to nitrocellulose membranes, and blotted using various antibodies. For RT-PCR assays, cDNA was made from 500ng of total RNA using the Superscript II kit (Invitrogen). Antibody sources and primer sequences used to amplify specific genes and amplification conditions are available in Supplemental Materials.
Chromatin Immunoprecipitations
ChIPs were performed according to the Upstate Biotechnology protocol with some modifications as described in Supplemental Materials.
Co-Immunoprecipitations
2mg of whole cell lysates were diluted in IP buffer with 0.5% Triton X-100. Protein mixes were precleared for 1-2 hours with protein G Sepharose after which the beads were removed and CTCF (Upstate) or anto-phosphotyrosine (Upstate) antibody added overnight at 4°C to capture complexes. Complexes were recovered with protein G Sepharose, washed 4 times in IP buffer and subsequently analyzed by SDS-PAGE.
In Vitro Binding of PARP-1 and CTCF
Reactions were performed such that PARP-1 was catalytically active in presence of 1mM β-NAD+. Reaction buffer contained 20mM Tris-HCl, pH 8.0, 1mM MgCl2, 1mM DTT, 50ng salmon sperm DNA, 50ng BSA. 250ng of recombinant CTCF (isolated from overexpressing NIH3T3 cells) or PARP-1 (Alexis Biochemicals) protein was added where appropriate. Binding was carried out at 30°C for 1 hour. At this time reactions were diluted in 0.5% Triton IP buffer and CTCF was immunoprecipitated as described.
CTCF and PARP-1 Knockdown
CTCF knockdown was achieved using pSHAG-MAGIC2 retroviral vectors (OpenBiosystem) and PARP-1 knockdown using Mission Lentiviral shRNAs (Sigma) as described in Supplemental Materials.
Supplementary Material
Acknowledgments
This work was supported by grants to B.M.E. from The Samuel Waxman Cancer Research Foundation and to M.W. from The Canadian Institute of Health Research. We thank Dr. S.V. del Rincon for technical assistance with retroviral packaging, Dr. Michal Krawczyk for help with CTCF constructs and Dr. Fernando Lopez-Diaz for assistance with Lentiviral packaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Bovenzi V, Le NL, Cote S, Sinnett D, Momparler LF, Momparler RL. DNA methylation of retinoic acid receptor beta in breast cancer and possible therapeutic role of 5-aza-2′-deoxycytidine. Anticancer Drugs. 1999;10:471–476. doi: 10.1097/00001813-199906000-00007. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher DT, Mancini-DiNardo DN, Archer TK, Rodenhiser DI. DNA binding sites for putative methylation boundaries in the unmethylated region of the BRCA1 promoter. Int J Cancer. 2004;111:669–678. doi: 10.1002/ijc.20324. [DOI] [PubMed] [Google Scholar]
- De La Rosa-Velazquez IA, Rincon-Arano H, Benitez-Bribiesca L, Recillas-Targa F. Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res. 2007;67:2577–2585. doi: 10.1158/0008-5472.CAN-06-2024. [DOI] [PubMed] [Google Scholar]
- Di Vinci A, Perdelli L, Banelli B, Salvi S, Casciano I, Gelvi I, Allemanni G, Margallo E, Gatteschi B, Romani M. p16(INK4a) promoter methylation and protein expression in breast fibroadenoma and carcinoma. Int J Cancer. 2005;114:414–421. doi: 10.1002/ijc.20771. [DOI] [PubMed] [Google Scholar]
- Egger G, Aparicio AM, Escobar SG, Jones PA. Inhibition of histone deacetylation does not block resilencing of p16 after 5-aza-2′-deoxycytidine treatment. Cancer Res. 2007;67:346–353. doi: 10.1158/0008-5472.CAN-06-2845. [DOI] [PubMed] [Google Scholar]
- Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213–7218. [PubMed] [Google Scholar]
- Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299:1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- Foster SA, Wong DJ, Barrett MT, Galloway DA. Inactivation of p16 in human mammary epithelial cells by CpG island methylation. Mol Cell Biol. 1998;18:1793–1801. doi: 10.1128/mcb.18.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande S. Chromatin (dis)organization and cancer: BUR-binding proteins as biomarkers for cancer. Curr Cancer Drug Targets. 2002;2:157–190. doi: 10.2174/1568009023333917. [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, Sanchez-Cespedes M, Mendez J, Antequera F, Serrano M. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calabrese L, Caiafa P. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem. 2008;283:21873–21880. doi: 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Holst CR, Nuovo GJ, Esteller M, Chew K, Baylin SB, Herman JG, Tlsty TD. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–1601. [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- McDermott KM, Zhang J, Holst CR, Kozakiewicz BK, Singla V, Tlsty TD. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4:e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Ng MH, Chung YF, Lo KW, Wickham NW, Lee JC, Huang DP. Frequent hypermethylation of p16 and p15 genes in multiple myeloma. Blood. 1997;89:2500–2506. [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- Otterson GA, Khleif SN, Chen W, Coxon AB, Kaye FJ. CDKN2 gene silencing in lung cancer by DNA hypermethylation and kinetics of p16INK4 protein induction by 5-aza 2′deoxycytidine. Oncogene. 1995;11:1211–1216. [PubMed] [Google Scholar]
- Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- Qi CF, Martensson A, Mattioli M, Dalla-Favera R, Lobanenkov VV, Morse HC., 3rd CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc Natl Acad Sci U S A. 2003;100:633–638. doi: 10.1073/pnas.0237127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev. 2006;16:119–124. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, Hong C, Fridlyand J, Costello JF, Tlsty TD. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281:24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- Shinozaki M, Hoon DS, Giuliano AE, Hansen NM, Wang HJ, Turner R, Taback B. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005;11:2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- Silva J, Garcia JM, Pena C, Garcia V, Dominguez G, Suarez D, Camacho FI, Espinosa R, Provencio M, Espana P, Bonilla F. Implication of polycomb members Bmi-1, Mel-18, and Hpc-2 in the regulation of p16INK4a, p14ARF, h-TERT, and c-Myc expression in primary breast carcinomas. Clin Cancer Res. 2006;12:6929–6936. doi: 10.1158/1078-0432.CCR-06-0788. [DOI] [PubMed] [Google Scholar]
- Strunnikova M, Schagdarsurengin U, Kehlen A, Garbe JC, Stampfer MR, Dammann R. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol. 2005;25:3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, Tang SH, Silva FJ, Tsark WM, Mann JR. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol. 2004;24:4791–4800. doi: 10.1128/MCB.24.11.4791-4800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xue L, Weng M, Sun Y, Zhang Z, Wang W, Tong T. Sp1 is essential for p16 expression in human diploid fibroblasts during senescence. PLoS ONE. 2007;2:e164. doi: 10.1371/journal.pone.0000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, et al. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–1110. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Rubenstein JN, Jang TL, Pins M, Javonovic B, Yang X, Kim SJ, Park I, Lee C. Insensitivity to transforming growth factor-beta results from promoter methylation of cognate receptors in human prostate cancer cells (LNCaP) Mol Endocrinol. 2005;19:2390–2399. doi: 10.1210/me.2005-0096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.