Summary
Molecular epidemiological investigations have uncovered the patterns of emergence and global spread of Plasmodium falciparum resistance to chloroquine and sulfadoxine-pyrimethamine. Malaria parasites highly resistant to chloroquine and pyrimethamine spread from Asian origins to Africa, at great cost to human health and life. If artemisinin-resistant falciparum malaria follows the same pattern, renewed efforts to eliminate and eradicate malaria will be gravely threatened. This paper, adapted from a talk given in honour of Professor Malcolm Molyneux in Liverpool in September 2008, reviews the rise and fall of clinically important forms of drug-resistant falciparum malaria and considers how lessons learned from studying the evolution of drug-resistant malaria can be applied to efforts to prevent and deter resistance.
Keywords: Malaria, Plasmodium falciparum, Drug resistance, Molecular epidemiology, Molecular evolution, Eradication
In field studies conducted by David Clyde in Tanzania in the 1950s, pyrimethamine was administered monthly to inhabitants of a rural village and the ability of the drug to cure Plasmodium falciparum infection was measured. Initially, there were no treatment failures but the third monthly dose failed to clear 8% of falciparum malaria infections and by the fifth month of prophylaxis 37% of infections persisted in the face of a curative dose of pyrimethamine. Increasing the frequency of dosing to once a week resulted, within a year, in a firmly established focus of pyrimethamine resistance at rates of 50% or higher.1 Clyde mapped the spread of this resistance over time: after 8 years, 25–40% of P. falciparum infections occurring within 10–15 miles of the original focus were resistant; the prevalence of resistance dropped to 3–15% at a radius of 12–55 miles from the focus; resistance was rare more than 55 miles away; and the furthest point of detectable resistance was 150 miles from the original site of drug pressure.
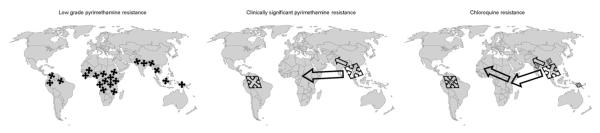
This pattern, of rapid focal emergence of pyrimethamine resistance in response to locally applied drug pressure followed by directly contiguous spread (Figure 1), was seen elsewhere in the context of mass pyrimethamine administration schemes and suggested that the underlying mechanism might be a relatively simple and common event, such as a point mutation in a single parasite gene. This suspicion was confirmed three decades later when pyrimethamine resistance was shown to be conferred by point mutations in P. falciparum dihydrofolate reductase (DHFR) adjacent to the active site of the enzyme where drug and substrate bind.2 Malaria parasites with a single mutation at DHFR amino acid position 108 were up to 300-fold more resistant to pyrimethamine in vitro, those with two additional mutations at positions 51 and 59 (the ‘triple mutant’) could be more than 2000-fold more resistant, and parasites carrying two or three of these mutations plus a fourth at DHFR position 164 were as much as 20 000-fold more resistant to pyrimethamine.
Figure 1.
Schematic representation of the patterns of origin and dissemination of low-grade resistance of Plasmodium falciparum to pyrimethamine caused by a single dihydrofolate reductase (DHFR) mutation that arose independently many times in response to local drug pressure (left); clinically significant pyrimethamine resistance caused by the ‘triple mutant’ DHFR, which may have arisen only twice, spreading from Asia to Africa (centre); and chloroquine resistance caused by the pfcrt K76T mutation on a variable background of other pfcrt mutations, which has arisen a handful of times, with the major Asian and African forms sharing a common Southeast Asian origin (right). Patterns of origin and spread are inferred from the chronology of historical reports of resistance and later molecular epidemiological analyses of the drug resistance-encoding genes and surrounding microsatellites and other genetic markers as described in references 7, 9 and 10 and other sources. Arrows represent approximate patterns of spread from original foci and are not an accurate representation of the current distribution of resistant parasites of different ancestral origins.
Early molecular epidemiological investigations suggested that the DHFR mutations occurred in a stepwise fashion, resulting in higher levels of resistance, based on the observation that each additional mutation associated with a higher level of resistance occurred only on the background of preceding mutations. A similar picture emerged for a stepwise accumulation of mutations in P. falciparum dihydropteroate synthase (DHPS) conferring resistance to sulfadoxine, the other sulfa drugs and sulfones.3 Molecular analyses of pre- and post-treatment parasites in clinical trials in Africa showed that a specific set of three DHFR and two DHPS mutations accounted for sulfadoxine-pyrimethamine treatment failure.4
By contrast with pyrimethamine resistance, chloroquine-resistant falciparum malaria seemed to have emerged just twice (Figure 1). Resistance was first reported in the late 1950s along both the Panama-Colombian and Thai-Cambodian borders and radiated slowly and inexorably outward from these two foci, taking 10 years to advance across Thailand to Burma, reaching central India by the late 1970s. Resistance more rapidly disseminated throughout the Amazon region, crossing southwards across Bolivia in the early 1960s and extending to the Atlantic coast of French Guiana and Surinam by the early 1970s.5 Chloroquine resistance appeared in East Africa in 1978 and moved westward across the continent in a less well-documented pattern owing to limited surveillance in Central and West Africa. Chloroquine efficacy was severely compromised throughout East Africa by the early 1990s, although chloroquine retained good efficacy in much of West Africa for another 10 years or more. The rare emergence and slow, contiguous spread of chloroquine resistance was interpreted as suggesting that the cause might be complex sets of genetic mutations that arose only very rarely.
This prediction was confirmed when chloroquine-resistant falciparum malaria was found to be caused by multiple mutations in a gene encoding a membrane transporter in the parasite’s food vacuole, pfcrt.6 Although genetic transformation experiments showed that a single lysine to threonine mutation at pfcrt codon 76 confers resistance, this mutation can only occur on a variable background of several other pfcrt mutations, which by themselves do not cause resistance but which must be present together for parasites with the resistance-causing mutation K76T to be viable. Analysis of the full sets of pfcrt mutations and surrounding microsatellites (non-coding variable-length repeat sequences) upstream and downstream from pfcrt demonstrated that South American and Southeast Asian pfcrt lineages were indeed distinct and that the Southeast Asian pfcrt had spread to Africa.7
Thus, the interpretation of the epidemiological observations of rare origin and contiguous spread as evidence of a rare and complex underlying genetic mechanism of chloroquine resistance appeared to have been correct. If Clyde’s early studies of the molecular epidemiology of antifolate-resistant malaria were also correctly interpreted to mean that antifolate-resistant malaria arose focally and locally in direct response to drug pressure, these different evolutionary patterns would have different implications for deterring resistance to chloroquine and the antifolates. Resistance that arose only rarely and spread globally, if detected early, might be stopped in its tracks by targeted efforts to eliminate a new focus of resistance before it disseminated. By contrast, containing forms of resistance that can emerge locally wherever drug pressure is applied would be virtually impossible.
Clyde’s field studies were repeated 40 years later with the added knowledge of the molecular basis of pyrimethamine resistance.8 Residents of a village in Mali were administered weekly pyrimethamine prophylaxis for 6 weeks. The prevalence of the DHFR 108 mutation in the study population rose from 10% at baseline to 100% in breakthrough infections occurring during the period of prophylaxis, and the prevalence of the DHFR triple mutant rose from 4% to 50%. Eight individuals, who had persistent asymptomatic infection with P. falciparum both at baseline and 1 week later, had only wild-type DHFR detectable at baseline but had at least two DHFR mutations just a week later, consistent with the possibility that the mutations arose spontaneously in response to drug pressure. However, microsatellite analyses demonstrated that these were complex, polyclonal infections, suggesting that the original infections might have contained small minorities of resistant DHFR that only reached a threshold of detection following their selection under drug pressure. But whether mutations arose spontaneously or, more likely, were present at very low levels and rapidly selected by pyrimethamine, this study seemed to support the idea that antifolate resistance could emerge rapidly and focally when drug pressure was applied.
It therefore came as a surprise when genetic analyses similar to those used to trace the global ancestry of chloroquine-resistant parasites were applied to study the dissemination of antifolate-resistant malaria throughout the Amazon region, and results showed that a single ancestral origin appeared to account for the most highly resistant forms of DHFR and DHPS. This suggested that regional spread from a single focus, akin to that seen with chloroquine, had occurred with sulfadoxine-pyrimethamine resistance in South America.9 Moreover, when Roper and colleagues used similar microsatellite methods to study the origins of Asian and African DHFR, they found that while the single-mutant DHFR A108N indeed occurred on many different genetic backgrounds in Africa and Asia, implying that these moderately resistant single-mutant forms of DHFR arose repeatedly and frequently in many locales, far fewer ancestral lineages of double-mutant DHFR were found and, remarkably, triple-mutant DHFR appeared to have arisen only twice, once in South America and once in Southeast Asia. In a striking parallel to the pattern of emergence and spread of chloroquine resistance, it was the Southeast Asian DHFR triple-mutant form that had spread to Africa, bringing with it clinically significant resistance to pyrimethamine that contributed to sulfadoxine-pyrimethemine treatment failure.10 These molecular epidemiological studies thus finally demonstrated that clinically important forms of drug-resistant malaria had very limited origins and then spread globally.
In an example of evolutionary forces driving resistance in the opposite direction, chloroquine resistance completely disappeared from southern Malawi just 8 years after chloroquine was replaced by sulfadoxine-pyrimethamine.11,12 Microsatellite analyses of the resurgent chloroquine-sensitive parasites indicate that this unexpected phenomenon represents an expansion of genetically diverse forms of wild-type sensitive pfcrt, not a backmutation or reversion of mutant pfcrt (M. Laufer, unpublished data). These observations suggest that chloroquine resistance carries a fitness cost and that sensitive parasites have a survival advantage in the absence of drug pressure.
One important public-health implication of these insights into the evolution of drug-resistant malaria is that if newly emerging resistance can be detected in time to block its regional and intercontinental spread, the useful life of important antimalarial drugs can potentially be prolonged. Unfortunately, this insight arrived too late to prevent the demise of chloroquine and sulfadoxine-pyrimethamine as first-line treatments for malaria in most of the world, but the lessons learned should be applied with alacrity to safeguard the efficacy of artemisinin-based combination therapies (ACT), as has been proposed through the creation of a World Antimalarial Resistance Network.13 While bona fide artemisinin resistance has not yet been proven, reports of waning ACT efficacy in Southeast Asia have raised the alarm14 and intensive efforts are underway to confirm the presence or absence of artemisinin resistance and, if present, to apply genomic approaches to identify the molecular mechanism and rapidly develop and deploy molecular resistance markers as tools for surveillance.15 If artemisinin resistance has emerged (or when it does) the historical lessons are clear — no effort should be spared in an aggressive, coordinated campaign to contain it before it disseminates globally, resulting in the stillbirth of the nascent global malaria eradication campaign. Mass screening and treatment of malaria is now being considered at the site of emergence of suspected artemisinin resistance in western Cambodia in the hope of eradicating resistance at its source. This could be the single most important public-health intervention in the history of malaria control, given the potentially catastrophic consequences of the global spread of artemisinin resistance just as headway against malaria is being made but before the pipeline of effective malaria drugs is primed.
Acknowledgments
Funding: The author is supported by the Howard Hughes Medical Institute and by a Distinguished Clinical Scientist Award from the Doris Duke Charitable Foundation. His current work on drug-resistant malaria is supported by grant 48821 from the Bill and Melinda Gates Foundation and by US National Institutes of Health grants U01AI44824, U01AI47858 and R34AI079284 from the National Institute of Allergy and Infectious Disease and D43TW01589 from the Fogarty International Center.
Footnotes
Conflicts of interest: None declared.
Ethical approval: Not required.
References
- 1.Clyde DF, Shute GT. Resistance of Plasmodium falciparum in Tanganyika to pyrimethamine administered at weekly intervals. Trans R Soc Trop Med Hyg. 1957;51:505–13. doi: 10.1016/0035-9203(57)90039-1. [DOI] [PubMed] [Google Scholar]
- 2.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–8. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plowe CV, Kublin JG, Doumbo OK. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist. 1998;1:389–96. doi: 10.1016/s1368-7646(98)80014-9. [DOI] [PubMed] [Google Scholar]
- 4.Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 5.Clyde DF. Genesis of chloroquine-resistant Plasmodium falciparum in the American region. Med Trop Cooperaz Sviluppo. 1987;3:41–4. [Google Scholar]
- 6.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–71. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–3. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 8.Doumbo OK, Kayentao K, Djimde A, Cortese JF, Diourte Y, Konaré A, et al. Rapid selection of Plasmodium falciparum dihydrofolate reductase mutants by pyrimethamine prophylaxis. J Infect Dis. 2000;182:993–6. doi: 10.1086/315787. [DOI] [PubMed] [Google Scholar]
- 9.Cortese JF, Caraballo A, Contreras CE, Plowe CV. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J Infect Dis. 2002;186:999–1006. doi: 10.1086/342946. [DOI] [PubMed] [Google Scholar]
- 10.Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 11.Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–5. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 12.Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–66. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 13.Plowe CV, Roper C, Barnwell JW, Happi CT, Joshi HH, Mbacham W, et al. World Antimalarial Resistance Network (WARN) III: molecular markers for drug resistant malaria. Malar J. 2007;6:121. doi: 10.1186/1475-2875-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alker AP, Lim P, Sem R, Shah NK, Yi P, Bouth DM, et al. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am J Trop Med Hyg. 2007;76:641–7. [PubMed] [Google Scholar]
- 15.Laufer MK, Djimde AA, Plowe CV. Monitoring and deterring drug-resistant malaria in the era of combination therapy. Am J Trop Med Hyg. 2007;77:160–9. [PubMed] [Google Scholar]