Abstract
Mitochondrial RNA turnover in yeast involves the degradosome, composed of DSS-1 exoribonuclease and SUV3 RNA helicase. Here, we describe a degradosome-like complex, containing SUV3 and DSS-1 homologues, in the early branching protozoan, Trypanosoma brucei. TbSUV3 is mitochondrially localized and co-sediments with TbDSS-1 on glycerol gradients. Co-immunoprecipitation demonstrates that TbSUV3 and TbDSS-1 associate in a stable complex, which differs from the yeast degradosome in that it is not stably associated with mitochondrial ribosomes. This is the first report of a mitochondrial degradosome-like complex outside of yeast. Our data indicate an early evolutionary origin for the mitochondrial SUV3/DSS-1 containing complex.
Keywords: RNA turnover, RNA processing, trypanosome, RNA helicase, exoribonuclease
1. Introduction
The degradation of RNA is an essential element in the regulation of gene expression. It both controls the abundance of mature RNAs and eliminates processing by-products and aberrant or defective molecules that form during RNA synthesis and maturation [1]. RNA degradation is of particular importance in mitochondria where transcriptional control is minimal and polycistronic transcription produces precursor RNAs that require extensive processing [2-5]. The machineries that catalyze RNA turnover in mitochondria exhibit substantial divergence between species [6]. In yeast, the mitochondrial degradosome is comprised of an RNR (RNase II/RNase R-like) family hydrolytic exoribonuclease, encoded by DSS-1 gene, and an NTP-dependent RNA helicase, encoded by the SUV3 gene. DSS-1 and SUV3 appear to be the sole components of the degradosome based on TAP-tagging studies in yeast, which also showed that the complex is exclusively associated with mitochondrial ribosomes [7]. The degradosome, which exhibits hydrolytic 3′ to 5′ exoribonuclease and RNA helicase activities, is the only known exoribonuclease involved in yeast mitochondrial RNA (mtRNA) turnover [8]. S. cerevisiae strains that are genetically inactivated for either DSS-1 or SUV3 have similar phenotypes, strongly accumulating excised introns as well as mRNA and rRNA precursors with abnormal 5′ and 3′ termini [9-11]. These cells also display decreased steady-state levels of mature transcripts along with disruption of translation [7,11,12]. Orthologues of the SUV3 helicase are present in the genomes of a wide spectrum of eukaryotes, and they have been shown to be at least partially mitochondrially localized in humans and plants [13-15]. In contrast to yeast, however, human and plant mitochondria lack the DSS-1 exoribonuclease. They do contain the phosphorolytic exoribonuclease polynucleotide phosphorylase (PNPase), although there is no evidence for its association with SUV3 [16,17]. A recent study demonstrated that human cells depleted of the SUV3 helicase accumulate shortened poly(A+) mtRNAs and are impaired in translation [18]. These studies indicate that SUV3 can profoundly affect mitochondrial RNA metabolism in the absence of a yeast-like degradosome complex.
Trypanosoma brucei is a protozoan parasite that has consistently been identified as one of the earliest branching mitochondria-containing eukaryotes [19]. Mitochondrial RNA metabolism in T. brucei is extraordinarily complicated, involving polycistronic transcription, extensively overlapping genes, and massive remodeling of mRNAs by guide RNA-directed uridine insertion/deletion editing [20]. We previously identified a gene encoding a homologue of DSS-1 in the T. brucei genome (termed TbDSS-1) [21], and a peptide originating from TbDSS-1 was recently detected in the mitochondrial proteome [22]. Targeted depletion of TbDSS-1 in insect stage T. brucei results in aberrant levels of several mitochondrial RNA species, including never edited, unedited and edited mRNAs as well as guide RNAs [21]. TbDSS-1 depleted cells also accumulate RNA maturation by-products originating from the region upstream of the first genes on the major and minor strands of the mitochondrial genome, and 12S rRNA processing intermediates with mature 3′ ends and unprocessed 5′ ends [23]. Overall, these studies suggest that TbDSS-1 represents at least one of the main exoribonucleases involved in RNA turnover and surveillance in T. brucei mitochondria. In the present study, we report a T. brucei homologue of the SUV3 RNA helicase (TbSUV3). To determine whether TbSUV3 interacts with TbDSS-1 in a mitochondrial degradosome-like complex, we created a T. brucei cell line expressing a PTP (ProtC-TEV-ProtA [24]) tagged TbSUV3 protein at an endogenous allele. We show that the TbSUV3-PTP fusion protein is properly expressed and targeted to the mitochondrion. Glycerol gradient fractionation suggests that TbSUV3 and TbDSS-1 co-sediment in a high-molecular-weight complex, and subsequent IgG purification of TbSUV3-PTP containing complexes shows that the two proteins interact in T. brucei mitochondria. These studies represent the first report of a core enzymatic complex that is likely involved in RNA turnover and surveillance in the mitochondria of T. brucei. Further, this is the first report of a mitochondrial degradosome-like complex in an organism other than yeast. Our data demonstrate an early evolutionary origin for the mitochondrial SUV3/DSS-1 containing complex.
2. Materials and methods
2.1. Oligonucleotides used for 5′ RACE analysis
The oligonucleotides used for 5′ RACE are listed as follows with restriction sites underlined. CSL-22 (5′-GCATCGATGCTATTATTAGAACAGTTTCTGTACTATATTG -3′), SUV3-8 (5′-GCGGATCCAACGCCGCGTGAGTCTTCC-3′), SUV3-9 (5′-GCGGATCCGTGCCTTCGGGTACCAGTC-3′).
2.2. Trypanosome cell culture, transfection, and cell fractionation
The procyclic form (PF) T. brucei brucei clone IsTAR1 stock EATRO 164 was grown as previously described [25]. Stable cell lines constitutively expressing a TbSUV3 C-terminal PTP tag fusion protein were generated via electroporation. To generate the pC-PTP-TbSUV3 construct, a 500-nucleotide fragment of TbSUV3 C-terminal coding region was PCR amplified using TbSUV3-PTP5′ (5′-GCCGGGGCCCAAGACCTCAGGTGTGGTGCC-3′) forward and TbSUV3-PTP3′ (ATAAGAATGCGGCCGCGGCAACCTCCGCAACAGCTC-3′) reverse primers and cloned into the Apal /Not l restriction sites of the pC-PTP-Neo vector [24] (a generous gift from Arthur Günzl, Univ. of Connecticut). For genomic integration, pTbSUV3-PTP-NEO was linearized within the TbSUV3 sequence at a unique Bcl I restriction site. For transfection, log-phase PF T. brucei clone IsTAR1 stock EATRO 164 cells were electroporated in the presence of twenty micrograms of Bcl I linearized TbSUV3-PTP. Transfections were carried out on ice in 2-mm cuvettes using a Bio-Rad electroporator with two pulses at the following settings: 800 V, 25 μF, and 400 Ω. Following transfection, cells were selected with 40 μg of G418/ml and clonal cell lines were generated by limiting dilution. Expression of PTP-tagged protein was analyzed by western blotting with PAP probe (Sigma), which detects the Protein A domain of the PTP tag.
Mitochondria were isolated by the procedure of Harris, et al. [26]. Whole cell and cytoplasmic fractionation was carried out using the procedure of Zeiner, et al. [27]. The degree of cytoplasmic contamination of the mitochondrial preparation was assessed by western blotting using antibodies against cytoplasmic TbHsp70.4.
2.3. Glycerol gradient sedimentation
Glycerol gradient fractionation of mitochondrial lysates was performed as previously described [28]. Mitochondrial lysate was obtained from 1010 PF T. brucei cell equivalents by adding 500 μl of mitochondrial lysis buffer (20mM Tris-HCl (pH 7.5), 50 mM KCl, 10mM MgCl2, 100 μM ATP, 0.2% NP-40, Complete EDTA-free protease inhibitors (Roche)) to purified mitochondria and incubating on ice for 10 min prior to centrifugation at 13,000 × g for 15 min. Five hundred μl of purified mitochondrial lysate was layered onto a 12-ml 5-20% linear glycerol gradient and centrifuged for 20 h at 4°C in Beckman SW-41 rotor at 35,000 rpm. Twenty-four 500 μl fractions were collected from the top of the tube, and 20 μl of each fraction was analyzed by western blotting with PAP reagent for the detection of TbSUV3 and polyclonal anti-TbDSS-1 antibodies [21] to detect endogenous TbDSS-1. Standards (cytochrome c, 1.9S; bovine serum albumin, 4S; yeast alcohol dehydrogenase, 7.4S; catalase, 11S; and thyroglobulin, 19S) were fractionated in a parallel gradient and analyzed by SDS-PAGE and Coomassie blue staining.
2.4. Immunoprecipitation of the TbSUV3 containing complex
For IgG purification of TbSUV3-PTP, peak glycerol gradient fractions (7-13) were pooled and fresh protease inhibitor tablet was added. Five hundred μl of pooled gradient fractions was incubated with 20 μl IgG Fastflow Sepharose beads (Amersham Biosciences) for 2 h at 4°C. The bound material was pelleted by centrifugation at 10,000 × g for 5 min and the unbound supernatant transferred to a separate tube. The Sepharose beads were then washed three times with PA-150 buffer (20mM Tris-HCl (pH 7.7), 150 mM KCl, 3 mM MgCl2, 0.5 mM DTT, 0.1% Tween20) and resuspended in Buffer A (10mM Tris-HCl (pH 8.0), 25mM KCl, 10mM MgCl2). The IgG Sepharose beads were boiled at 95°C for 5 min prior to loading on SDS-PAGE. Ten percent of each fraction was analyzed by western blot to detect TbDSS-1 and TbSUV3-PTP using anti-TbDSS1 antibodies [21] and PAP reagent, respectively.
Immunoprecipitations using anti-ProtC antibodies were performed as described above with the following modifications. Ten μg of anti-ProtC antibody (Roche monoclonal HPC4) was incubated with 500 μl of pooled gradient fractions in 2 mM CaCl2 with rocking at 4°C for 2 h. Control reactions in the absence of antibody were processed in parallel. Twenty μl of Protein G Sepharose beads were then added to each tube and the slurry was incubated with rocking for an additional 2 h at 4°C. The bound material was pelleted by centrifugation at 10,000 × g for 5 min and unbound supernatant transferred to a separate tube. The Sepharose beads were then washed three times with PC-150 buffer (20mM Tris-HCl (pH 7.7), 150 mM KCl, 3 mM MgCl2, 1mM CaCl2, 0.1% Tween20), resuspended in buffer A and boiled at 95°C for 5 min prior to loading on SDS-PAGE. Ten percent of each fraction was analyzed by western blot as described above.
3. Results and discussion
3.1 TbSUV3 encodes a putative ATP-dependent RNA helicase that localizes to the mitochondria
We previously described the role of the mitochondrial exoribonuclease, TbDSS-1, in RNA stability and as part of an RNA surveillance system that eliminates stalled 12S rRNA processing intermediates and maturation by-products from the system [21,23]. To determine if T. brucei possesses an SUV3 homologue that might act in concert with TbDSS-1, analogous to the yeast mitochondrial degradosome, we searched T. brucei GeneDB with the S. cerevisiae SUV3 sequence. We identified a predicted protein highly homologous to SUV3, which we term TbSUV3 (Tb927.4.1990). To confirm its expression and characterize the corresponding mRNA, we performed 5′ RACE on oligo(dT)-primed PF RNA using a gene-specific primer in combination with a primer homologous to the 39 nt spliced leader sequence present on all T. brucei nuclear encoded mRNAs. This analysis indicated the presence of a mature trans spliced TbSUV3 mRNA with an 18 nt 5′ untranslated region. To confirm the open reading frame (ORF) identified in silico, the predicted ORF was PCR amplified from PF T. brucei cDNA, cloned, and sequenced. The 1,881-nt ORF is identical to the sequence in T. brucei Gene DB with the exception of an alanine to valine substitution at amino acid 617, and predicts a protein with a molecular mass of 70.6 kDa and a pI of 9.01. The predicted protein exhibits 32-38% identity and 50-54% similarity with SUV3 homologues from Homo sapiens, Saccharomyces cerevisiae Arabidopsis thaliana, and Caenorhabditis elegans [15] over the conserved central region (Fig. 1A). SUV3-like proteins form a distinct and conserved Ski2p family of DExH/D RNA helicases that possess seven conserved motifs within their central region. Depicted in Fig. 1B is a sequence alignment of Motifs I to VI of SUV3 proteins from T. brucei, H. sapiens, S. cerevisiae, A. thaliana, and C. elegans. Motifs I and II, also known as the Walker A and B box, are crucial for ATPase and helicases activities [29]. Motifs Ia and Ib are thought to be involved in RNA binding in association with motifs IV and V, although this has only been demonstrated for eIF4A [30]. Interestingly, Motif III is characteristically absent in all SUV3 helicases, including TbSUV3. Together, these analyses indicate that T. brucei expresses an SUV3 homologue.
Fig. 1.
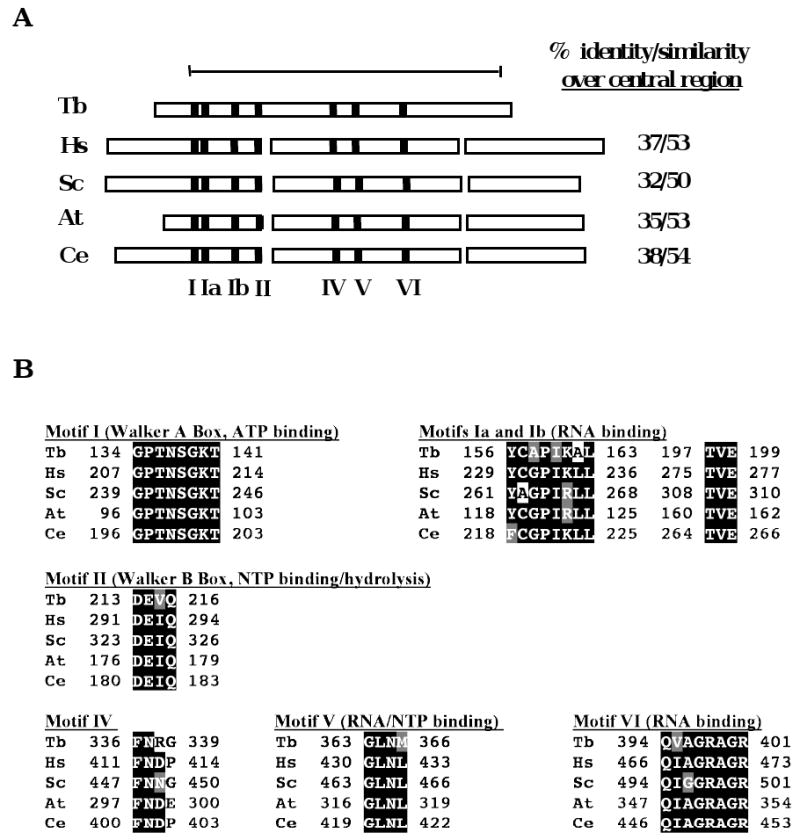
TbSUV3 is a putative ATP-dependent RNA helicase with conserved motifs. (A) Schematic representation of the structure of SUV3 homologues from T. brucei (Tb; accession no. XP844349), H. sapiens (Hs; accession no. NP_003162), S. cerevisiae (Sc; accession no. NP015296), A. thaliana (At; accession no. BAF01552), and C. elegans (Ce; accessin no. NP001040911) illustrating the presence of conserved motifs (Motifs I, Ia, Ib, II, IV, V, and VI) and variable N- and C-termini. The percent amino acid identity/similarity over the conserved central region (encompassing amino acids 115 to 602 of TbSUV3) is shown. (B) A multiple sequence alignment of Motifs I-VI of SUV3 homologues shown in panel A was generated using ClustalW. Identical amino acids are indicated by white letters on a black background; similar amino acids are indicated by white letters on a gray background.
We next wanted to confirm the expected mitochondrial localization of TbSUV3. TbSUV3 is predicted to contain a mitochondrial import sequence at its N-terminus by the PSORTII program (http://psort.ims.u-tokyo.ac.jp/). In addition, the N-terminus of the TbSUV3 ORF exhibits characteristic regions of homology to known trypanosome mitochondrial import sequences [21,31,32], including tandem arginine residues followed by multiple hydrophobic amino acids with interspersed and flanking serine and threonine residues (Fig. 2A). To determine whether TbSUV3 is mitochondrially localized, we used epitope tagging to create a TbSUV3 fusion protein in T. brucei. We utilized a construct designed for stable integration of a PTP tag into an endogenous allele [24], which allows PTP-tagged proteins to be expressed at normal levels, potentially alleviating problem of nonspecific protein-protein interactions due to overexpression of the tagged protein. To this end, a 500-bp fragment of the TbSUV3 C-terminal coding region was cloned into pC-PTP-Neo vector [24]. The resulting vector was linearized within the TbSUV3 sequence at a unique restriction site and transfected into T. brucei EATRO 164 strain. Cultures that had integrated the plasmid construct into an endogenous TbSUV3 allele were selected by G418 treatment, and clonal lines subsequently isolated by limiting dilution. Western blot analysis using the PAP reagent, which recognizes the Protein A domain in the PTP tag, confirmed that a protein of the correct size (∼84 kDa) was expressed in TbSUV3-PTP cells (Fig. 2B, top panel, compare lanes 1 and 2). Total protein, cytosolic, and mitochondrial extracts were prepared from TbSUV3-PTP cells, and equal microgram amounts of protein were analyzed using the PAP reagent. TbSUV3-PTP was detected only in the total protein and mitochondrial fractions (Fig. 2B, lanes 2 and 4), and not in the cytosolic fraction (Fig. 2B, lane 3). Moreover, TbSUV3-PTP was enriched in mitochondria compared to whole cells, demonstrating that it is targeted to the mitochondrion. Western blotting using anti-TbHsp70.4 (Fig. 2B, bottom panel) confirmed the efficiency of the mitochondrial fractionation. The presence of a putative mitochondrial import sequence and enrichment in mitochondrial extracts, lead us to conclude that TbSUV3 is a nuclear encoded, mitochondrially localized protein. Consistent with this conclusion, a recent survey of the T. brucei mitochondrial proteome identified several TbSUV3 peptides [22].
Fig. 2.
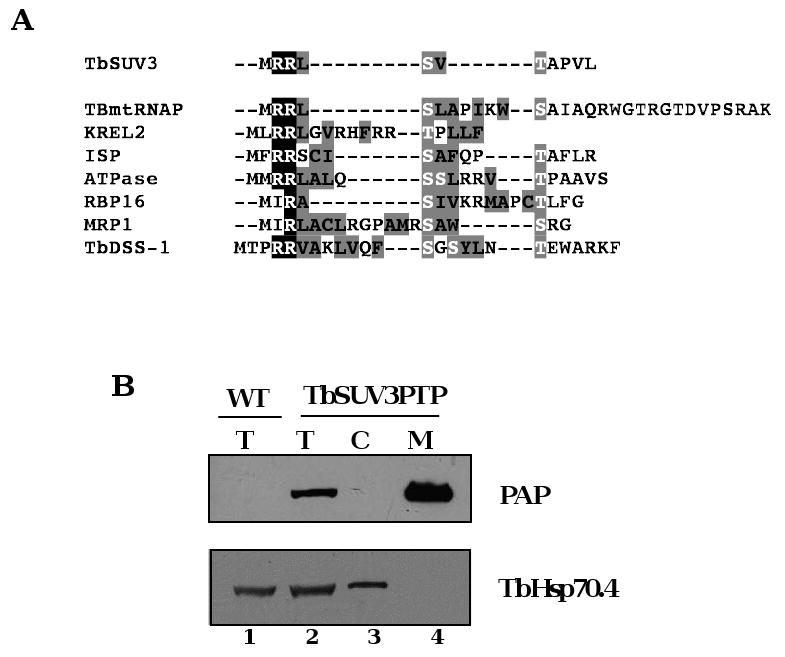
TbSUV3 is mitochondrially localized. (A) Sequence alignment of the TbSUV3 N terminal amino acid sequence with known and predicted targeting sequences from T. brucei mitochondrial proteins TBmtRNAP, KREL2, ISP, ATPase subunit 9, RBP16, MRP1, and TbDSS-1. The characteristic one or two N-terminal arginine residues are shown by white letters on black background. Serine and threonine residues (white letters on gray background) flank hydrophobic residues that are shown by black letters on gray background. (B) Mitochondrial localization of TbSUV3-PTP fusion protein. Wild type (WT) or TbSUV3-PTP expressing cells were analyzed by western blot with the PAP reagent to detect TbSUV3-PTP (top) or with antibody against TbHSP70.4 as a cytoplasmic marker (bottom). Each lane contains 10 μg protein. Total cell extracts (T, lanes 1 and 2); cytoplasmic extract (C, lane 3); mitochondrial extract (M, lane 4).
3.2 TbSUV3 is present in higher order complexes
To address whether TbSUV3 is present in a macromolecular complex, we subjected mitochondrial extract from cells expressing TbSUV3-PTP to glycerol gradient fractionation. Fractions from a 5 to 20% glycerol gradient were first resolved by SDS-PAGE and analyzed by western blot with the PAP reagent (Fig. 3, top panel). TbSUV3-PTP exhibited a broad distribution in fractions 5 to 19, sedimenting predominantly between 8 and 11.3S. Monomeric TbSUV3-PTP was expected to exhibit sedimentation values of less than 7.4S, suggesting the presence of TbSUV3-containing macromolecular complexes.
Fig. 3.
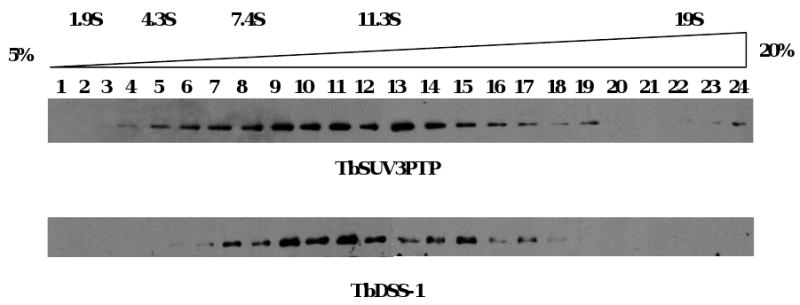
TbSUV3 and TbDSS-1 co-sediment in glycerol gradients. Mitochondrial extracts from TbSUV3-PTP expressing cells were fractionated on a 5 to 20% glycerol gradient. Aliquots of each fraction were analyzed by western blot using the PAP reagent to detect TbSUV3-PTP (top panel) and antibodies against TbDSS-1 (bottom panel). Sedimentation coefficients were determined by sedimentation of markers in a parallel gradient.
Previous glycerol gradient analysis demonstrated that TbDSS-1 is also present in higher order complexes, but unlike yeast DSS-1, is not stably associated with ribosomes [21]. To determine whether the sedimentation profile of TbDSS-1 overlaps or coincides with that of TbSUV3-PTP, we analyzed the same gradient fractions with antibodies against TbDSS-1. HRP conjugated goat anti-rabbit antibodies, which would not be expected to interact with the Protein A portion of the TbSUV3-PTP protein [33], were utilized as secondary antibodies. As shown in Fig. 3 (bottom panel), TbDSS-1 exhibits a broad sedimentation profile in fractions 7-17, corresponding to 8 to 10S, similar to TbSUV3-PTP. These results suggest that TbSUV3 and TbDSS-1 may be associated in mitochondrial complexes.
3.3 TbSUV3 interacts with TbDSS-1 in a mitochondrial degradosome-like complex
To determine whether TbSUV3 and TbDSS-1 are stably associated, we performed co-immunoprecipitation experiments. Peak glycerol gradient fractions (fractions 7-13, Fig. 3) were pooled and incubated with IgG Sepharose beads, which bind the Protein A moiety of the PTP tag. After washing, ten percent of the input, unbound, and bound proteins were analyzed by western blot with the PAP reagent to confirm TbSUV3-PTP precipitation and with anti-TbDSS-1 antibodies to assess their interaction. As shown in Fig. 4A, the majority of both TbSUV3 and TbDSS-1 were identified in the pellet, demonstrating an in vivo association of the two proteins. To confirm this interaction, we performed co-immunoprecipitation experiments with the same pooled gradient fractions and anti-ProtC antibodies directed against the ProtC moiety of the PTP tag. TbSUV3-PTP was precipitated using the anti-ProtC antibodies bound to Protein G Sepharose beads, and negative control experiments were performed in the absence of anti-ProtC antibodies to account for non-specific binding of TbDSS-1 to the beads. In these experiments, we detected both TbSUV3-PTP and TbDSS-1 in the pellet in the presence of anti-ProtC antibody, but not in controls performed in the absence of primary antibody (Fig. 4B). Collectively, these results demonstrate that TbSUV3 associates with TbDSS-1 in T. brucei mitochondria.
Fig. 4.
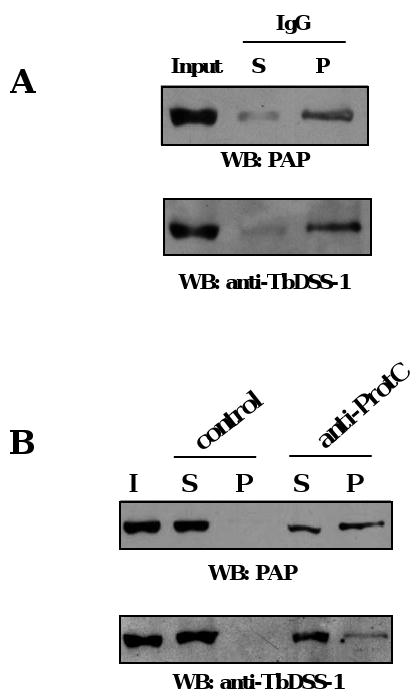
In vivo interaction between TbSUV3 and TbDSS-1. (A) Peak glycerol gradient fractions from TbSUV3-PTP expressing cells were pooled and incubated with IgG Sepharose beads. Beads were washed, and ten percent of input (I), supernatant (S), and pellet (P) fractions were analyzed by SDS-PAGE and western blot with the PAP reagent (top) and anti-TbDSS-1 (bottom). (B) TbSUV3-PTP was isolated from pooled glycerol gradient fractions by immunoprecipitation with anti-ProtC antibodies. Reactions lacking primary antibodies (control) were performed as a negative control. Ten percent of input (I), supernatant (S), and pellet (P) fractions were analyzed by western blot as in panel A.
3.4. Concluding remarks
This study reports the expression of an SUV3 RNA helicase homologue, TbSUV3, in the early branching eukaryote, T. brucei. We further demonstrate that TbSUV3 is localized to mitochondria where it associates with the TbDSS-1 exoribonuclease in a complex analogous to the S. cerevisiae mitochondrial degradosome [7]. This constitutes the first report of a DSS-1/SUV3 containing complex in an organism other than yeast. While we were unable to isolate enzymatically active TbSUV3 or TbDSS-1 from trypanosomes or bacteria, presumably due to the lability of these proteins, both proteins possess all of the residues necessary for their predicted enzymatic functions, indicative of a role in mitochondrial RNA metabolism. We were also unable to obtain TbSUV3 depleted cells for functional studies despite exhaustive attempts. Nevertheless, TbDSS-1 knockdown cells display pleitropic effects on trypanosome mitochondrial RNA metabolism similar to those described in DSS-1 or SUV3 null yeast cells [7,21,23]. Thus, our data reveal an early evolutionary origin for the mitochondrial degradosome. This complex has apparently been lost during evolution, as human SUV3 functions in mitochondrial RNA metabolism in the absence of any DSS-1 homologue [18]. It will be of future interest to determine the spectrum of organisms that possess a mitochondrial DSS-1/SUV3 containing complex, and to better understand the myriad functions of this important complex in vivo.
Acknowledgments
We thank Jay Bangs for antibodies and Arthur Günzl for the PTP vector. This work was supported by NIH R56AI047329 to LKR. JLM was supported in part by NIH T32AI07614.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Housely J, Tollervery D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Koslowsky DJ, Yahampath G. Mitochondrial mRNA 3′ cleavage/polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol Biochem Parasitol. 1997;90:81–94. doi: 10.1016/s0166-6851(97)00133-3. [DOI] [PubMed] [Google Scholar]
- 3.Montoya J, Gaines GL, Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983;34:151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- 4.Schafer B. RNA maturation in mitochondria of S. cerevisiae and S. pombe. Gene. 2005;354:80–85. doi: 10.1016/j.gene.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 5.Binder S, Brennicke A. Gene expression in plant mitochondria: transcriptional and post-transcriptional control. Philos Trans R Soc Lond B Biol Sci. 2003;358:181–188. doi: 10.1098/rstb.2002.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its componsition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 8.Malecki M, Jedrzejczak R, Stepien PP, Golik P. In vitro reconstitution and chracterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J Mol Biol. 2007;372:23–36. doi: 10.1016/j.jmb.2007.06.074. [DOI] [PubMed] [Google Scholar]
- 9.Margossian SP, Li H, Zassenhaus HP, Butow RA. The DexH box protein Suv3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell. 1996;84:199–209. doi: 10.1016/s0092-8674(00)80975-7. [DOI] [PubMed] [Google Scholar]
- 10.Stepien PP, Kokot L, Leski T, Bartnik E. The suv3 nuclear gene product is required for the in vivo processing of the yeast mitochondrial 21S rRNA transcripts containing the R1 intron. Curr Genet. 1995;27:234–238. doi: 10.1007/BF00326154. [DOI] [PubMed] [Google Scholar]
- 11.Golik P, Szczepanek T, Bartnik E, Stepien PP, Lazowska J. The S. cerevisiae nuclear gene SUV3 encoding a putative RNA helicase is necessary for the stability of mitochondrial transcripts containing multiple introns. Curr Genet. 1995;28:217–224. doi: 10.1007/BF00309780. [DOI] [PubMed] [Google Scholar]
- 12.Rogowska AT, Puchta O, Czarnecka AM, Kaniak A, Stepien PP, Golik P. Balance between transcription and RNA degradation is vital for Saccharomyces cerevisiae mitochondria: reduced transcription rescues the phenotype of deficient RNA degradation. Mol Biol Cell. 2006;17:1184–1193. doi: 10.1091/mbc.E05-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minczuk M, Piwowarski J, Papworth MA, Awiszus K, Schalinski S, Dziembowski A, Dmochowska A, Bartnik E, Tokatlidis K, Stepien PP, Borowski P. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucl Acids Res. 2002;30:5074–5086. doi: 10.1093/nar/gkf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagliardi D, Kuhn J, Spadinger U, Brennicke A, Leaver CJ, Binder S. An RNA helicase (AtSUV3) is present in Arabidopsis thaliana mitochondria. FEBS Lett. 1999;458:337–342. doi: 10.1016/s0014-5793(99)01168-0. [DOI] [PubMed] [Google Scholar]
- 15.Dmochowska A, Kalita K, Krawczyk M, Golik P, Mroczek K, Lazowska J, Stepien PP, Bartnik E. A human putative Suv3-like RNA helicase is conserved between Rhodobacter and all eukaryotes. Acta Biochem Pol. 1999;46:155–162. [PubMed] [Google Scholar]
- 16.Holec S, Lange H, Kuhn K, Alioua M, Borner T, Gagliardi D. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol Cell Biol. 2006;26:2869–2876. doi: 10.1128/MCB.26.7.2869-2876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slomovic S, Schuster G. Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khidr L, Wu G, Davila A, Procaccio V, Wallace D, Lee WH. Role of SUV3 helicase in maintaining mitochondrial homeostatis in human cells. J Biol Chem. 2008;283:27064–27073. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dacks JB, Doolittle WF. Reconstructing/deconstructing the earliest eukaryotes: how comparative genomics can help. Cell. 2001;107:419–425. doi: 10.1016/s0092-8674(01)00584-0. [DOI] [PubMed] [Google Scholar]
- 20.Lukes J, Hashimi H, Zikova A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- 21.Penschow JL, Sleve DA, Ryan CM, Read LK. TbDSS-1, an essential Trypanosoma brucei exoribonuclease homolog that has pleiotropic effects on mitochondrial RNA metabolism. Eukaryot Cell. 2004;3:1206–1216. doi: 10.1128/EC.3.5.1206-1216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9:434–50. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattiacio JL, Read LK. Roles for TbDSS-1 in RNA surveillance and decay of maturation by-products from the 12S rRNA locus. Nucl Acids Res. 2008;36:319–329. doi: 10.1093/nar/gkm690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schimanski B, Nguyen TN, Gunzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brun R, Schonenberger R. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- 26.Harris ME, Moore DR, Hajduk SL. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- 27.Zeiner GM, Sturm NR, Campbell DA. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot Cell. 2003;2:222–230. doi: 10.1128/EC.2.2.222-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goulah CC, Read LK. Differential effects of arginine methylation on RBP16 mRNA binding, guide RNA (gRNA) binding, and gRNA-containing ribonucleoprotein complex (gRNP) formation. J Biol Chem. 2007;282:7181–7190. doi: 10.1074/jbc.M609485200. [DOI] [PubMed] [Google Scholar]
- 29.Shu Z, Vijayakumar S, Chen CF, Chen PL, Lee WH. Purified human SUV3pexhibits multiple-substrate unwinding activity upon conformational change. Biochemistry. 2004;43:4781–4790. doi: 10.1021/bi0356449. [DOI] [PubMed] [Google Scholar]
- 30.Rogers GW, Jr, Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucl Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 31.Hausler T, Stierhof YD, Blattner J, Clayton C. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma, and Trichomonas. Eur J Cell Biol. 1997;73:240–251. [PubMed] [Google Scholar]
- 32.McManus MT, Shimamura M, Grams J, Hajduk SL. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonifacino JS, Esteban CD, Springer TA. Immunoprecipitation. In: Ausubel FM, et al., editors. Current Protocols in Molecular Biology. Vol. 2. John Wiley & Sons, Inc.; 1999. Chapter 10.16.1. [Google Scholar]


