Abstract
Changes in intracellular calcium in pea root hairs responding to Rhizobium leguminosarum bv. viciae nodulation (Nod) factors were analyzed by using a microinjected calcium-sensitive fluorescent dye (dextran-linked Oregon Green). Within 1–2 min after Nod-factor addition, there was usually an increase in fluorescence, followed about 10 min later by spikes in fluorescence occurring at a rate of about one spike per minute. These spikes, corresponding to an increase in calcium of ≈200 nM, were localized around the nuclear region, and they were similar in terms of lag and period to those induced by Nod factors in alfalfa. Calcium responses were analyzed in nonnodulating pea mutants, representing seven loci that affect early stages of the symbiosis. Mutations affecting three loci (sym8, sym10, and sym19) abolished Nod-factor-induced calcium spiking, whereas a normal response was seen in peas carrying alleles of sym2A, sym7, sym9, and sym30. Chitin oligomers of four or five N-acetylglucosamine residues could also induce calcium spiking, although the response was qualitatively different from that induced by Nod factors; a rapid increase in intracellular calcium was not observed, the period between spikes was lower, and the response was not as sustained. The chitin-oligomer-induced calcium spiking did not occur in nodulation mutants (sym8, sym10, and sym19) that were defective for Nod-factor-induced spiking, suggesting that this response is related to nodulation signaling. From our data and previous observations on the lack of mycorrhizal infection in some of the sym mutants, we propose a model for the potential order of pea nodulation genes in nodulation and mycorrhizal signaling.
A nitrogen-fixing root nodule is the culmination of a developmental program that begins as a sequence of signal exchanges between plant and bacterial symbionts. In leguminous plants, the first step of this process is rhizobial perception of plant metabolites (typically flavonoids) present in the rhizosphere (1). This stimulates bacterial synthesis and secretion of signaling molecules called Nod factors (2, 3), which are lipochitin oligosaccharides usually containing four or five β-1,4-linked N-acetylglucosamine residues with an N-acyl group at the nonreducing end (2). Modifications made to this basic structure are a major factor in determining which legume species will be nodulated by a given rhizobial strain. Thus, for example, the Nod factors made by Sinorhizobium meliloti (which nodulates alfalfa) differ from those of Rhizobium leguminosarum bv. viciae (which nodulates peas and vetch) in the length and unsaturation of their acyl chains and have an O-linked sulfate group at the reducing end (4, 5). R. leguminosarum bv. viciae produces a mixture of four Nod factors in which a tetrameric or pentameric sugar backbone carries a C18:1 or C18:4 N-linked acyl group and an O-acetyl group on the acylated sugar (5).
Nod factors induce several characteristic biochemical, genetic, and morphological responses when recognized by the appropriate legume (6). Changes in root-hair morphology, activation of cortical cell division, and the formation of cytoplasmic bridges occur over a period of hours to days and are readily correlated with the nodulation process (4, 7, 8). More rapid responses, such as ion fluxes at the root-hair plasma membrane and associated partial membrane depolarization, are seen less than a minute after Nod-factor addition (9–12). As yet, there is relatively little evidence about the precise role of these rapid responses, and there is, indeed, no formal proof that they are implicitly required for nodulation to proceed. Their potential significance can be inferred because of their induction by very low levels of Nod factor and their specificity for Nod-factor structure. Another response, seen in the root hairs of alfalfa and other legumes after application of Nod factor, is calcium spiking (10, 13).
The term calcium spiking has been used in mammalian systems to describe the repetition of sharp transient oscillations in intracellular calcium concentration ([Ca2+]int) (14, 15). Many nonexcitable mammalian cells exhibit calcium spiking, induced in response to the perception of an agonist (typically a hormone). This phenomenon has been linked to the activation of gene expression (16, 17). There are few examples of calcium spiking in plant cells, although it appears to play a role in regulating stomatal guard cell aperture (18), and calcium oscillations are prominent in pollen tubes where pulses in [Ca2+]int at the tip are apparently coincident with growth (19–21). In alfalfa root hairs, calcium spiking starts about 10 min after the addition of S. meliloti Nod factor, with increases in [Ca2+]int occurring mostly around the nuclei (13). This spiking response was specific for the Nod-factor structure (Nod factors lacking the O-linked sulfate at the reducing end were inactive) and did not occur in a nonnodulating mutant of alfalfa, indicating a role in Nod-factor signal transduction (13).
There is indirect evidence to suggest a G protein-mediated signaling system may operate in Nod-factor signal transduction. Mastoparan (a G protein agonist widely used in mammalian studies) and Nod factors stimulate expression of the early nodulin gene MtENOD12-GUS in transgenic alfalfa in a similar way (22). Additionally, pertussis toxin (a G protein antagonist) blocked Nod-factor- and mastoparan-induced expression, and the phospholipase C antagonists neomycin and U73122 interfered with this activity (22). These results are compatible with a model analogous to that described in mammalian cells, where recognition of an agonist by a receptor stimulates G protein activation, which, in turn, activates phospholipase C. Phospholipase C cleaves the lipid phosphatidylinositol 4,5-bisphosphate to yield the secondary messengers inositol 1,4,5-trisphosphate (InsP3) and diacylglycerol. InsP3-sensitive channels present on intracellular calcium stores (organelles) are activated, stimulating the release of Ca2+.
We have investigated the intracellular calcium responses of pea root hairs responding to Nod factors and N-acetylglucosamine oligomers and have investigated calcium spiking in various pea nodulation mutants to relate the spiking response to the sequence of events in Nod-factor signal transduction.
Materials and Methods
Plant Material.
The pea lines used have all been described. E69 and R25 are derived from var. Sparkle and carry mutations affecting sym7 and sym8, respectively (23). Mutant lines P4, P5, P53, P54, P55, and P56 are derivatives of var. Frisson (24, 25). P4 and P55 belong to complementation group c (24, 25), which corresponds to the sym19 locus (26). P5 and P56 are in complementation group d (24, 25) and carry alleles of sym10. P54 is in complementation group b (24, 25) and carries an allele of sym9. P53 is in complementation group a (24, 25) and carries an allele of sym30 (M. Sagan and G. Duc, personal communication).
To facilitate the use of microscope lenses with short working distances, lateral (secondary) roots were used. Pea seeds were surface-sterilized and germinated on FP agar as described (27). After 4 days of growth, the primary root tip was removed, and the seedling was transferred to the edge of a 10-cm square Petri dish containing FP agar and 1 μM aminoethoxyvinyl glycine. A hole had been cut in the side of the dish next to the seedling to allow outgrowth of the shoot. The plates were covered with black plastic and held at a 45° angle at room temperature in ambient light conditions to allow the formation of lateral roots. After 5 days, lateral roots were aseptically removed (2.5 cm from the tip) and fixed to the base of 3.5-cm diameter Petri dishes with semimolten FP agar. Root sections were then immersed in 1 ml of buffer containing 6.8 mM CaCl2⋅2H2O, 7.4 mM KH2PO4, 10.6 mM Na2HPO4, 0.15 mM FeC6H6O7⋅5H2O, and 150 mM sorbitol at pH 7.2. This buffer was derived from the major components of FP liquid (in which peas nodulate normally), with the optimum concentration of sorbitol and pH derived empirically (data not shown). Dishes containing root sections were then attached to microscope slides that could be held on the microscope stage. Nod factors, N-acetylglucosamine oligomers (Sigma), and mastoparan (Sigma) were added directly to the buffer covering the root section. Nod factors were prepared and quantitatively estimated essentially as described (5). The stock preparation used contained NodRlv-IV (Ac, C18:4) at a concentration of 10−5 M. There was also some NodRlv-V(Ac, C18:1) present in this stock (corresponding to 10% of the total amount of Nod factor).
Root-hair deformation assays were done with root-tip-excised seedlings that were grown aeroponically in liquid FP medium inoculated with R. leguminosarum bv. vicae strain A34 carrying the lacZ expression plasmid pXLGD4 (28) to enhance detection of infection threads. Aeroponic growth was achieved by threading the seedling roots through holes in a layer of aluminum foil covering a 1-liter beaker containing 400 ml of liquid medium, which was bubbled with filtered air passed through a scintered glass sparger. Plants were scored 24 and 72 h after inoculation.
Fluorescence Imaging.
Oregon Green-dextran (Mr = 10,000) or Texas Red-dextran (Mr 10,000) from Molecular Probes was dissolved in sterile water to a final concentration of 5 mM and frozen in 1-μl aliquots. For each experiment, an aliquot was made up to a final volume of 10 μl with sterile water and 2 μl of injection buffer (13) and centrifuged at 11,000 × g in a microcentrifuge for 5 min before removing the upper 5 μl for injection. For most measurements, only Oregon Green was used, but for ratiometric analysis, 1-μl aliquots of both dyes were mixed before making the final volume up to 10 μl. Needles were loaded via capillary action, and root hairs were impaled and pressure-microinjected under a Nikon Optiphot-2 microscope equipped with a ×40 Nikon superlong working distance lens, essentially as described (21). Briefly, micropipettes were pulled from nonfilamented capillary tubes (Clark Electromedical Instruments, Pangbourne, U.K.) with a micropipette puller (Campden Instruments, Loughborough, U.K.). The micropipette was front-loaded with dye by capillary action. A Narishige four-dimensional micromanipulator (model MO-204 coupled to MN-3) was used to position the micropipette and impale root hairs that were visualized with an upright Nikon Optiphot-2 microscope and a ×40 Nikon superlong working distance lens. Dye was introduced into root hairs by increasing the pressure in the micropipette with an oil-filled hydraulic pressure injector (Micro Instruments, Witney, U.K.). Cytoplasmic streaming was used as a measure of cell viability, and only cells actively streaming were used for imaging.
Cells loaded with dye were imaged with a Bio-Rad MRC-1024 confocal microscope built around the Nikon Optiphot-2 microscope. A Zeiss ×40 variable-media immersion lens was used to view the injected cells. Oregon Green was excited with the 488-nm spectral line from a 100-mW argon-ion laser using a Bio-Rad UBHS filter block (351-to 365-nm/488-nm polychroic) with the power typically set at 3% or less. Emitted fluorescent light was directed to a photomultiplier tube passing through a 515-nm long-pass filter. Ratiometric experiments using Oregon Green and Texas Red required the 488-nm and 568-nm spectral lines from a 25-mW argon-krypton laser and Bio-Rad T1 (488-nm/568-nm/647-nm polychroic) and E2 (562-nm long-pass) filter blocks. Emitted fluorescent light was directed to two photomultiplier tubes passing through 585-nm long-pass and 522-nm/535-nm emission filters for the Texas Red and Oregon Green fluorescence, respectively. The short working distance of the Zeiss ×40 variable-media immersion lens meant that it had to be lowered into the buffer covering the root section. To prevent carry-over of Nod factors, the lens was covered in a single layer of clingfilm (Wrapfilm Systems, Telford, U.K.), which was replaced after each experiment. During an experiment, sets of images were usually taken at the following times before or after addition of Nod factor: −6, 0, +6, +12, +20, +30, and + 40 min. Each set had 60 images taken 5 s apart, and each image was a scan of 512 × 512 pixels. This approach was taken to allow minor adjustments in focus between sets and to limit the file sizes of the data generated.
Image Analysis.
Fluorescence intensities were measured retrospectively by using Bio-Rad TIMECOURSE software. Regions of interest were outlined on the images, and the average pixel intensity value was measured for that region for each picture in the data set. Changes in fluorescence were calculated essentially as described by Ehrhardt et al. (13). Quantitative ratiometric measurements for images taken with the Texas Red/Oregon Green dye mix were calculated by using Bio-Rad software. Regions of interest were highlighted on the Oregon Green and Texas Red images, and the ratio of the average pixel intensity between the two was calculated. Values were exported into EXCEL and referenced against a calibration series generated in vitro with buffered calcium standards (Molecular Probes), to give an approximate concentration of calcium for the region of interest.
Results
Calcium Spiking in Pea Root Hairs Is Induced by R. leguminosarum bv. viciae Nod Factors.
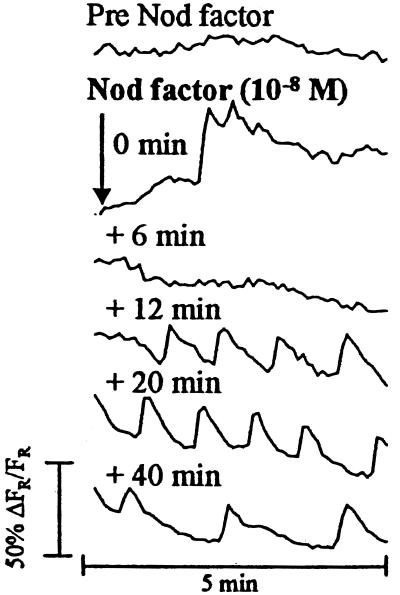
Root hairs of Pisum sativum L. var. Frisson were injected with the calcium-sensitive fluorescent dye Oregon Green, and changes in fluorescence were monitored with a confocal microscope. R. leguminosarum bv. viciae Nod factors initiated a response as shown in Fig. 1. Between 1 and 2 min after Nod-factor application, a 30–50% increase in fluorescence was typically seen (80% showing increase, number of cells = 10 from 10 root sections; Fig. 1, 0-min trace). A further 5–15 min after this event, a spiking response was established in all cells imaged (n = 10; Fig. 1, +12-min trace). The pattern of this response is very similar to that described in alfalfa (13), with spikes in fluorescence averaging one spike per minute and with the rising phase of the spike steeper than the falling phase. After 40–60 min, the frequency of spikes dropped to an average of one spike every 2 min (n = 10; Fig. 1, +40-min trace). These less frequent spikes usually continued for the duration of the experiment (up to 3 h).
Figure 1.
Calcium-spiking patterns in a pea root hair injected with Oregon Green and exposed to R. leguminosarum bv. viciae Nod factors. The traces are taken from one root-hair experiment and show the typical pattern observed (10 individual cells examined from 10 root sections). Measurements were taken from the nuclear region of the hair. Each trace represents data collected from 60 images, one image taken every 5 s. The time label on each trace indicates when the first image for that set was taken relative to the addition of Nod factor. Nod factor was added immediately after the last image of the pre-Nod-factor data set was taken.
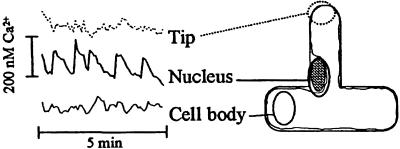
To confirm that the observed changes in fluorescence were caused by changes in the intracellular calcium, ratiometric measurements were made with coinjected Oregon Green and Texas Red. Thus it was possible to get quantitative spatial information for the calcium spikes. Fig. 2 shows the traces obtained when these ratiometric data are plotted against time for different regions of the cell. There was a rapid increase in the calcium concentration around the nuclear region, followed by a slower decay. The maximal increase in the calcium concentration, averaged over the nuclear region during a typical spike, was around 200 nM. There is relatively little change in [Ca2+]int in other parts of the cell (Fig. 2). However, it should be recognized that the majority of the cytoplasm (and therefore dye) is localized around the cell nucleus, which may distort the detection of the true response. The characteristics of the calcium spikes in pea root hairs are similar to those seen in alfalfa, although the peaks in calcium concentration around the nuclear region are apparently smaller in pea root hairs (200 nM) than those measured in alfalfa with a different dye (500 nM) (13).
Figure 2.
Change in [Ca2+]int in a pea root hair 10 min after exposure to R. leguminosarum bv. viciae Nod factors. Concentrations were measured by use of an Oregon Green/Texas Red dye mixture and calibrated in vitro with buffered calcium solutions. Traces show the change in calcium concentration for the particular region indicated.
Calcium Spiking Is Blocked by Mutations at Three sym Loci in Pea.
If Nod-factor-induced calcium spiking is involved in nodulation signaling, then some of the pea mutants unable to form nodules should be defective for calcium spiking. We selected pea mutants completely defective for nodulation and chose another mutant affected at a later stage. Thus we selected mutants representing six loci (sym7, sym8, sym9, sym10, sym19, and sym30) in which no nodulation had been observed (23, 24, 25). In addition, we analyzed cv. Afghanistan, which carries the sym2A allele that blocks nodulation by most European isolates of R. leguminosarum bv. viciae, although a few infection threads are formed (29).
The root-hair phenotypes of some of these mutants have been described (25, 29, 30). These results were confirmed and others tested for root-hair deformation and curling by using secondary roots. The wild-type and cv. Afghanistan lines had, as expected (29), root-hair deformation and curling (Table 1); infection threads were observed in cv. Afghanistan, although they were reduced in number. The sym7 mutant (E69) induced root-hair deformation but did not induce hair curling or infection. Mutations in the other five genes (sym8, sym9, sym10, sym19, and sym30) blocked root-hair deformation induced by R. leguminosarum bv. viciae (Table 1).
Table 1.
Summary of phenotypes of pea nodulation mutants
| Allele | Line | Ca2+ spike | Had | Hac | Inf | Myc |
|---|---|---|---|---|---|---|
| WT | Frisson | + | + | + | + | + |
| sym2A | cv. Afg. | + | + | + | + | NA |
| sym7 | E69 | + | + | − | − | NA |
| sym8 | R25 | − | − | − | − | − |
| sym9 | P54 | + | − | − | − | − |
| sym10 | P56 | − | − | − | − | + |
| sym10 | P5 | − | − | − | − | + |
| sym19 | P55 | − | − | − | − | − |
| sym19 | P4 | − | − | − | − | − |
| sym30 | P53 | + | − | − | − | − |
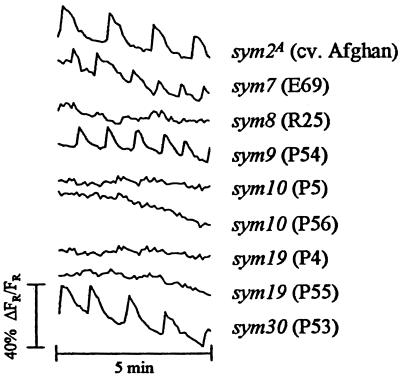
Nod factors induced calcium spiking in root hairs of cv. Afghanistan (sym2A), E69 (sym7), P54 (sym9), and P53 (sym30) (Fig. 3). The timing, frequency, and shape of the spikes were indistinguishable from those seen with wild-type Frisson or Sparkle. Three mutants, R25 (sym8), P56 (sym10), and P55 (sym19), consistently failed to induce calcium spiking (a minimum of six root hairs from six plants for each mutant were tested). Other alleles of sym10 and sym19 have been described in the mutants P5 and P4, respectively (24). The Nod-factor preparation also failed to induce a response in these mutants (again, six root hairs from six plants for each mutant were tested). These results demonstrate that mutations affecting at least three loci in pea can block Nod-factor-induced calcium spiking in root hairs.
Figure 3.
Changes in intracellular calcium in the root hairs of pea nodulation mutants. Traces are from individual root hairs 10 min after addition of 10−8 M Nod factor and are representative of the typical responses seen (see Table 1). The label after each trace indicates the mutant gene of interest, with the line number in parentheses. Each trace represents data collected from 60 images, one image taken every 5 s. Measurements were taken from the nuclear regions of the cells. At least six cells were examined from six root sections for mutants showing no response.
Calcium Spiking Can Be Induced in Root Hairs by Chitin Oligomers.
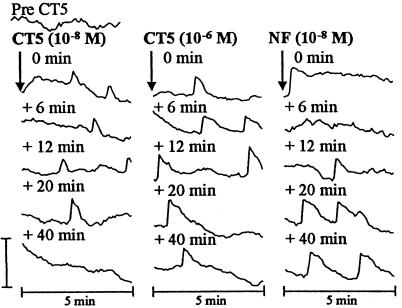
To identify the minimal structural requirements necessary to elicit a calcium-spiking response, we first tested the most basic Nod-factor components. N,N′,N′′,N′′′-tetraacetylchitotetraose (CT4) and N,N′,N′′,N′′′,N′′′′-pentaacetylchitopentaose (CT5) are chitin oligomers that represent the two unsubstituted backbones of R. leguminosarum bv. viciae Nod factors. When added to a final concentration of 10−8 M, CT5 induced the appearance of one or more spikes in each 5-min data set (Fig. 4). These spikes were somewhat sporadic, with a frequency lower than that induced by Nod factor, with an average of one spike every 3.3 min (number of cells tested = 4, 100% showing spikes). However, by increasing the concentration of CT5 100-fold, this could be increased to an average of one spike every 1.8 min. Similar results were found with CT4 (data not shown). The occurrence of CT4- and CT5-induced spikes is surprising, because similar experiments in alfalfa and bean failed to elicit a response (10, 13). In none of the tests with CT4 or CT5 did we observe the rapid step up of fluorescence that was usually seen after Nod-factor addition. However, as illustrated in Fig. 4, the addition of Nod factor after CT5 induced an apparent increase in [Ca2+]int, which occurred less than a minute after Nod-factor addition. Sustained spiking then followed, although the frequency of spikes was lower than that seen in root hairs treated with Nod factor alone (Fig. 1). However, this low frequency of spiking may not reflect an effect of prior treatment with CT4 or CT5, because root hairs injected with Oregon Green and then left for 2 h before treatment with Nod factor alone often showed a similar low-frequency spiking response (data not shown). If the calcium-spiking response induced by CT4 and CT5 is related to that induced by Nod factors, it should be blocked by mutations at the sym8, sym10, and sym19 loci. No calcium spiking response to CT5 (at 10−6 M) was seen with root hairs of the mutants R25 (sym8), P56 (sym10), or P55 (sym19) (a minimum of four root hairs from four different plants of each of the mutants were tested).
Figure 4.
Calcium spiking in a pea root hair injected with Oregon Green and exposed to CT5. The trace shown is representative of four experiments and shows the response of the same root hair for all points. Measurements were taken from the nuclear region of the root hair. Each trace represents data collected from 60 images, one image taken every 5 s. The time labels indicate when the first image for that set was taken relative to the addition (or increase in concentration) of CT5 and Nod factor. The time taken to increase the CT5 concentration or add Nod factor was ≈30 s. Similar results were obtained with CT4. The vertical bar represents 40% ΔFR/FR for all traces.
Mastoparan Did Not Induce Calcium Spiking.
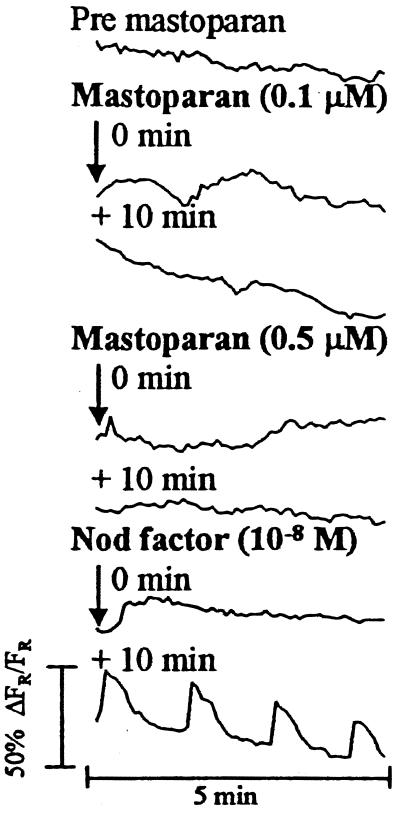
Pingret et al. (22) showed that MtEnod12A expression may be mediated by a G protein-regulated pathway. A key observation was that mastoparan could induce expression of a MtEnod12A-GUS transgene in Medicago truncatula independently of Nod factor (22). Mastoparan is a 14-amino acid peptide isolated from wasp venom that, in mammalian cells, has been shown to activate G proteins independently of their regulatory receptor (31). Addition of mastoparan at 0.01 μM, 0.1 μM, or 0.5 μM did not induce any calcium responses in pea, with root-hair cells remaining competent to spike when Nod factor was subsequently added (Fig. 5). When applied to a final concentration of 1.0 μM (the concentration that was seen to give maximal MtEnod12A-GUS expression in M. truncatula; ref. 22), mastoparan was apparently toxic to pea root hairs, irreversibly inhibiting cytoplasmic streaming.
Figure 5.
Calcium response of a pea root hair injected with Oregon Green and exposed to various concentrations of mastoparan. The trace is taken from one root-hair experiment and represents the typical response seen (n = 5). Measurements were taken from the nuclear region of the hair. Each trace represents data collected from 60 images, one image taken every 5 s. The time label on each trace indicates when the first image for that set was taken relative to the addition of Nod factor or mastoparan
Discussion
Nod factors elicit calcium spiking in the root hairs of pea, alfalfa (13), M. truncatula (32), Phaseolus bean (10), Vicia hirsuta, Vicia sativa, and Lotus japonicus (S. Long, D. Ehrhardt, and R. Wais, personal communication), suggesting this response may be conserved among nodulating legumes. In pea root hairs, Oregon Green may detect two responses to Nod factors: a step up in intracellular calcium, followed several minutes later by calcium spiking. However, the calcium step up was not seen in all cases, even though calcium spiking was initiated later. Similar observations were made with alfalfa (13), suggesting that the two responses need not be coupled. The step up of intracellular calcium occurs at about the same time membrane depolarization occurs in alfalfa at the root-hair tip after an influx of Ca2+ and efflux of Cl− and K+ (11). It remains to be established if the step up of fluorescence observed in this case is caused by an influx of extracellular calcium (11) that was concluded to be required for Nod-factor signal transduction in alfalfa (33). Calcium spiking appears to be necessary for nodulation signaling because nodulation-defective mutants of pea, alfalfa (13), and M. truncatula (32) are defective for Nod-factor-induced calcium spiking.
We were surprised to observe that chitin oligomers could induce some calcium spiking. On no occasion have we observed any chitin-oligomer-induced root-hair deformation on vetch or pea. The observation that the calcium response induced by chitin oligomers is blocked by mutations at three loci suggests that it is indeed related to nodulation signaling rather than a defense response. No calcium response was seen when chitin oligomers were added to alfalfa or bean roots (10, 13). Unsulfated Nod factors also failed to induce calcium spiking in alfalfa (13). The difference between pea and alfalfa may be caused by the O-linked sulfate group on the S. meliloti Nod factor, which is crucial for Nod-factor-inducible events in alfalfa (4, 7, 13). It would be interesting to determine whether sulfated chitin oligomers can induce calcium spiking in alfalfa.
The calcium responses induced by chitin oligomers in pea root hairs were different from those induced by Nod factors in the following ways. (i) No rapid step up in intracellular calcium was observed (this could be induced by the subsequent addition of Nod factors). (ii) The frequency of spiking was lower and was not as sustained. (iii) The concentration required was much higher. This may suggest that the CT5-induced effect could result from the activation of only one part of a multicomponent pathway. Alternatively, partial activation of the spiking response may result from poor association of the chitin oligomer with a Nod-factor receptor. In soybean roots, chitin oligomers induced transient expression of the early nodulin gene ENOD40 and acted synergistically with Nod factors to sustain ENOD40 expression and to induce ENOD2 expression (34, 35). These results and work on infection by bacterial mutants defective for host-specific modification of Nod factors (28, 36) have led to the suggestion that there may be more than one type of recognition event in Nod-factor detection.
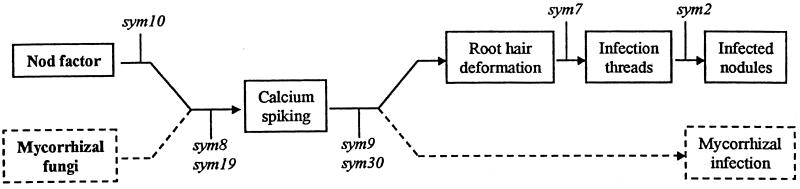
The phenotypes of the nodulation-defective pea mutants provide insight into the sequence of some nodulation-signaling events. The sym7 and sym2 genes influence events that occur relatively late (Fig. 6), because lines carrying the sym7 and sym2A alleles retain root-hair deformation and calcium spiking. Mutations at five loci (sym8, sym9, sym10, sym19, and sym30) affect early events because they block root-hair deformation. Mutations at sym30 and sym9 did not affect calcium spiking, and so these loci act downstream of calcium spiking but upstream of root-hair deformation (Fig. 6). There is some preliminary evidence to suggest that P54 (sym9) may belong to the sym30 complementation group (S.A.W. and J.A.D., unpublished observations). If this were correct, then P53 (sym30) and P54 could simply be considered to contain different alleles at the same locus. Mutations at the sym8, sym10, and sym19 loci block root-hair deformation and calcium spiking (Table 1), and therefore, it is probable that their gene products play a role in establishing calcium spiking (Fig. 6)
Figure 6.
Model relating pea sym loci to steps in nodulation. Mutations in sym8 and sym19 block both Nod-factor-induced calcium spiking and mycorrhizal infection. Because mutation of sym10 blocks calcium spiking but not mycorrhizal infection, we postulate that sym10 acts upstream of the common steps to nodulation and mycorrhizal infection. Mutation of sym9 and sym30 blocks root-hair deformation and mycorrhizal infections but not calcium spiking. Mutation of sym7 blocks infection thread formation induced by Rhizobium but not root-hair deformation or calcium spiking The sym2A locus inhibits nodulation by the strain of R. leguminosarum bv. viciae used herein, but some infections are found. The data on mycorrhizal infection were as described (37, 38).
It is instructive to consider the mycorrhizal phenotypes of the various Nod− mutants (Table 1). P5 and P56, carrying mutations at sym10, form a normal mycorrhizal symbiosis, as do other mutants within this complementation group (37). Our interpretation of this observation is that the sym10 gene product is required for a step upstream of calcium spiking but on a part of a pathway that is not required to establish a mycorrhizal symbiosis (Fig. 6). Mutations at the sym8, sym19, and sym30 loci block both nodulation and mycorrhization (37, 38), and a reassessment of the phenotype of P54 (sym9) suggests that it is also Myc− (M. Sagan and G. Duc, personal communication). In view of the observation (Table 1) that some Nod− Myc− mutants induce calcium spiking but others do not, we speculate that calcium spiking may be common to both mycorrhizal and nodulation signaling and that the pathways could diverge after calcium spiking (Fig. 6).
Several predictions can be made based on the model presented in Fig. 6: (i) The sym10 gene product may be a Nod-factor receptor or transduce the Nod-factor-induced signal. (ii) The initial stages of mycorrhizal signaling probably uses a different receptor from the Nod-factor receptor. (iii) Mycorrhizal fungi may induce a calcium-spiking response. (iv) Calcium spiking may be necessary for nodulation signaling. The order of events proposed in Fig. 6 is intended to be a working model and does not try to take into account the possible existence of two types of Nod-factor detection. Other interpretations of the results are possible. For example, calcium spiking could be a secondary event that is not directly on the signaling pathway but is blocked by mutations that affect the signaling pathway. More complex models can be formulated if it is assumed that the sym gene products can function in independent parallel pathways, but at this stage we have kept the model relatively simple. The results and model presented herein are substantially similar to those of Wais et al. (32), based on the analysis of M. truncatula mutants. It seems likely that the sym8 and sym19 mutants in pea could correspond to the dmi1 and dmi2 mutants of M. truncatula, whereas dmi3 could correspond to sym9 or sym30. Genetic mapping and gene identification will be required to confirm this suggestion.
The nature of the pathway leading from Nod-factor recognition to the induction of calcium spiking remains elusive. Similarities found with the responses identified in mammalian cells (39) suggest the involvement of a receptor-linked G protein, phospholipase C, and inositol trisphosphate (22). There is circumstantial evidence that components of such a pathway operate in Nod-factor signaling in alfalfa and that mastoparan can induce early nodulation gene expression (22). Stimulating calcium spiking by mastoparan would have provided continuity between observations. However, we found that addition of mastoparan at concentrations giving maximal responses in alfalfa (22) adversely affected the integrity of pea root hairs.
By identifying Nod-factor-induced calcium spiking in pea root hairs and demonstrating that it does not occur in Nod− mutants affected at three loci, we have provided good evidence that calcium spiking plays a role in Nod-factor signal transduction. Future work should concentrate on identification of the components of this pathway, which may indicate the degree of divergence between plant and mammalian signaling systems.
Acknowledgments
We thank Karen Wilson for preparing and analyzing Nod factors, Grant Calder for advice on confocal microscopy and microinjection, and Dave Ehrhardt and Sharon Long and their colleagues for advice and encouragement on the calcium-imaging experiments and communicating results before publication. Gerard Duc, Muriel Sagan, Tom LaRue, and Ton Bisseling gave us pea mutants; and Muriel Sagan and Gerard Duc generously communicated unpublished information about genetic complementation tests. Mike Ambrose helped with stocks of seeds, and Nick Brewin and Bjorn Drøbak made constructive comments during the course of the work. The research was supported by the Biotechnology and Biological Sciences Research Council and the European Union (Contract FMRX-CT98–0243).
Abbreviations
- [Ca2+]int
intracellular calcium concentration
- CT4
N,N′,N′′,N′′′-tetraacetylchitotetraose
- CT5
N,N′,N′′,N′′′,N′′′′-pentaacetylchitopentaose
Note Added in Proof.
Recently, Cárdenas et al. (40) thoroughly reviewed data on ion changes in legume root hairs responding to Nod factors. They point out that in some legumes such as Phaseolus bean, increases in calcium clearly occur at the root hair tip prior to, and concomitant with, the transient changes observed around the nuclear region (10). At this stage, we do not know why we did not observe such Nod-factor-induced changes in calcium in the root hair tip region of pea. Possibly, the difference could be species-specific; the observations with alfalfa (13) and M. truncatula (32) are broadly similar to those seen with pea. Alternatively, the difference may be due to different methods of sample preparation or analysis.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230440097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230440097
References
- 1.Schlaman H R M, Phillips D A, Kondorosi É. In: The Rhizobiaceae. Spaink H P, Kondorosi Á, Hooykaas P J J, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 361–386. [Google Scholar]
- 2.Dénarié J, Debellé F, Promé J C. Annu Rev Biochem. 1996;65:503–535. doi: 10.1146/annurev.bi.65.070196.002443. [DOI] [PubMed] [Google Scholar]
- 3.Long S R. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé J C, Dénarié J. Nature (London) 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 5.Spaink H P, Sheeley D M, van Brussel A A N, Glushka J, York W S, Tak T, Geiger O, Kennedy E P, Reinhold V N, Lugtenberg B J J. Nature (London) 1991;354:125–130. doi: 10.1038/354125a0. [DOI] [PubMed] [Google Scholar]
- 6.Downie J A, Walker S A. Curr Opin Plant Biol. 1999;2:483–489. doi: 10.1016/s1369-5266(99)00018-7. [DOI] [PubMed] [Google Scholar]
- 7.Truchet G, Roche P, Lerouge P, Vasse J, Camut S, de Billy F, Promé J C, Dénarié J. Nature (London) 1991;351:670–673. [Google Scholar]
- 8.van Brussel A A N, Bakhuizen R, van Spronsen P C, Spaink H P, Tak T, Lugtenberg B J J, Kijne J W. Science. 1992;257:70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]
- 9.Ehrhardt D W, Atkinson E M, Long S R. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 10.Cárdenas L, Feijó J A, Kunkel J G, Sánchez F, Holdaway-Clarke T, Hepler P, Quinto C. Plant J. 1999;19:347–352. doi: 10.1046/j.1365-313x.1999.00517.x. [DOI] [PubMed] [Google Scholar]
- 11.Felle H H, Kondorosi É, Kondorosi Á, Schultze M. Plant J. 1998;13:455–463. [Google Scholar]
- 12.Kurkdjian A C. Plant Physiol. 1995;107:783–790. doi: 10.1104/pp.107.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrhardt D W, Wais R, Long S R. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 14.Meyer T, Stryer L. Annu Rev Biophys Biophys Chem. 1991;20:153–174. doi: 10.1146/annurev.bb.20.060191.001101. [DOI] [PubMed] [Google Scholar]
- 15.Fewtrell C. Annu Rev Physiol. 1993;55:427–454. doi: 10.1146/annurev.ph.55.030193.002235. [DOI] [PubMed] [Google Scholar]
- 16.Dolmetsch R E, Xu K, Lewis R S. Nature (London) 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Llopis J, Whitney M, Zlokarnik G, Tsien R Y. Nature (London) 1998;392:936–941. doi: 10.1038/31965. [DOI] [PubMed] [Google Scholar]
- 18.Staxén I, Pical C, Montgomery L T, Gray J E, Hetherington A M, McAinsh M R. Proc Natl Acad Sci USA. 1999;96:1779–1784. doi: 10.1073/pnas.96.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Messerli M, Robinson K R. J Cell Sci. 1997;110:1269–1278. doi: 10.1242/jcs.110.11.1269. [DOI] [PubMed] [Google Scholar]
- 20.Franklin-Tong V E, Drobak B K, Allan A C, Watkins P A C, Trewavas A J. Plant Cell. 1996;8:1305–1321. doi: 10.1105/tpc.8.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calder G M, Franklin-Tong V E, Shaw P J, Drøbak B K. Biochem Biophys Res Commun. 1997;234:690–694. doi: 10.1006/bbrc.1997.6514. [DOI] [PubMed] [Google Scholar]
- 22.Pingret J L, Journet E P, Barker D G. Plant Cell. 1998;10:659–671. doi: 10.1105/tpc.10.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kneen B E, Weeden N F, LaRue T A. J Hered. 1994;85:129–133. doi: 10.1093/oxfordjournals.jhered.a111432. [DOI] [PubMed] [Google Scholar]
- 24.Duc G, Messager A. Plant Sci. 1989;60:207–213. [Google Scholar]
- 25.Sagan M, Huguet T, Duc G. Plant Sci. 1994;100:59–70. [Google Scholar]
- 26.Schneider A, Walker S A, Poyser S, Sagan M, Ellis T H N, Downie J A. Mol Gen Genet. 1999;262:1–11. doi: 10.1007/s004380051053. [DOI] [PubMed] [Google Scholar]
- 27.Knight C D, Rossen L, Robertson J G, Wells B, Downie J A. J Bacteriol. 1986;166:552–558. doi: 10.1128/jb.166.2.552-558.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker S A, Downie J A. Mol Plant–Microbe Interact. 2000;13:754–762. doi: 10.1094/MPMI.2000.13.7.754. [DOI] [PubMed] [Google Scholar]
- 29.Geurts R, Heidstra R, Hadri A E, Downie J A, Franssen H, van Kammen A, Bisseling T. Plant Physiol. 1997;115:351–359. doi: 10.1104/pp.115.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markwei C M, LaRue T A. Can J Microbiol. 1992;38:548–554. [Google Scholar]
- 31.Ross E M, Higashijima T. Methods Enzymol. 1994;237:26–37. doi: 10.1016/s0076-6879(94)37050-8. [DOI] [PubMed] [Google Scholar]
- 32.Wais R J, Galera C, Oldroyd G, Catiora R, Penmesta R V, Cook D, Gough C, Dénarié J, Long S R. Proc Natl Acad Sci USA. 2000;97:13407–13412. doi: 10.1073/pnas.230439797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felle H H, Kondorosi É, Kondorosi Á, Schultze M. Plant Physiol. 1999;121:273–279. doi: 10.1104/pp.121.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minami E, Kouchi H, Cohn J, Ogawa T, Stacey G. Plant J. 1996;10:23–32. doi: 10.1046/j.1365-313x.1996.10010023.x. [DOI] [PubMed] [Google Scholar]
- 35.Minami E, Kouchi H, Carlson R W, Cohn J, Kolli V J, Day R B, Ogawa T, Stacey G. Mol Plant–Microbe Interact. 1996;9:574–583. doi: 10.1094/mpmi-9-0574. [DOI] [PubMed] [Google Scholar]
- 36.Ardourel M, Demont N, Debellé F D, Maillet F, de Billy F, Promé J C, Dénarié J, Truchet G. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duc G, Trouvelot A, Gianinazzi-Pearson V, Gianinazzi S. Plant Sci. 1989;60:215–222. [Google Scholar]
- 38.Albrecht C, Geurts R, Lapeyrie F, Bisseling T. Plant J. 1998;15:605–614. doi: 10.1046/j.1365-313x.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- 39.Petersen O H, Wakui M. J Membr Biol. 1990;118:93–105. doi: 10.1007/BF01868467. [DOI] [PubMed] [Google Scholar]
- 40.Cárdenas L, Holdaway-Clarke T L, Sánchez F, Quinto C, Feijó J A, Kunkel J G, Hepler P K. Plant Physiol. 2000;123:443–451. doi: 10.1104/pp.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]