Abstract
We report the first thermodynamic analysis of parallel β-sheet formation in a model system that folds in aqueous solution. NMR chemical shifts were used to determine β-sheet population, and van’t Hoff analysis provided thermodynamic parameters. Our approach relies upon the D-prolyl-1,1-dimethyl-1,2-diaminoethane unit to promote parallel β-sheet formation between attached peptide strands. The development of a macrocyclic reference molecule to provide chemical shift data for the fully folded state was crucial to the quantitative analysis.
Protein folding patterns are dominated by two regular substructures, α-helix and β-sheet. The origins of conformational stability for these secondary structures, a subject of intense interest, can be explored with peptides that form a helix or sheet in the absense of a tertiary context. Design rules for medium-length peptides that form autonomous α-helices in aqueous solution were delineated in the 1980s,1 and comparable achievements for autonomous antiparallel β-sheets were reported in the 1990s.2 In both cases, the development of strategies for determining folded populations has allowed thermodynamic analysis of secondary structure formation.3
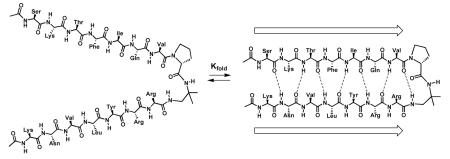
An autonomously folding parallel β-sheet (in contrast to an autonomous antiparallel β-sheet or α-helix) cannot be created exclusively from α-amino acid residues because the N-terminus of one strand in a parallel β-sheet does not lie near the C-terminus of a neighboring strand. Many groups have explored non-peptide units that promote parallel sheet interactions, most commonly by linking C-termini with a short turn-forming diamine.4 We have previously reported that the D-prolyl-1,1-dimethyl-1,2-diaminoethane (D-Pro-DADME) supports formation of a two-stranded parallel β-sheet in water, as indicated by 2D NMR data.5 Here we show that the thermodynamics of parallel β-sheet formation can be evaluated in such a model system. This accomplishment provides a foundation for exploring sequence-stability relationships in the parallel β-sheet structural manifold, which would fill a significant gap in our understanding of proteins and protein aggregates.
NMR chemical shifts provide the most reliable insight on folded populations among antiparallel β-sheet model systems,3,6 and we therefore aimed for a chemical shift-based analysis of parallel β-sheet folding. Most autonomous helix or sheet systems are not fully folded under accessible conditions; therefore, chemical shifts for such systems are population-weighted averages of the contributions from the limiting folded and unfolded states. To analyze 1 we have built upon the strategy previously developed for antiparallel β-sheets by constructing model compounds intended to provide empirical estimates of chemical shifts in the fully folded and fully unfolded states.3b The unfolded state is represented by diastereomer 2; changing proline configuration from D to L completely disrupts parallel β-sheet formation.5,7 The folded state is represented by cyclic molecule 3, an analogue of 2 in which the N-termini of the strand segments have been linked via a succinyl-glycine unit. Cyclization is intended to enhance the propensity for parallel β-sheet formation in 3 relative to linear molecule 2. Among antiparallel model systems, two-stranded β-sheet conformations have been stabilized by cyclization strategies involving either the backbone (capping a β-hairpin with a β-turn) or side chains (disulfide formation between terminal Cys residues).3a-b,8 We focused on backbone cyclization because Cys disulfide crosslinks are not compatible with parallel β-sheet secondary structure.9 The succinyl-glycine linker in 3 allows formation of a 10-membered ring H-bond [C=O(Succ-18) to H-N (Ser-2)], which is analogous to the H-bond common among β-turns.
NMR analysis indicated that 3 adopts the intended parallel β-sheet conformation in aqueous solution, and the data suggest that the extent of folding is greater for cyclic 3 than for linear analogue 1. Four unambiguous interstrand NH--CαH NOEs were observed in the center of the intended β-sheet region of 3 (Figure 1); the Val-8/Arg-11 NOE, if present, would have been obscured by the residual solvent resonance. These NOEs are consistent with the expected parallel β-sheet hydrogen bonding registry.10 The lack of interstrand NH--CαH NOEs near the succinyl-glycine linker, however, suggests that this region is not as well folded as the rest of molecule. A large set of side chain-side chain NOEs was observed for 3, all consistent with the intended parallel β-sheet conformation. For linear molecule 1, a comparable set of cross-strand NOEs was seen for only the eight strand residues nearest to the D-Pro-DADME unit (Phe-5 to Val-8 and Arg-11 to Leu-14), but not for the three residues at each strand terminus.7 The difference in interstrand NOE patterns observed for 1 vs. 3 suggests that the parallel β-sheet secondary structure encompasses nearly the entire strand length for 3, but only the strand residues closer to the turn segment in 1. The behavior of 1 is consistent with evidence that the termini of antiparallel β-sheet model systems tend to be unfolded (“frayed”) in aqueous solution.2 As intended, macrocyclization, in 3, strongly discourages terminal fraying.
Figure 1.
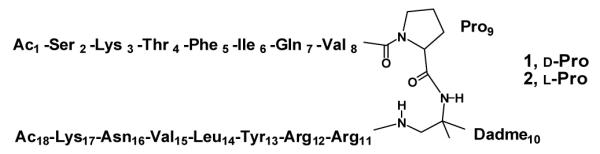
Chemical structures of linear compounds 1 and 2. The numbering scheme was chosen to allow easy comparison between the linear and cyclic molecules.
Downfield shifts in α-proton chemical shifts (δCαH), relative to a random coil reference value, indicate participation in β-sheet secondary structure.6 We use 2 to provide the “random coil” δCαH values for assessment of folding in the strand regions of 1 and 3 because 2 shows no sign of folding (no NOEs between sequentially non-adjacent residues),7 and the δCαH values measured for 2 account for the effects of sequence context. Nearly all strand residues in 3 show ΔδCαH [= δCαH(3) - δCαH(2)] ≥ +0.1 ppm in aqueous solution at 287 K, which suggests extensive β-sheet formation along the entire length of each strand segment. This conclusion matches that reached from the interstrand NOEs observed for 3. In contrast, the outermost residues of linear molecule 1, Ser-2 to Thr-4 and Val-15 to Lys-17, display random coil-like ΔδCαH values. The strand segments nearer the linker in 1, Phe-5 to Val-8 and Arg-11 to Leu-14, display ΔδCαH values consistent with β-sheet formation, although each ΔδCαH value is smaller than the corresponding value for 3. Thus, parallel β-sheet structure is well-developed only for the segments of 1 near the linker, and this region is only partially folded. Based on these observations, we regard strand segments Phe-5 to Val-8 and Arg-11 to Leu-14 as the folded core of 1, and we focus on this core in the analysis below.
We examined the effect of 2,2,2-trifluoroethanol (TFE) on ΔδCαH for 3 in an effort to determine whether 3 is fully folded in aqueous buffer. Addition of increasing proportions of TFE to aqueous solutions has been shown to induce progressively larger extents of antiparallel β-sheet folding in several designed peptides.11 The ΔδCαH value for each strand residue of 3 became larger as the TFE content was raised from 0% to 30%,7 suggesting that the parallel β-sheet conformation is not fully populated in purely aqueous solution. Further increases in TFE proportion to 40% or 50% had relatively little effect,7 suggesting that β-sheet population is maximal at 30% TFE. We used δCαH values obtained with 3 in 50% TFE to represent the fully folded state in our population analysis of 1. Parallel β-sheet population at a particular residue of 1 in aqueous solution at a given temperature was estimated by interpolating the observed δCαH value between the δCαH value for the corresponding residue in unfolded reference compound 2 under the same conditions and the δCαH for folded reference compound 3 at 287 K in 50% TFE. For each of the eight residues in the parallel β-sheet core of 1 we compared folded populations determined at 287 K and 354 K in aqueous buffer.7 The apparent population change is reasonably consistent across this set of residues, which suggests that the eight-residue core can be analyzed in terms of a two-state model, unfolded vs. parallel β-sheet. We used the nonlinear fitting method developed by Searle et al.3a for van't Hoff analysis of two-state folding (Figure 3), which provided the following thermodynamic parameters for parallel β-sheet formation at 298 K: ΔH° = - 1.1±0.1 kcal/mol, ΔS° = -2.5±0.2 cal/mol K, ΔCp° = -73±2 cal/mol K.12 The thermodynamic signature for parallel β-sheet folding in 1 is qualitatively similar to that observed for a number of antiparallel β-hairpins in that β-sheet formation is enthalpically favorable and entropically unfavorable near room temperature.3,13 This signature differs from that of a classical hydrophobic effect, but the observation of a significant and negative heat capacity change upon folding suggests that there is a hydrophobic contribution to the drive for folding, presumably from interstrand side chain-side chain interactions.
Figure 3.
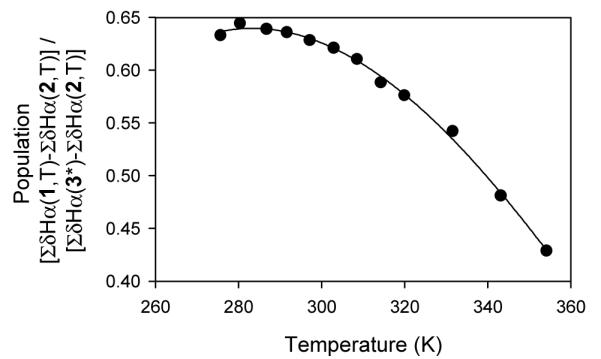
Change in folded population of 1 as a function of temperature, calculated from δCαH data by the method of ref. 3a. See Supporting Information for details. *: Data for 3 at 287 K and 50% TFE.
The results reported here lay the groundwork for thermodynamic analysis of the factors that control parallel β-sheet folding preferences. Such studies should provide fundamental insight on a structural motif that is very common in proteins and in protein aggregates associated with human diseases.14
Supplementary Material
Figure 2.
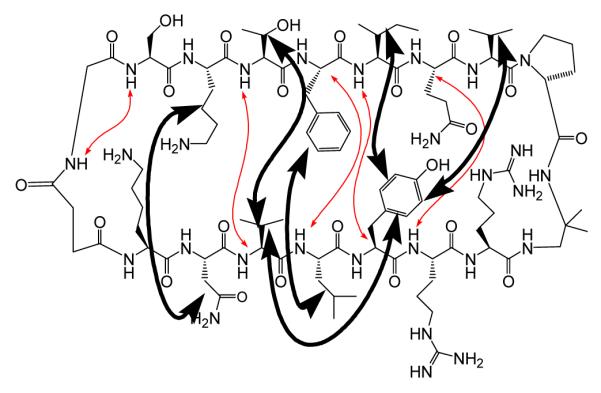
NOEs observed in 3 between residues non-adjacent in sequence. Light arrows (red) indicate backbone-backbone NOEs. Heavy arrows indicate multiple NOEs between side chain pairs (at least 3 NOEs for each pair).
Acknowledgements
This research was supported by the National Institutes of Health (GM-61238). J.D.F. and M.A.S. were supported in part by a Biophysics Training Grant from NIGMS. NMR spectrometers were purchased in part with funds from the NIH and NSF.
Footnotes
Supporting Information Available: Information concerning the synthesis of 3; chemical shift and NOE information on 1, 2, and 3; TFE titration data for 3 and the van’t Hoff analysis of folding. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Chakrabartty A, Baldwin RL. Adv. protein chem. 1995;46:141. [PubMed] [Google Scholar]
- (2) (a).Gellman SH. Curr. Opin Chem. Biol. 1998;2:717. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]; (b) Searle MS, Ciani B. Curr. Opin. Struct. Biol. 2004;14:458. doi: 10.1016/j.sbi.2004.06.001. [DOI] [PubMed] [Google Scholar]
- (3) (a).Maynard AJ, Sharman GJ, Searle MS. J. Am. Chem. Soc. 1998;120:1996. [Google Scholar]; (b) Syud FA, Espinosa JF, Gellman SH. J. Am. Chem. Soc. 1999;121:11577. [Google Scholar]; (c) Tatko CD, Waters ML. J. Am. Chem. Soc. 2002;124:9372. doi: 10.1021/ja0262481. [DOI] [PubMed] [Google Scholar]; (d) Russell SJ, Blandl T, Skelton NJ, Cochran AG. J. Am. Chem. Soc. 2003;125:388. doi: 10.1021/ja028075l. [DOI] [PubMed] [Google Scholar]; (e) Fesinmeyer RM, Hudson FM, Andersen NH. J. Am. Chem. Soc. 2004;126:7238. doi: 10.1021/ja0379520. [DOI] [PubMed] [Google Scholar]
- (4) (a).Selected examples: Wagner G, Feigel M. Tetrahedron. 1993;49:10831.Nowick JS, Insaf S. J. Am. Chem. Soc. 1997;119:10903.Chitnumsub P, Fiori WR, Lashuel HA, Diaz H, Kelly JW. Bioorg. Med. Chem. 1999;7:39. doi: 10.1016/s0968-0896(98)00222-3.Fisk JD, Powell DR, Gellman SH. J. Am. Chem. Soc. 2000;122:5443–5447.
- (5).Fisk JD, Gellman SH. J. Am. Chem. Soc. 2001;123:343. doi: 10.1021/ja002493d. [DOI] [PubMed] [Google Scholar]
- (6) (a).Wishart DS, Sykes BD, Richards FM. J. Mol. Biol. 1991;222:311. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]; (b) Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- (7).Please see Supporting Information
- (8).McDonnell JM, Fushman D, Cahill SM, Sutton BJ, Cowburn D. J. Am. Chem. Soc. 1997;119:5321. [Google Scholar]
- (9).Cootes AP, Curmi PM, Cunningham R, Donnelly C, Torda AE. Proteins. 1998;32:175. [PubMed] [Google Scholar]
- (10).Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley-Interscience; New York: 1986. [Google Scholar]
- (11).Ramirez-Alvarado M, Blanco FJ, Serrano L. Nature Struct. Biol. 1996;3:604. doi: 10.1038/nsb0796-604. [DOI] [PubMed] [Google Scholar]
- (12).The uncertainties shown arise from the fitting and do not reflect the potentially larger but unquantifiable uncertainties that stem from the assumptions behind this analysis. The thermodynamic parameters did not change significantly if data from subsets of residues within the eight-residue core were used (i.e., only those residues H-bonded to the partner strand, only those residues not H-bonded to the partner strand, or omission of the two residues closest to the D-Pro-DADME linker).
- (13) (a).Searle MS, Griffiths-Jones SR, Skinner-Smith H. J. Am. Chem. Soc. 1999;121:11615. [Google Scholar]; (b) Espinosa JF, Gellman SH. Angew. Chem. Int. Ed. 2000;39:2330. doi: 10.1002/1521-3773(20000703)39:13<2330::aid-anie2330>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]; (c) Cochran AG, Skelton NJ, Starovasnik MA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5578. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Parallel β-sheet structure in amyloid: Burkoth TS, Benzinger TLS, Urban V, Morgan DM, Gregory DM, Thiyagarajan P, Botto RE, Meredith SC, Lynn DG. J. Am. Chem. Soc. 2000;122:7883.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio R, Tycko R. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16742. doi: 10.1073/pnas.262663499.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.