SUMMARY
Fungi are a rich source of bioactive secondary metabolites and mushroom-forming fungi (Agaricomycetes) are especially known for the synthesis of numerous bioactive and often cytotoxic sesquiterpenoid secondary metabolites. Compared to the large number of sesquiterpene synthases identified in plants, less than a handful of unique sesquiterpene synthases have been described from fungi. Here we describe the functional characterization of six sesquiterpene synthases (Cop1 to Cop6) and two terpene oxidizing cytochrome P450 monooxygenases (Cox1 and Cox2) from Coprinus cinereus. The genes were cloned and, except for cop5, functionally expressed in Escherichia coli and/or Saccharomyces cerevisiae. Cop1 and Cop2 each synthesize germacrene A as the major product. Cop3 was identified as a α-muurolene synthase, an enzyme that has not been described previously, while Cop4 synthesizes δ-cadinene as its major product. Cop6 was originally annotated as a trichodiene synthase homolog, but instead was found to catalyze highly specific the synthesis of α-cuprenene. Co-expression of cop6 and the two monooxygenase genes next to it yields oxygenated α-cuprenene derivatives, including cuparophenol, suggesting that these genes encode the enzymes for the biosynthesis of antimicrobial quinone sesquiterpenoids (known as lagopodins) that were previously isolated from C. cinereus and other Coprinus species.
INTRODUCTION
Fungi produce many well-known bioactive secondary metabolites including terpenoids which represent the largest class of natural products (Keller et al., 2005, Misiek & Hoffmeister, 2007, Spiteller, 2008, Davis & Croteau, 2000). The diversity of terpene structures is in part the result of a biosynthetic process where prenyl diphosphate chains with ten, fifteen or twenty carbons undergo numerous cyclization and rearrangement reactions catalyzed by terpene synthases (aka. cyclases) that generate and then guide the procession of a reactive carbocation through a prenyl chain. Terpene synthases differ in their prenyl diphosphate chain length specificity (mono- (C10), sesqui- (C15) and diterpene (C20) synthases), cyclization mechanism and whether they synthesize a narrow or broad range of cyclization products. Subsequent modifications of the terpene scaffolds further increase the structural diversity of this group of compounds (Davis & Croteau, 2000, Sacchettini & Poulter, 1997, Christianson, 2008, Christianson, 2006).
Many terpene synthases have been described from plant species (Tholl, 2006), but relatively few microbial enzymes have been functionally characterized (Lesburg et al., 1997b, Caruthers et al., 2000, Rynkiewicz et al., 2001, Shishova et al., 2007, Pinedo et al., 2008, Kawaide, 2006, Toyomasu et al., 2007, Toyomasu et al., 2004, Hamano et al., 2002, Dairi et al., 2001, Kawaide et al., 1997). The availability of genome sequences recently led to the discovery of several sesquiterpene synthases from actinomycetes and cyanobacteria (Cane et al., 2006, Gust et al., 2003, Cane & Watt, 2003, Komatsu et al., 2008, Zhao et al., Agger et al., 2008, Giglio et al., 2008). However, so far only three genes encoding fungal sesquiterpene synthases have been cloned and functionally characterized. These enzymes are trichodiene synthase, aristolochene synthase and very recently, presiliphiperfola-8β-ol synthase which generate the sesquiterpene scaffolds of the trichothecene mycotoxins produced by Fusarium species, the PR-toxin made by Penicillium roqueforti and the botyrane phytotoxins synthesized by Botrytis cinerea, respectively (Cane et al., 1995, Cane & Kang, 2000, Hohn & Beremand, 1989, Hohn et al., 1995, Hohn & Plattner, 1989, Caruthers et al., 2000, Rynkiewicz et al., 2001, Pinedo et al.). Like their bacterial homologs (Zhao et al., 2008, Tetzlaff et al., 2006, Agger et al., 2008), fungal terpene synthases are frequently part of a biosynthetic cluster that includes additional terpene modifying genes such as cytochrome P450 monooxygenases. Trichothecene biosynthetic cluster, for example, have been studied in great detail (Tokai et al., 2007) and more recently, botrydial biosynthesis has been characterized (Pinedo et al., 2008, Collado et al., 2007, Siewers et al., 2005).
No terpene biosynthetic enzymes have so far been described from mushroom-forming higher fungi of the class Agaricomycetes (Homobasidiomycetes (sensu (Hibbett et al., 2007))), despite the fact that this class of fungi is known to produce numerous bioactive and often cytotoxic sesquiterpenoid secondary metabolites (reviewed in (Abraham, 2001)). This may in part be due to a general lack of genetic tools and genomic sequence information for this group of fungi. Unlike the many genome sequences released for filamentous fungi, only one Agaricales genome sequence, that of the model species Coprinus cinereus (aka. Coprinopsis cinerea) was known until very recently (Broad Institute). The genome sequence of another Agaricales species, Laccaria bicolor (Joint Genome Institute) (Martin et al., 2008) was released in early 2008 and a genomic survey of Moniliophthora perniciosa (LGE-UNICAMP, Brazil) (Mondego et al., 2008) was published in late 2008. Genome sequences of the following non-mushroom forming Basidiomycota have been completed and published: Cryptococcus neoformans (Loftus et al., 2005), Phanerochaete chrysosporium (Martinez et al., 2004), Ustilago maydis (Kamper et al., 2006) and Postia placenta (Martinez et al., 2009)).
In this study we describe the identification and cloning of six sesquiterpene synthase and two associated cytochrome P450 monooxygenase genes from C. cinereus. Expression in E. coli and S. cerevisiae allowed functional characterization of the corresponding enzymes. Major sesquiterpene hydrocarbons produced by these enzymes correspond to those detected in small quantities in cultures of C. cinereus. One of the characterized terpene synthases, a homolog of trichodiene synthase, produces α-cuprenene and appears to catalyze the first step in the biosynthesis of the antimicrobial lagopodins. Lagopodins are quinone sesquiterpenoids that were identified in C. cinereus cultures in a manuscript dating two decades ago (Bu'Lock & Darbyshire, 1976).
RESULTS
Sesquiterpene synthase homologs in fungi
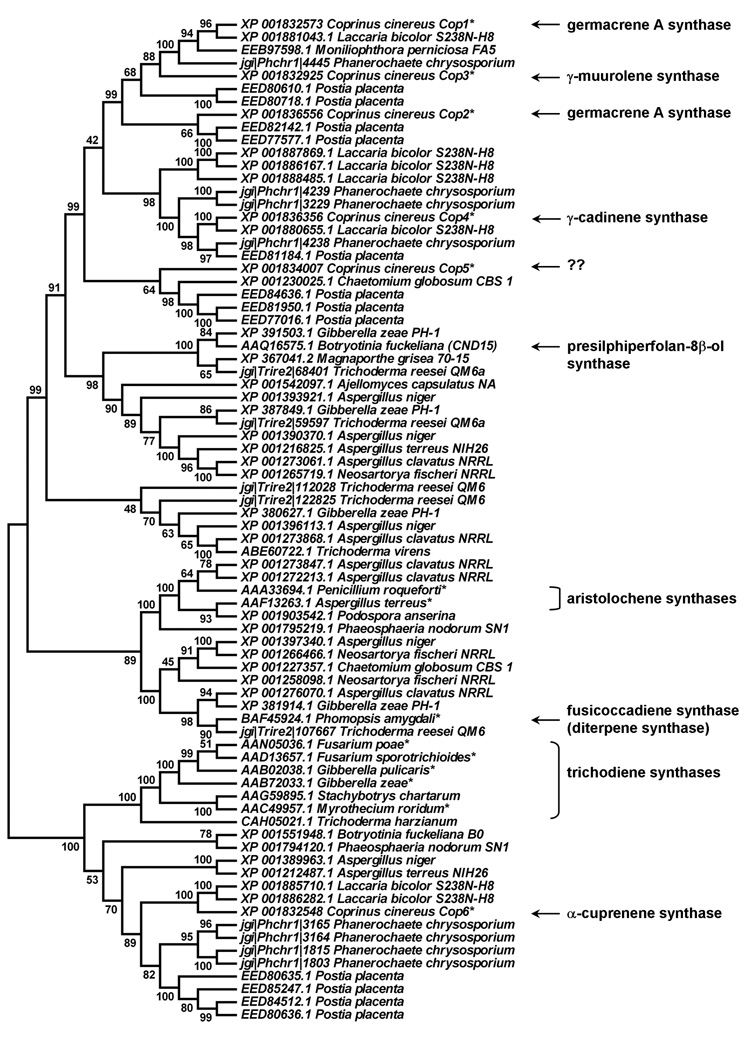
A BLAST search of fungal genome sequences at NCBI, JGI and Broad Institute with experimentally characterized microbial sesquiterpene synthases resulted in the identification of a large number of sesquiterpene synthase homologs, of which only a handful have so far been functionally characterized (Fig. 1). Most fungi have several sesquiterpene synthase homologs. Fungi belonging to the genera Aspergillus, Gibberella, Phanerochaete, Postia, Coprinus and Laccaria have four or more putative sesquiterpene synthases, while sesquiterpene synthase genes appear to have undergone extensive gene duplication in Phanerochaete chrysosporium, Postia placenta and Laccaria bicolor.
Fig. 1. Unrooted neighbor-joining tree of fungal sesquiterpene synthase homologs.
Protein sequences of known bacterial sesquiterpene sequences were used for homology searches in NCBI’s non-redundant protein sequence database and in fungal genome sequences obtained from the Broad Institute and JGI. Sequences that did not contain the first conserved metal binding domain (DDXXDD motif) of terpene synthases or appeared to be incorrectly annotated were not included in the alignment. Only a few representatives of fungal genera with several sequenced genomes (e.g. Aspergillus) were included in the search. Branches are labeled with their respective bootstrap values, gene accession numbers and strain names. Functionally characterized sesquiterpene synthases (including the Cop enzymes in this study) are also indicated. Question marks indicate that the function of Cop5 could not be determined in this study.
Phylogenetic analysis reveals that several terpene synthase homologs, including proteins from Phanerochaete, Laccaria and one protein from Coprinus, cluster together with the experimentally characterized trichodiene synthases (Hohn & Desjardins, 1992), suggesting that they may catalyze the same or a related cyclization reaction (Fig. 1). Other putative terpene synthases cluster together with the aristolochene synthases from Penicillium and Aspergillus (Cane & Kang, 2000, Proctor & Hohn, 1993), the recently characterized presilphiperfolan-8β-ol synthase (Pinedo et al., 2008) and fusicoccadiene synthase (Toyomasu et al., 2007). The majority of identified putative terpene synthase sequences cluster into clades without experimentally characterized representatives. These include several terpene synthase homologs from Coprinus, Laccaria and Phanerochate located at the top of the phylogenetic tree shown in Fig. 1.
To gain insight into the functions of the multiple terpene synthases present in many fungi, the six sesquiterpene synthase homologs (named Cop1 to Cop6) identified in C. cinereus were selected for cloning and functional characterization (see Supplementary Table 1 for accession numbers and genome locus tags of the six cop genes). Three of the six putative terpene synthases (Cop1 to Cop3) cluster in one clade of the phylogenetic tree, while Cop4 and Cop5 are located in separate clades and Cop6 is part of a large clade that also includes the known trichodiene synthases (Fig. 1). The cop6 gene is the only putative sesquiterpene synthase gene identified in C. cinereus that appears to be part of a biosynthetic gene cluster. The cop6 ORF is flanked on either side by two ORFs encoding putative cytochrome P450 monooxygenases (Cox1 and Cox2).
Analysis of sesquiterpenes produced by C. cinereus
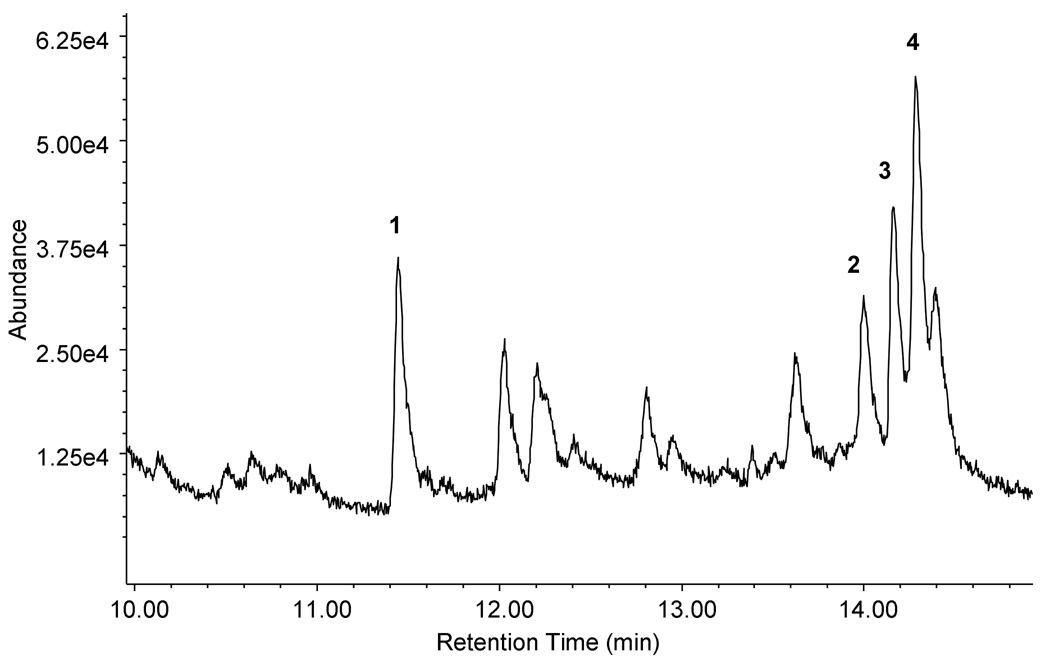
C. cinereus is not generally considered a terpenoid producer, although one study describes the isolation and structural identification of quinone sesquiterpenoids from this fungus (Bu'Lock & Darbyshire, 1976). Therefore to determine whether any of the putative terpene synthase homologs identified in the C. cinereus genome sequence are functionally expressed under laboratory growth conditions, C. cinereus was cultured for several weeks in the dark and terpene accumulation was analyzed periodically in the culture broth and headspace by solid phase microextraction (SPME) and GC/MS. Very low levels of volatile terpenoid compounds were detected in the culture headspace after 1 month of growth, suggesting that C. cinerus expresses terpene synthases, albeit at very low levels under standard laboratory growth conditions (Fig. 2). Four major peaks with mass fragmentation pattern typical for sesquiterpenes hydrocarbons and with parent ions at 204 m/z were identified. Comparison of their mass spectra and retention indices with those from published reference spectra and authentic compounds identified them as pentalenene 1, α-muurolene 2, α-cuprenene 3 and δ-cadinene 4 (numbers in bold text placed after compound names correspond to structures, if known, shown in Fig. 3). In addition, several minor sesquiterpenes were present that yielded spectra that did not match any reference spectra.
Fig. 2. GC/MS analysis of C. cinereus culture.
The culture was grown for 1 month in the dark at 28 °C with shaking at 125 rpm. Several different sesquiterpenes were detected including pentalenene 1, α-muurolene 2, α-cuprenene 3 and δ-cadinene 4.
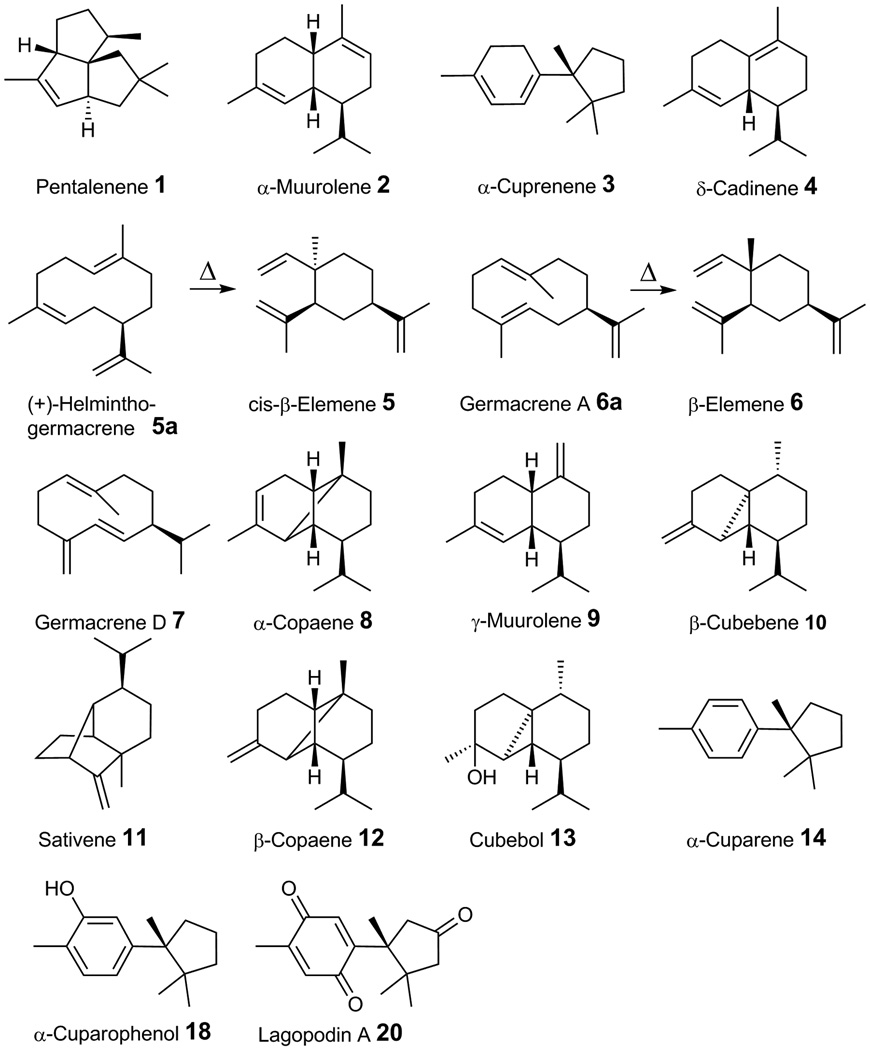
Fig. 3. Structures and names of sesquiterpenes described in this study.
Cloning and characterization of sesquiterpene synthases Cop1 to Cop6
To determine the products of the putative terpene synthases Cop1 to Cop6 from C. cinerea, their predicted coding sequences were amplified from cDNA using gene specific primers for the gene models predicted by the Broad Institute (Supplementary Table). PCR products were cloned into the E. coli expression vector pUCmodRBS and sequenced. PCR amplification products of the expected sizes and sequences were cloned for cop1, cop3 and cop4, while amplification products of cop2 and cop5 contained predicted introns despite several attempts at obtaining spliced products. Predicted introns in cop2 and cop5 (using the Broad Institute gene models) were therefore subsequently removed by overlap extension PCR.
The resulting plasmids containing the cop1 to cop6 sequences were transformed into E. coli JM109 for gene expression and subsequent analysis of terpene compounds produced by the recombinant cultures. For this, 50 ml of cultures were grown to mid log phase before analysis of the culture headspace by SPME and GC/MS for the presence of terpene products. Terpene compounds were identified by comparing retention indices and mass spectra to published reference spectra and authentic standard compounds.
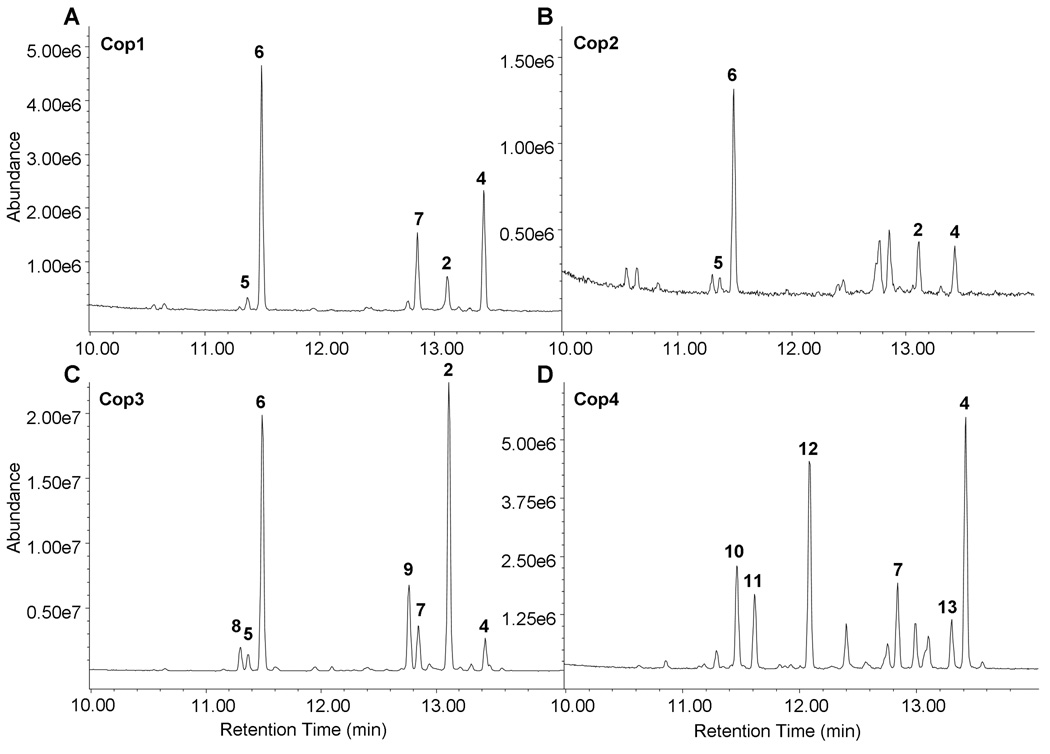
The culture headspaces of E. coli cells expressing either the terpene synthase Cop1 or Cop2 contained one major product along with several minor compounds. The major volatile compound in both cultures had a mass fragmentation pattern typical for a sesquiterpene with a parent ion at 204 m/z and characteristic daughter ions at 189, 147, 107, 93 and 81 m/z (Fig. 4A and 4B, see Supplementary Fig. 2 for mass spectra). The retention index and fragmentation pattern identified this compound as β-elemene 6 which is the heat-induced Cope rearrangement (thermal isomerization of a 1,5-diene) product of germacrene A 6a (see Fig. 3 for structures) (de Kraker et al., 1998, Faraldos et al., 2007). Cis-β-elemene 5, the Cope rearrangement product of the 4Z-somer of germacrene A, (+)-helminthogermacrene A 5a, was also detected as a minor peak eluting just before β-elemene 6. Minor compounds produced by both Cop1 and Cop2 included α-muurolene 2 and δ-cadinene 4. Germacrene D 7 was detected only in the culture head space of E. coli cultures expressing Cop1 but not in Cop 2 cultures.
Fig. 4. GC/MS analysis of volatile organic compounds produced by E. coli transformants expressing sesquiterpene synthases from C. cinereus.
E. coli cells expressing Cop1 (A) or Cop2 (B) produced as a major compound germacrene A 6a which undergoes Cope rearrangement to β-elemene 6 under the high temperatures of the injection port. E. coli cultures expressing Cop3 (C) accumulated significant quantities of α-muurolene 2, while δ-cadinene 4 was the major product of E. coli cells transformed with Cop4 (D). Peaks are labeled with numbers that correspond to their identified structures shown Fig. 3. Mass spectra for individual peaks are shown in Supplementary Fig. 2. Peak assignments were confirmed by comparing retention indices with that of authentic compounds.
E. coli strains expressing either Cop3 or Cop4 produced several volatile sesquiterpene compounds (Fig. 4C and 4D). α-muurolene 2 (major ions m/z 204, 161, 105) was the major terpenoid compound detected in Cop3 cultures, accounting for about 30% of the total sesquiterpenes detected. In addition, significant amounts of β-elemene 6, γ-muurolene 9, germacrene D 7, and δ-cadinene 4 were detected in the headspace of Cop3 cultures (Fig. 4C). E. coli strains expressing Cop4 accumulated δ-cadinene (major ions m/z 204, 161, 134, 119, 105) as the major terpenoid, accounting for about 40% of the total sesquiterpenes detected (Fig. 4D). However, unlike E. coli cultures that expressed Cop1, Cop2 or Cop3, cultures expressing Cop4 did not produce detectable levels of germacrene A 6a (β-elemene 6). Instead, Cop4 cultures produced several sesquiterpene compounds (β-cubebene 10, sativene 11, β-copaene 12, cubebol 13) not synthesized by any of the other three enzymes.
No terpene compounds were detected in E. coli cultures expressing the cop5 gene that was generated from an intron-containing PCR amplification product by overlap extension PCR based on the gene model annotated by the Broad Institute. Realizing that the cop5 transcript may be spliced differently than the annotated gene model in the genome sequence, two alternative gene models for cop5 were predicted using the Augustus gene prediction program (Stanke et al., 2008, Stanke et al., 2006, Stanke et al., 2004) (Supplementary Fig. 1). Splice site predictions between the three gene models differ in intron 2 and at the 3’ end of the transcript. The two alternative gene models were synthesized by overlap extension PCR from the original, intron-containing cop5 clone. In addition, a chimeric cop5 gene construct was generated that combines predictions of both alternative gene models (Supplementary Fig. 1). However, none of the three designed cDNA constructs led to the production of sesquiterpenoid compounds in E. coli cultures transformed with the genes cloned into pUCmodRBS. In a further attempt to detect Cop5 enzyme activity, gene constructs were subcloned into a pET expression vector to achieve higher expression levels. However, while protein expression was detected by SDS-PAGE analysis (not shown), none of the expressed recombinant proteins were functional either in vivo in E. coli or under in vitro reaction conditions using E. coli cell lysates.
E. coli cultures expressing trichodiene synthase homolog Cop6 accumulated one major sesquiterpene, accounting for 99% of total products detected (Supplementary Fig. 3). The mass fragmentation pattern and retention index identified this compound as α-cuprenene. α-Cuprenene has previously been isolated and structurally characterized from Hypericum perforatum (St. John’s wort) (Weyerstahl et al., 1995) and an extract of this plant was used to obtain an authentic standard for α-cuprenene. The retention time of the Cop6 product was identical to the authentic α-cuprenene and therefore identifies Cop6 as a α-cuprenene synthase (Supplementary Fig. 3).
Characterization of P450 monooxygenases Cox1 and Cox2 located adjacent to Cop6
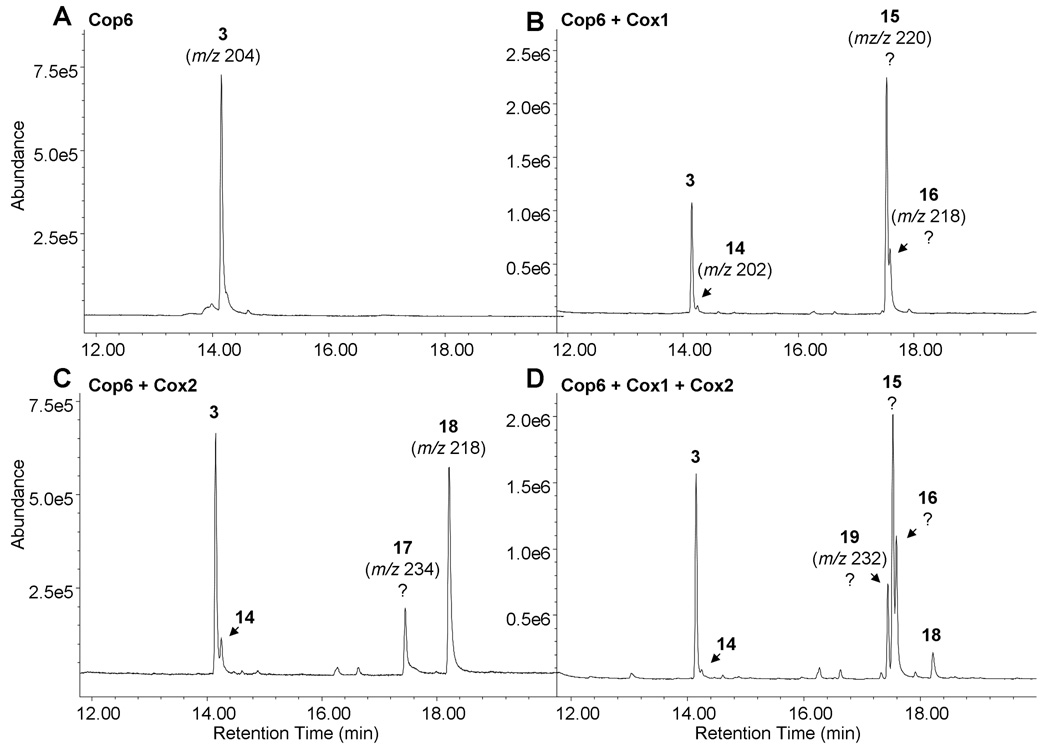
The cop6 gene is flanked by two putative cytochrome P450 monooxygenase genes (cox1 and cox2, “cox” stands for α-cuprenene oxidase). Biosynthetic genes are frequently clustered in fungi so it was reasonable to assume that the P450 enzymes encoded by these two genes modify α-cuprenene to produce oxygenized sesquiterpenes. To characterize the activities of the two enzymes, S. cerevisiae was chosen for co-expression of cop6 and cox genes. S. cerevisiae expresses a NADPH cytochrome P450 reductase needed for functional expression of fungal microsomal P450 monooxygenases that is not present in E. coli (Tokai et al., 2007). The cuprenene synthase gene, cop6, was subcloned from pUCmodRBS into the galactose inducible yeast expression vector pESC-trp (Supplementary Table). cox1 and cox2 were first amplified from cDNA using gene specific primers and directly cloned into the yeast plasmids pESC-leu (pESC-Cox1) and pESC-ura (pESC-Cox2), respectively (Supplementary Table). Expression vector pESC-Cop6 was then transformed into S. cerevisiae alone, along with plasmid pESC-Cox1 or pESC-Cox2 and with both P450 expression vectors. Recombinant yeast cultures were grown up and induced with galactose. After 48 hours post induction, culture media were analyzed for the accumulation of α-cuprenene oxidation products using SPME followed by GC/MS analysis (Fig. 5).
Fig. 5. GC/MS analysis of recombinant S. cerevisiae cultures expressing Cop6 and P450s Cox1 and Cox2.
Culture media of S. cerevisiae expressing sesquiterpene synthase Cop6 alone (A) or together with Cox1 (B), or Cox2 (C), or both (D), were analyzed for the accumulation of sesquiterpene compounds. Recombinant yeast cultures transformed with Cop6 made almost exclusively α-cuprenene 3. α-cuparene 14 is detected as a minor compound in all P450 co-expressing yeast cultures. Cultures co-expressing Cox2 make α-cuparophenol 18, while mass spectra of other new peaks (numbers correspond to mass spectra in Supplementary Fig. 2) detected in P450 and Cop6 co-expressing cultures yielded no matches in perused spectral libraries (indicated by questions marks). Parent ions for peaks are shown.
S. cerevisiae cultures expressing only Cop6 produced as expected α-cuprenene 3 (Fig. 5A). However, when Cop6 and Cox1 were co-expressed, three new compound peaks (compounds 14, 15 and 16) in addition to α-cuprenene 3 were detected in the culture medium (Fig. 5B, Supplementary Fig. 2 for mass spectra). Compound 14 had a parent ion of m/z 202 and, based on retention index and mass fragmentation pattern, was identified as α-cuparene, an aromatized derivative of α-cuprenene (m/z 204) (see Fig. 3 for structures). The parent ion of m/z 220 of the most abundant compound 15 suggests the addition of one oxygen atom to α-cuprenene, while the parent ion of m/z 218 of peak 14 indicates a similar oxidation reaction for α-cuparene. Comparison of retention indices and mass fragments of compounds 14 and 15 with reference data in the MassFinder and NIST databases yielded no match for structural assignment.
Co-expression of Cop6 with Cox2 resulted in the appearance of two new sesquiterpene peaks 17 and 18 in addition to α-cuprenene 3 and α-cuparene 14 in the culture medium (Fig. 5C, Supplementary Fig. 2 for mass spectra). The retention index and mass fragmentation pattern of the more abundant compound 18 (m/z 218) matched those of the aromatic sesquiterpene alcohol cuparophenol 18 (Fig. 3). The mass of compound 17 (m/z 234) indicates a α-cuprenene derivative containing two oxygen atoms, but its mass fragmentation pattern and retention index do not give any matches to compounds in reference databases.
When Cox1 and Cox2 together were co-expressed with Cop6, one new peak 19 occurred with a m/z of 232, suggesting again synthesis of a α-cuprenene derivative with two additional oxygen atoms but with slightly longer retention time compared to compound 17 (m/z 234) which is produced by S. cerevisiae co-expressing Cop6 and only Cox2 (Fig. 5D, Supplementary Fig. 2 for mass spectra). Peaks corresponding to the sesquiterpene hydrocarbons α-cuprenene 3 and α-cuparene 14, the Cox2 oxidation product cuparophenol 18 and the two unknown Cox1 products 15 and 16 were also detected in yeast cultures expression all three genes of the cop6 gene cluster.
DISCUSSION
Analysis of sequence data shows that sesquiterpene synthase homologs are widespread among fungi and that this class of enzymes appears to have undergone extensive gene duplication in some fungal species (Fig. 1). However, despite the preponderance of sesquiterpene synthases homologs in fungi, relatively little is known about their activities and biological functions. The few enzymes that have been characterized catalyze the first committed step in the biosynthesis of mycotoxins. Aristolochene synthase, characterized from Penicillium and Aspergillus (Caruthers et al., 2000, Shishova et al., 2007), synthesizes the hydrocarbon precursor of a number of eremophilane mycotoxins (Capasso et al., 1984, Moule et al., 1977, Proctor & Hohn, 1993). Another well-characterized fungal sesquiterpene synthase, trichodiene synthase (Rynkiewicz et al., 2001), makes the hydrocarbon scaffold of the trichothecene mycotoxins produced by e.g. Trichothecium, Stachybotrys and Fusarium; infection of grain crops with Fusarium species, for example, represents a major human and animal health concern (Brown et al., 2004, Desjardins & Proctor, 2007). Presilphiperfolan-8β-ol synthase from the gray mold fungus Botrytis cinerea has recently been functionally characterized (Pinedo et al., 2008) and shown to produce the hydrocarbon scaffold that is modified to the phytotoxic bicyclic sesquiterpene botrydial and its derivatives (Collado et al., 2007).
Some Coprinus species are known to produce bioactive sesquiterpene compounds (Reina et al., 2004, Gonzalez del Val et al., 2003), but sesquiterpene synthases have not been characterized from these species. C. cinereus has long been used as model for fungal development, but is generally not described as a sesquiterpene producer except for a study published in the 1970s (see below). In the current study, analysis of liquid culture and culture headspace revealed that this species produces sesquiterpenes at very low levels. Major peaks in the headspace analysis corresponded to pentalenene 1, α-muurolene 2, α-cuprenene 3 and δ-cadinene 4, suggesting the presence of sesquiterpene synthases with corresponding specificities. These sesquiterpenes have also been identified in fruiting bodies of wood rotting basidiomycetes (Rosecke et al., 2000). Enzymes specific for α-cuprenene and α-muurolene have not yet been identified, and pentalenene synthase is only known from bacteria (Lesburg et al., 1997a).
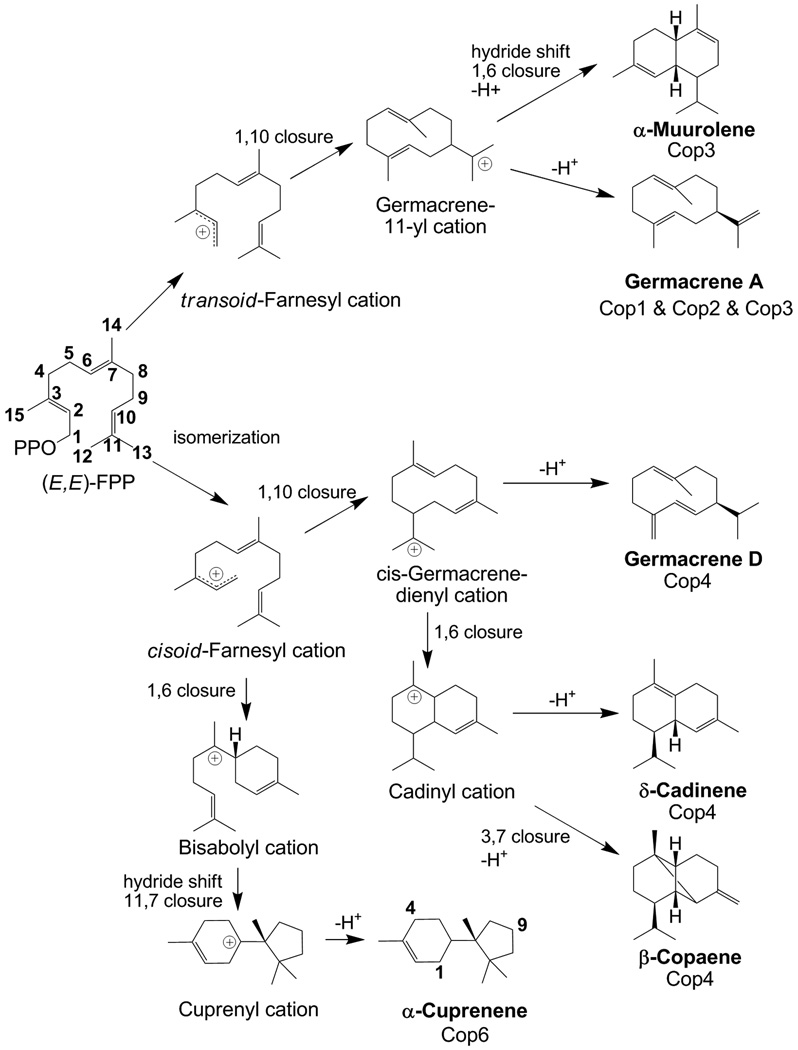
Among the six sesquiterpene synthase homologs identified in C. cinereus, Cop1, Cop2 and Cop3 are most closely related to one another (∼ 45% amino acid sequence identity, Fig. 1), suggesting that they may cyclize all-trans (E,E)-FPP via the same carbocation intermediate. Indeed, formation of the major products of all three enzymes can be proposed to proceed through a germacren-11-yl cation generated via 1,10 ring closure from a primary transoid-farnesyl cation (Fig. 6). Germacrene A 6a, the major product of Cop1 and Cop2, is then derived from this cation by deprotonation. This sesquiterpenoid is the product of several known plant sesquiterpene synthases and many plant natural products are derived from germacrene A by subsequent oxidation steps (Bertea et al., 2006, Bouwmeester et al., 2002, de Kraker et al., 1999, de Kraker et al., 2001, de Kraker et al., 1998, Prosser et al., 2004, Prosser et al., 2002). Surprisingly, neither germacrene A 6a nor its heat induced Cope-rearrangement product β-elemene 6 was detected in C. cinereus cultures, suggesting that transcription levels of their genes are very low under the cultivation conditions chosen. In fact, a correctly spliced Cop2 transcript could not be amplified from cDNA preparations.
Fig. 6. Proposed reaction mechanisms for the formation of major products of C. cinereus sesquiterpene synthases Cop1-4 and Cop6.
Shown cyclization pathways are based on previous investigations on cyclization reactions catalyzed by various sesquiterpene synthases reported in the literature (Steele et al., 1998, Tholl et al., 2005, Vedula et al., 2008).
Cop3 also accumulates germacrene A 6a, but this enzyme is proposed to catalyze a hydride shift and ring closure of the germacren-11-yl cation to produce, after deprotonation, α-muurolene 2 as its major product. A dedicated muurolene synthase has not been previously characterized to the best of our knowledge. Several plant species are known to produce this sesquiterpene, including for example Cedrela odorata (Brunke et al.) and Cucumis melo (Portnoy et al., 2008). A sesquiterpene synthase was recently cloned from the latter species and shown to produce α-muurolene 2 as a minor compound when expressed in E. coli (Portnoy et al., 2008).
Cop4 and Cop6, which share only ∼ 10–25% sequence identity with Cop1, Cop2 and Cop3, are proposed to catalyze a different reaction mechanism that requires first the isomerization of all-trans (E,E)-FPP at the 2,3 double bond to generate a cisoid-farnesyl cation. Cop4 then catalyzes a 1,10 ring closure of this farnesyl cation isomer to form the secondary cis-germacrene-dienyl cation, while Cop6 catalyzes a 1,6 ring closure to generate the secondary bisabolyl cation. Both enzymes subsequently generate tertiary cations (Fig. 6). Cop4 generates a tertiary cadinyl cation which upon deprotonation yields its major cyclization productδ-cadinene 4. The cadinyl cation reacts further to yield other cyclization products (β-cubebene 10, cubebol 13, β-copaene 12, sativene 11) observed for Cop4 (Fig. 6). The detection of δ-cadinene 4 and germacrene D 7 (derived from cis-germacrenyl dienyl cation by deprotonation) in GC/MS traces of Cop1 and Cop2 suggest that these enzymes too must catalyze to some extent the isomerization of all-trans (E, E)-FPP to generate a cadinyl cation. Its major cyclization product identifies Cop4 as a δ-cadinene synthase. Sesquiterpene synthases with this activity have been cloned and characterized from cotton (Gossypium) where they catalyze the first committed step in the biosynthesis of the phytoalexin gossypol (Yoshikuni et al., 2006, Benedict et al., 2001, Chen et al., 1996, Chen et al., 1995).
Cop6 generates the tertiary cuprenyl cation which yields α-cuprenene after deprotonation (Fig. 6). Unlike all the other Coprinus sesquiterpene synthases characterized in this study, Cop6 is a highly specific enzyme that catalyzes the cyclization of all-trans (E,E)-FPP into only one product: α-cuprenene (accounting for greater than 99% of all terpenoid products detected in the culture head space). A α-cuprenene synthase has to the best of our knowledge not been described previously; although α-cuprenene is made as a very minor product by the Arabidopsis sesquiterpene synthase At5g44630 (Tholl et al., 2005). Cop6 was originally annotated as a trichodiene synthase and does share a similar reaction mechanism with trichodiene synthase. Both enzymes generate a cuprenyl cation which in the case of Cop6 is protonated, while trichodiene synthase catalyzes an additional methyl shift before releasing the final deprotonated cyclization product. Site-directed mutagenesis of trichodiene synthase supported by structural information has resulted in a variant that makes α-cuprenene as a minor cyclization product (Cane et al., 1996, Rynkiewicz et al., 2002).
The cop6 gene is the only C. cinerea sesquiterpene synthase gene that appears to be part of a biosynthetic gene cluster consisting of cop6 and two predicted genes, cox1 and cox2, encoding predicted P450 monooxygenases. Co-expression of Cop6 with the two P450s yielded six new oxidized α-cuprenene derivatives. Two of the new compounds could be identified as α-cuparene 14 and cuparophenol 18, while the unidentified compounds contained one (peaks 15, 16) or two (peaks 17, 19) oxygen atoms. Oxidized cuparene-type sesquiterpenoids have been described from several fungi and include the antimicrobial enokipodins from the edible mushroom Flammulina velutipes (Ishikawa et al., 2001, Ishikawa et al., 2000), the helicobasidin pigments of the violet root rot fungus Helicobasidium mompa (Srikrishna & Ravikumar, 2005, Srikrishna & Ravikumar, 2006, Natori et al., 1967, Natori et al., 1964) and the antibiotic lagopodins from several Coprinus species (Bu'Lock & Darbyshire, 1976, Bottom & Siehr, 1975, Srikrishna et al., 2007, Srikrishna et al., 2006). Lagopodin A and B have been isolated from C. cinereus shake flask cultures and structurally characterized by NMR (Bu'Lock & Darbyshire, 1976). Lagopodin A 20 can be derived from α-cuprenene by first oxidation of its cylohexadiene ring to yield an aromatic ring as in α-cuparene, followed by two additional oxidation reactions at positions 1 and 4 (see sesquiterpenoid numbering shown in Fig. 6) to yield a quinone ring. The pentane ring of lagopodin A 20 at position 9 is also oxidized to the corresponding ketone. Lagopodin A is likely the primary biosynthetic product of C. cinereus. Other lagopodins (e.g. lagopodin B containing additional hydroxy groups) that were isolated from C. cinereus cultures are suspected to be artifacts of the isolation procedure (Bu'Lock & Darbyshire, 1976).
The oxidation pattern of lagopodin A 20 allows us to make assumptions as to the function of the two P450s Cox1 and Cox2. The identification of α-cuparene 14 and α-cuparophenol 18 and the detection of another peak with a parent ion of m/z 234 in yeast cultures co-expressing Cop6 and Cox2, suggests that Cox2 oxidizes the cyclohexadiene ring of α-cuprenene at positions 1 and 4. The resulting oxidation will first give α-cuparene 14 (m/z 202, ring-oxidation followed by H2O elimination upon ring-aromatization), followed by α-cuparophenol 18 (m/z 218, one oxygen atom added) and compound 17 (m/z 234, two oxygen atoms added). Cox1 will then likely catalyze the oxidation at position 9 of the pentane ring of α-cuprenene to give the corresponding hydroxy (m/z 220, compound 15) or ketone (m/z 218, compound 16) derivatives. Co-expression of Cop6 with Cox1 and Cox2 results in one new peak 19 with a parent ion of m/z 232. Considering the proposed activities of the two P450s, peak 19 likely corresponds to an oxidized α-cuparophenol derivative containing a keto-group at the pentane ring. Additional optimization of expression and cultivation conditions will be necessary to fully explore the catalytic activities of the two P450 in the heterologous expression host S. cerevisiae. Larger scale synthesis of the more oxidized lagopodin A 20 will be required for structural confirmation of oxidized α-cuprenene derivatives by NMR.
Pentalenene is the most abundant volatile sesquiterpene produced by C. cinereus, but the sesquiterpene synthase responsible for its synthesis has not yet been identified. The cop5 ORF is a good candidate to encode this sesquiterpene synthase, but despite several attempts we have been unable to obtain a cDNA that leads to the expression of a functional protein. Nevertheless, this study shows that basidiomycetes represent a rich source for the identification of new terpene synthases and associated biosynthetic genes which may lead to the discovery and subsequent recombinant production of new bioactive compounds; especially in light of the many bioactive sesquiterpenes already isolated from these fungi (Abraham, 2001). Fungal genome sequences with their numerous and often duplicated putative terpene synthase genes give us a glance into a potentially diverse terpenoid metabolism present in this class of organism. Multiple terpene synthase homologs can be identified in otherwise well-studied commercial fungi such as some Aspergillus species. It can be expected that these uncharacterized terpene synthases participate in the synthesis of yet unidentified bioactive terpenes. Experimental evidence supports a role of fungal secondary metabolites as antifeedants that provide protection against insect, amoebae, nematode and other invertebrate feeding (Fox & Howlett, 2008). Consequently, many of these secondary metabolites show potent bioactivities against higher eukaryotic organisms, underlining the potential of fungal natural products, including terpene derived compounds, for the discovery of new drugs.
EXPERIMENTAL PROCEDURES
Strains and growth conditions
Coprinus cinereus 9/55 was obtained from University of Minnesota mycological culture collection in St. Paul, Minnesota. C. cinereus was maintained on potato dextrose agar (PDA) and grown for 3 weeks in the dark prior to inoculating liquid cultures. To start liquid cultures, a small agar plug containing mycelium was cut from a plate and ground into small particles using a sterile blender. The slurry was then added to fungal medium containing 1 g L −1 of potassium phosphate (monobasic), 3 g L−1 of sodium nitrate, 0.5 g L−1 of potassium chloride, 0.5 g L−1 of magnesium sulfate (heptahydrate), 5 g L−1 of corn steep powder and 40 g L−1 of glucose and grown in Fernbach flasks containing 1 L of medium at 28 °C and 150 rpm in the dark for three weeks.
All cloning and investigations of sesquiterpene biosynthesis were carried out in E. coli strain JM109 or S. cerevisiae strain YPH 500 (ura3–52 lys2–801amber ade2–101ochre trp1-Δ63 his3-Δ200 leu2-Δ1). E. coli cultures were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 µg ml−1) at 30 °C and 250 rpm. S. cerevisiae was grown in synthetic dextrose minimal medium (SD) containing 6.7 g L−1 of yeast nitrogen base without amino acids, 20 g L−1 of dextrose, 1.3 g L−1 of amino acid dropout powder. Protein expression was induced in synthetic galactose minimal medium (SG) which has the same composition as SD medium except that dextrose is replaced with galactose. The specific drop out medium used corresponded to the auxotrophic strain requirements for the specific plasmids transformed into the yeast (Supplementary Table).
Homology searches and phylogenetic tree construction
Homology searches were performed using the NCBI BLAST software based on protein sequences of experimentally characterized microbial sesquiterpene synthases from bacteria (Lesburg et al., 1997b, Caruthers et al., 2000, Rynkiewicz et al., 2001, Shishova et al., 2007, Pinedo et al., 2008, Kawaide, 2006, Toyomasu et al., 2007, Toyomasu et al., 2004, Hamano et al., 2002, Dairi et al., 2001, Kawaide et al., 1997, Cane et al., 2006, Gust et al., 2003, Cane & Watt, 2003, Komatsu et al., 2008, Zhao et al., Agger et al., 2008, Giglio et al., 2008) and fungi (Cane et al., 1995, Cane & Kang, 2000, Hohn & Beremand, 1989, Hohn et al., 1995, Hohn & Plattner, 1989, Caruthers et al., 2000, Rynkiewicz et al., 2001, Pinedo et al.). Putative fungal terpene synthase sequences were identified in NCBI’s non-redundant protein sequence database and in fungal genome sequences obtained from the Broad Institute and Joint Genome Institute. Sequence alignments were computed using ClustalW (Thompson et al., 2002) and the Mega 4.1 software interface (Tamura et al., 2007). For phylogenetic tree construction, alignments were manually inspected to eliminate sequences that either did not contain the first conserved metal binding domain (DDXXDD motif) of terpene synthases, or seemed to be incorrectly annotated (e.g. sequences appeared to be too short or long). Phylogenetic analysis was conducted in Mega 4.1 (Tamura et al., 2007) using the Neighbor-Joining method (Saitou & Nei, 1987) with a bootstrap test of phylogeny (2000 replicates) (Felsenstein, 1992).
Cloning of sesquiterpene synthase and cytochrome P450 monooxygenase genes
For the cloning of the six sesquiterpene synthase genes (cop1 to cop6) and two cytochrome P450 monooxygenase genes (cox1 and cox2), approximately 1 g of freeze dried mycelium of C. cinereus was ground with a mortar and pestle and mRNA was extracted using the Qiagen RNAeasy Plant Mini Kit (Valencia, CA) followed by DNase I digestion with the DNA-free Kit (Ambion Foster City, CA). mRNA was annealed to oligo dT (Invitrogen Carlsbad, CA) followed by RT-PCR using Transcriptor Reverse Transcriptase (Roche, Balsa, Switzerland). The genes were then amplified from cDNA by PCR with Vent polymerase (New England Biolabs Ipswich, MA) using gene-specific primers with added restriction sites. The PCR products were digested with the appropriate restriction enzymes, gel purified, and ligated into one or more of the vectors pUCmodRBS (Agger et al., 2008), pHIS8 (Jez et al., 2000) (E. coli expression vector)or pESC (Stratagene, La Jolla, CA) (yeast expression vector) or for overexpression in either S. cerevisiae or E. coli (Supplementary Table). Cloned genes were verified by DNA sequencing. Predicted introns in amplification products obtained for cop2 and cop5 were removed by overlap-extension PCR (Pogulis et al., 1996).
Analysis of sesquiterpenes produced by recombinant E. coli
To investigate sesquiterpene production, single colonies of E. coli JM109 transformants harboring the putative sesquiterpene synthases genes on pUCmodRBS were grown for 12 hrs in 4 ml of LB medium supplemented with ampicillin at 30 °C. A 50 ml culture was then inoculated with 1 ml from the seed culture and allowed to shake for 20 hrs at 30 °C. The culture headspace was then sampled for 10 min by solid phase microextration (SPME) using a 100 µm polydimethlysiloxane fiber (Supelco Bellefonte, PA). The fiber was inserted through a tin foil seal into the flask headspace (gas phase) and, after absorption, inserted into the injection port of a GC/MS for thermal desorption.
Analysis of sesquiterpenes produced by recombinant S. cerevisiae co-expressing Cop6 and cytochrome P450 genes
The sesquiterpene synthase gene cop6 from C. cinereus was subcloned from pUCmodRBS into pESC-trp under the control of the gal1–10 promoter. The two putative cytochrome P450 genes cox1 and cox2 flanking the cop6 ORF, were cloned into pESC-leu and pEC-ura, respectively (Supplementary Table). Plasmid pESC-Cop6 was transformed into S. cerevisiae strain YPH500 alone, or co-transformed with pESC-Cox1 or pESC-Cox2 or with both constructs by electroporation and plated onto selective SD minimal medium plates. Recombinant yeast strains were allowed to grow for 48 hrs at 30 °C on agar plates at which time they were scraped off and resuspended to an OD600 of 1 in 50 ml of SG broth containing galactose for induction of protein expression. Yeast cultures were then grown at 30 °C and 250 rpm for 48 hrs before analysis of culture medium and headspace for the presence of sesquiterpene hydrocarbons and oxidized sesquiterpenes. Culture headspace was analyzed by SPME as described above for recombinant E. coli cultures. Cells were removed by centrifugation prior to the analysis of terpene products in culture medium. For this, a SPME fiber was submerged into slowly stirring medium for 10 min before thermal desorption in the GC/MS injection port.
Gas chromatography-mass spectrometry (GC/MS) analysis
GC/MS analysis was carried out on a Varion 3800 GC coupled to an ion-trap mass spectrometer (Saturn, Palo Alto, CA). Separation was carried out on a HP-1ms capillary column (30 m x 0.25 mm inner diameter X 1.5 µm) with an injection port temperature of 250 °C and helium as a carrier gas. Mass spectra were recorded in electron impact ionization mode. Volatiles were absorbed from the headspace over 10 minutes, and the fiber was desorbed for 5 min in the injection port. The temperature program started at 60 °C and ramped up 8 °C min−1 to a final oven temperature of 250 °C. Mass spectra were scanned in the range of 5–300 atomic mass units at 1 s intervals.
Structural identification of sesquiterpene compounds
Sesquiterpene compounds were identified by comparison of their mass spectra with those of published reference terpene spectra in MassFinder’s (software version 3) terpene library (Konig et al., 1999) and in the National Institute of Standards and Technology (NIST) MS database. In addition, retention indices (RI) of sesquiterpene peaks (derived by calibration of GC runs with n-alkane standards) were compared to RI values of terpenoid compounds in MassFinder’s terpene library. Essential oils with known terpene compositions served as authentic standards to further confirm the identity of sesquiterpenoid compounds produced by recombinant cultures. The following essential oils with relevant sesquiterpenoid compositions were purchased from Liberty Natural Products: Cedrela woods oil (α-muurolene (1% w/w), δ-cadinene (11.7 % w/w), α-copaene (15.6% w/w); Cubeb oil (germacrene D (1 % w/w), γ-muurolene (4.2 % w/w), β-cubebene (4.4 % w/w), cubebol (15.2 % w/w); Amyris wood oil (β-elemene (germacrene A) (0.1% w/w). St. John’s wort oil (alcohol extract of young flowering tops of Hypericum perforatum) was purchased from Natural Answers Inc. to provide an authentic standard for α-cuprenene (Weyerstahl et al., 1995).
ACKNOWLEDGEMENTS
This research was supported by the Academic Health Center of the University of Minnesota (grant #2005-12) and the National Institute of Health (grant GM080299).
REFERENCES
- Abraham WR. Bioactive sesquiterpenes produced by fungi: are they useful for humans as well? Curr Med Chem. 2001;8:583–606. doi: 10.2174/0929867013373147. [DOI] [PubMed] [Google Scholar]
- Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC 7120. J Bacteriol. 2008;190:6084–6096. doi: 10.1128/JB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict CR, Lu JL, Pettigrew DW, Liu J, Stipanovic RD, Williams HJ. The cyclization of farnesyl diphosphate and nerolidyl diphosphate by a purified recombinant delta-cadinene synthase. Plant Physiol. 2001;125:1754–1765. doi: 10.1104/pp.125.4.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertea CM, Voster A, Verstappen FW, Maffei M, Beekwilder J, Bouwmeester HJ. Isoprenoid biosynthesis in Artemisia annua: cloning and heterologous expression of a germacrene A synthase from a glandular trichome cDNA library. Arch Biochem Biophys. 2006;448:3–12. doi: 10.1016/j.abb.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Bottom CB, Siehr DJ. Hydroxylagopodin B: Sesquiterpenoid quinone from a mutant strain of Coprinus macrorhizus var microsporus. Phytochemistry. 1975;14:1433–1433. [Google Scholar]
- Bouwmeester HJ, Kodde J, Verstappen FWA, Altug IG, de Kraker JW, Wallaart TE. Isolation and characterization of two germacrene A synthase cDNA clones from chicory. Plant Physiol. 2002;129:134–144. doi: 10.1104/pp.001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, Dyer RB, McCormick SP, Kendra DF, Plattner RD. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet Biol. 2004;41:454–462. doi: 10.1016/j.fgb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Brunke EJ, Hammerschmidt EJ, Koste FH. Essential oil of Cedrela odorata L. (Meliaceae) from Brazil: revised list of constituents. In: Brunke EJ, editor. Progress in Essential Oil Research. Berlin: Walter de Gruyter; 1986. pp. 117–122. [Google Scholar]
- Bu'Lock JD, Darbyshire J. Lagopodin metabolites and artifacts in cultures of Coprinus. Phytochemistry. 1976;15:2004. [Google Scholar]
- Cane DE, He X, Kobayashi S, Omura S, Ikeda H. Geosmin biosynthesis in Streptomyces avermitilis. Molecular cloning, expression, and mechanistic study of the germacradienol/geosmin synthase. J Antibiot (Tokyo) 2006;59:471–479. doi: 10.1038/ja.2006.66. [DOI] [PubMed] [Google Scholar]
- Cane DE, Kang I. Aristolochene synthase: purification, molecular cloning, high-level expression in Escherichia coli, and characterization of the Aspergillus terreus cyclase. Arch Biochem Biophys. 2000;376:354–364. doi: 10.1006/abbi.2000.1734. [DOI] [PubMed] [Google Scholar]
- Cane DE, Shim JH, Xue Q, Fitzsimons BC, Hohn TM. Trichodiene synthase. Identification of active site residues by site-directed mutagenesis. Biochemistry. 1995;34:2480–2488. doi: 10.1021/bi00008a011. [DOI] [PubMed] [Google Scholar]
- Cane DE, Watt RM. Expression and mechanistic analysis of a germacradienol synthase from Streptomyces coelicolor implicated in geosmin biosynthesis. Proc Natl Acad Sci USA. 2003;100:1547–1551. doi: 10.1073/pnas.0337625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane DE, Xue Q, Fitzsimons BC. Trichodiene synthase. Probing the role of the highly conserved aspartate-rich region by site-directed mutagenesis. Biochemistry. 1996;35:12369–12376. doi: 10.1021/bi961344y. [DOI] [PubMed] [Google Scholar]
- Capasso R, Iacobellis NS, Bottalico A, Randazzo G. Structure toxicity relationships of the eremophilane phomenone and PR-toxin. Phytochemistry. 1984;23:2781–2784. [Google Scholar]
- Caruthers JM, Kang I, Rynkiewicz MJ, Cane DE, Christianson DW. Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti. J Biol Chem. 2000;275:25533–25539. doi: 10.1074/jbc.M000433200. [DOI] [PubMed] [Google Scholar]
- Chen XY, Chen Y, Heinstein P, Davisson VJ. Cloning, expression, and characterization of (+)-delta-cadinene synthase: a catalyst for cotton phytoalexin biosynthesis. Arch Biochem Biophys. 1995;324:255–266. doi: 10.1006/abbi.1995.0038. [DOI] [PubMed] [Google Scholar]
- Chen XY, Wang M, Chen Y, Davisson VJ, Heinstein P. Cloning and heterologous expression of a second (+)-delta-cadinene synthase from Gossypium arboreum. J Nat Prod. 1996;59:944–951. doi: 10.1021/np960344w. [DOI] [PubMed] [Google Scholar]
- Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- Christianson DW. Unearthing the roots of the terpenome. Curr Opin Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado IG, Sanchez AJ, Hanson JR. Fungal terpene metabolites: biosynthetic relationships and the control of the phytopathogenic fungus Botrytis cinerea. Nat Prod Rep. 2007;24:674–686. doi: 10.1039/b603085h. [DOI] [PubMed] [Google Scholar]
- Dairi T, Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J Bacteriol. 2001;183:6085–6094. doi: 10.1128/JB.183.20.6085-6094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EM, Croteau R. Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Topics Curr Chem. 2000;209:53–95. [Google Scholar]
- de Kraker JW, Bouwmeester HJ, Franssen MCR, de Groot A. (+)-Germacrene A synthesis in chicory (Cichorium intybus L.); the first step in sesquiterpene lactone biosynthesis. Acta Bot. Gall. 1999;146:111–115. [Google Scholar]
- de Kraker JW, Franssen MC, Dalm MC, de Groot A, Bouwmeester HJ. Biosynthesis of germacrene A carboxylic acid in chicory roots. Demonstration of a cytochrome P450 (+)-germacrene a hydroxylase and NADP+-dependent sesquiterpenoid dehydrogenase(s) involved in sesquiterpene lactone biosynthesis. Plant Physiol. 2001;125:1930–1940. doi: 10.1104/pp.125.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kraker JW, Franssen MC, de Groot A, Konig WA, Bouwmeester HJ. (+)-Germacrene A biosynthesi: the committed step in the biosynthesis of bitter sesquiterpene lactones in chicory. Plant Physiol. 1998;117:1381–1392. doi: 10.1104/pp.117.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins AE, Proctor RH. Molecular biology of Fusarium mycotoxins. Int J Food Microbiol. 2007;119:47–50. doi: 10.1016/j.ijfoodmicro.2007.07.024. [DOI] [PubMed] [Google Scholar]
- Faraldos JA, Wu S, Chappell J, Coates RM. Conformational analysis of (+)-germacrene A by variable-temperature NMR and NOE spectroscopy. Tetrahedron. 2007;63:7733–7742. doi: 10.1016/j.tet.2007.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet Res. 1992;60:209–220. doi: 10.1017/s0016672300030962. [DOI] [PubMed] [Google Scholar]
- Fox EM, Howlett BJ. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol. 2008;11:481–487. doi: 10.1016/j.mib.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Giglio S, Jiang J, Saint CP, Cane DE, Monis PT. Isolation and characterization of the gene associated with geosmin production in cyanobacteria. Environ Sci Technol. 2008;42:8027–8032. doi: 10.1021/es801465w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez del Val A, Platas G, Arenal F, Orihuela JC, Garcia M, Hernandez P, Royo I, De Pedro N, Silver LL, Young K, Vicente MF, Pelaez F. Novel illudins from Coprinopsis episcopalis (syn. Coprinus episcopalis), and the distribution of illudin-like compounds among filamentous fungi. Mycol Res. 2003;107:1201–1209. doi: 10.1017/s0953756203008487. [DOI] [PubMed] [Google Scholar]
- Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H, Dairi T. Functional analysis of eubacterial diterpene cyclases responsible for biosynthesis of a diterpene antibiotic, terpentecin. J Biol Chem. 2002;277:37098–37104. doi: 10.1074/jbc.M206382200. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lucking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Koljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schussler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hohn TM, Beremand PD. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene. 1989;79:131–138. doi: 10.1016/0378-1119(89)90098-x. [DOI] [PubMed] [Google Scholar]
- Hohn TM, Desjardins AE. Isolation and gene disruption of the Tox5 gene encoding trichodiene synthase in Gibberella pulicaris. Mol Plant Microbe Interact. 1992;5:249–256. doi: 10.1094/mpmi-5-249. [DOI] [PubMed] [Google Scholar]
- Hohn TM, Desjardins AE, McCormick SP. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol Gen Genet. 1995;248:95–102. doi: 10.1007/BF02456618. [DOI] [PubMed] [Google Scholar]
- Hohn TM, Plattner RD. Purification and characterization of the sesquiterpene cyclase aristolochene synthase from Penicillium roqueforti. Arch Biochem Biophys. 1989;272:137–143. doi: 10.1016/0003-9861(89)90204-x. [DOI] [PubMed] [Google Scholar]
- Ishikawa NK, Fukushi Y, Yamaji K, Tahara S, Takahashi K. Antimicrobial cuparene-type sesquiterpenes, enokipodins C and D, from a mycelial culture of Flammulina velutipes. J Nat Prod. 2001;64:932–934. doi: 10.1021/np000593r. [DOI] [PubMed] [Google Scholar]
- Ishikawa NK, Yamaji K, Tahara S, Fukushi Y, Takahashi K. Highly oxidized cuparene-type sesquiterpenes from a mycelial culture of Flammulina velutipes. Phytochemistry. 2000;54:777–782. doi: 10.1016/s0031-9422(00)00189-8. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- Kamper J, Kahmann R, Bolker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O, Perlin MH, Wosten HA, de Vries R, Ruiz-Herrera J, Reynaga-Pena CG, Snetselaar K, McCann M, Perez-Martin J, Feldbrugge M, Basse CW, Steinberg G, Ibeas JI, Holloman W, Guzman P, Farman M, Stajich JE, Sentandreu R, Gonzalez-Prieto JM, Kennell JC, Molina L, Schirawski J, Mendoza-Mendoza A, Greilinger D, Munch K, Rossel N, Scherer M, Vranes M, Ladendorf O, Vincon V, Fuchs U, Sandrock B, Meng S, Ho EC, Cahill MJ, Boyce KJ, Klose J, Klosterman SJ, Deelstra HJ, Ortiz-Castellanos L, Li W, Sanchez-Alonso P, Schreier PH, Hauser-Hahn I, Vaupel M, Koopmann E, Friedrich G, Voss H, Schluter T, Margolis J, Platt D, Swimmer C, Gnirke A, Chen F, Vysotskaia V, Mannhaupt G, Guldener U, Munsterkotter M, Haase D, Oesterheld M, Mewes HW, Mauceli EW, DeCaprio D, Wade CM, Butler J, Young S, Jaffe DB, Calvo S, Nusbaum C, Galagan J, Birren BW. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 2006;444:97–101. doi: 10.1038/nature05248. [DOI] [PubMed] [Google Scholar]
- Kawaide H. Biochemical and molecular analyses of gibberellin biosynthesis in fungi. Biosci Biotechnol Biochem. 2006;70:583–590. doi: 10.1271/bbb.70.583. [DOI] [PubMed] [Google Scholar]
- Kawaide H, Imai R, Sassa T, Kamiya Y. Ent-kaurene synthase from the fungus Phaeosphaeria sp. L487. cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- Keller NP, Turner G, Bennett JW. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H. Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA. 2008;105:7422–7427. doi: 10.1073/pnas.0802312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig WA, Bulow N, Saritas Y. Identification of sesquiterpene hydrocarbons by gas phase analytical methods. Flavour Frag. J. 1999;14:367–378. [Google Scholar]
- Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997a;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- Lesburg CA, Zhai GZ, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: Mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997b;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen JE, Bosdet IE, Brent MR, Chiu R, Doering TL, Donlin MJ, D'Souza CA, Fox DS, Grinberg V, Fu J, Fukushima M, Haas BJ, Huang JC, Janbon G, Jones SJ, Koo HL, Krzywinski MI, Kwon-Chung JK, Lengeler KB, Maiti R, Marra MA, Marra RE, Mathewson CA, Mitchell TG, Pertea M, Riggs FR, Salzberg SL, Schein JE, Shvartsbeyn A, Shin H, Shumway M, Specht CA, Suh BB, Tenney A, Utterback TR, Wickes BL, Wortman JR, Wye NH, Kronstad JW, Lodge JK, Heitman J, Davis RW, Fraser CM, Hyman RW. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buee M, Brokstein P, Canback B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbe J, Lin YC, Legue V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kues U, Lucas S, Van de Peer Y, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouze P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P, Hammel KE, Vanden Wymelenberg A, Gaskell J, Lindquist E, Sabat G, Bondurant SS, Larrondo LF, Canessa P, Vicuna R, Yadav J, Doddapaneni H, Subramanian V, Pisabarro AG, Lavin JL, Oguiza JA, Master E, Henrissat B, Coutinho PM, Harris P, Magnuson JK, Baker SE, Bruno K, Kenealy W, Hoegger PJ, Kues U, Ramaiya P, Lucas S, Salamov A, Shapiro H, Tu H, Chee CL, Misra M, Xie G, Teter S, Yaver D, James T, Mokrejs M, Pospisek M, Grigoriev IV, Brettin T, Rokhsar D, Berka R, Cullen D. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA. 2009;106:1954–1959. doi: 10.1073/pnas.0809575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Larrondo LF, Putnam N, Gelpke MD, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. [DOI] [PubMed] [Google Scholar]
- Misiek M, Hoffmeister D. Fungal genetics, genomics, and secondary metabolites in pharmaceutical sciences. Planta Med. 2007;73:103–115. doi: 10.1055/s-2007-967104. [DOI] [PubMed] [Google Scholar]
- Mondego JM, Carazzolle MF, Costa GG, Formighieri EF, Parizzi LP, Rincones J, Cotomacci C, Carraro DM, Cunha AF, Carrer H, Vidal RO, Estrela RC, Garcia O, Thomazella DP, de Oliveira BV, Pires AB, Rio MC, Araujo MR, de Moraes MH, Castro LA, Gramacho KP, Goncalves MS, Moura Neto JP, Goes Neto A, Barbosa LV, Guiltinan MJ, Bailey BA, Meinhardt LW, Cascardo JC, Pereira GA. A genome survey of Moniliophthora perniciosa gives new insights into Witches' Broom Disease of cacao. BMC Genomics. 2008;9:548. doi: 10.1186/1471-2164-9-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule Y, Moreau S, Bousquet JF. Relationships between the chemical structure and the biological properties of some eremophilane compounds related to PR toxin. Chem Biol Interact. 1977;17:185–192. doi: 10.1016/0009-2797(77)90083-7. [DOI] [PubMed] [Google Scholar]
- Natori S, Inoue Y, Nishikawa H. The structures of mompain and deoxyhelicobasidin and the biosynthesis of helicobasidin, quinonoid metabolites of Helicobasidium mompa Tanaka. Chem Pharm Bull. 1967;15:380–390. doi: 10.1248/cpb.15.380. [DOI] [PubMed] [Google Scholar]
- Natori S, Nishikawa H, Ogawa H. Structure of helicobasidin, a novel benzoquinone from helicobasdium from Helicobasidium mompa Tanaka. Chem Pharm Bull. 1964;12:236–243. doi: 10.1248/cpb.12.236. [DOI] [PubMed] [Google Scholar]
- Pinedo C, Wang CM, Pradier JM, Dalmais B, Choquer M, Le Pecheur P, Morgant G, Collado IG, Cane DE, Viaud M. Sesquiterpene synthase from the botrydial biosynthetic gene cluster of the phytopathogen Botrytis cinerea. ACS Chem Biol. 2008;3:791–801. doi: 10.1021/cb800225v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogulis RJ, Vallejo AN, Pease LR. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol Biol. 1996;57:167–176. doi: 10.1385/0-89603-332-5:167. [DOI] [PubMed] [Google Scholar]
- Portnoy V, Benyamini Y, Bar E, Harel-Beja R, Gepstein S, Giovannoni JJ, Schaffer AA, Burger J, Tadmor Y, Lewinsohn E, Katzir N. The molecular and biochemical basis for varietal variation in sesquiterpene content in melon (Cucumis melo L.) rinds. Plant Mol Biol. 2008;66:647–661. doi: 10.1007/s11103-008-9296-6. [DOI] [PubMed] [Google Scholar]
- Proctor RH, Hohn TM. Aristolochene synthase. Isolation, characterization, and bacterial expression of a sesquiterpenoid biosynthetic gene (Ari1) from Penicillium roqueforti. J Biol Chem. 1993;268:4543–4548. [PubMed] [Google Scholar]
- Prosser I, Altug IG, Phillips AL, Konig WA, Bouwmeester HJ, Beale MH. Enantiospecific (+)- and (-)-germacrene D synthases, cloned from goldenrod, reveal a functionally active variant of the universal isoprenoid-biosynthesis aspartate-rich motif. Arch Biochem Biophys. 2004;432:136–144. doi: 10.1016/j.abb.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Prosser I, Phillips AL, Gittings S, Lewis MJ, Hooper AM, Pickett JA, Beale MH. (+)-(10R)-Germacrene A synthase from goldenrod, Solidago canadensis; cDNA isolation, bacterial expression and functional analysis. Phytochemistry. 2002;60:691–702. doi: 10.1016/s0031-9422(02)00165-6. [DOI] [PubMed] [Google Scholar]
- Reina M, Orihuela JC, Gonzalez-Coloma A, de Ines C, de la Cruz M, Gonzalez del Val A, Torno JR, Fraga BM. Four illudane sesquiterpenes from Coprinopsis episcopalis. Phytochemistry. 2004;65:381–385. doi: 10.1016/j.phytochem.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Rosecke J, Pietsch M, Konig WA. Volatile constituents of wood-rotting basidiomycetes. Phytochemistry. 2000;54:747–750. doi: 10.1016/s0031-9422(00)00138-2. [DOI] [PubMed] [Google Scholar]
- Rynkiewicz MJ, Cane DE, Christianson DW. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc Natl Acad Sci USA. 2001;98:13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz MJ, Cane DE, Christianson DW. X-ray crystal structures of D100E trichodiene synthase and its pyrophosphate complex reveal the basis for terpene product diversity. Biochemistry. 2002;41:1732–1741. doi: 10.1021/bi011960g. [DOI] [PubMed] [Google Scholar]
- Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shishova EY, Di Costanzo L, Cane DE, Christianson DW. X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate. Biochemistry. 2007;46:1941–1951. doi: 10.1021/bi0622524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V, Viaud M, Jimenez-Teja D, Collado IG, Gronover CS, Pradier JM, Tudzynski B, Tudzynski P. Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. Mol Plant Microbe Interact. 2005;18:602–612. doi: 10.1094/MPMI-18-0602. [DOI] [PubMed] [Google Scholar]
- Spiteller P. Chemical defence strategies of higher fungi. Chemistry. 2008;14:9100–9110. doi: 10.1002/chem.200800292. [DOI] [PubMed] [Google Scholar]
- Srikrishna A, Babu RR, Ravikumar PC. A regioselective total synthesis of the fungal sesquiterpene (+/-)-lagopodin A. Synlett. 2007;4:655–657. [Google Scholar]
- Srikrishna A, Lakshmi BV, Ravikumar PC. The first total synthesis of (+/-)-lagopodin A. Tetrahedron Lett. 2006;47:1277–1281. [Google Scholar]
- Srikrishna A, Ravikumar PC. Total synthesis of HM-1 and HM-2, aromatic sesquiterpenes isolated from the phytopathogenic fungus Helicobasidium mompa. Structure revision of HM-2. Tetrahedron Lett. 2005;46:6105–6109. [Google Scholar]
- Srikrishna A, Ravikumar PC. Structure revision of HM-3, an aromatic sesquiterpene isolated from the phytopathogenic fungus Helicobasidium mompa. First total syntheses of HM-3 and HM-4. Tetrahedron. 2006;62:9393–9402. [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, Croteau R. Sesquiterpene synthases from grand fir (Abies grandis). Comparison of constitutive and wound-induced activities, and cDNA isolation, characterization, and bacterial expression of delta-selinene synthase and gamma-humulene synthase. J Biol Chem. 1998;273:2078–2089. doi: 10.1074/jbc.273.4.2078. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H. A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry. 2006;45:6179–6186. doi: 10.1021/bi060419n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol. 2006;9:297–304. doi: 10.1016/j.pbi.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 2005;42:757–771. doi: 10.1111/j.1365-313X.2005.02417.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2: Unit 2 3. [DOI] [PubMed] [Google Scholar]
- Tokai T, Koshino H, Takahashi-Ando N, Sato M, Fujimura M, Kimura M. Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem Biophys Res Commun. 2007;353:412–417. doi: 10.1016/j.bbrc.2006.12.033. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Nakaminami K, Toshima H, Mie T, Watanabe K, Ito H, Matsui H, Mitsuhashi W, Sassa T, Oikawa H. Cloning of a gene cluster responsible for the biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase alpha. Biosci Biotechnol Biochem. 2004;68:146–152. doi: 10.1271/bbb.68.146. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Tsukahara M, Kaneko A, Niida R, Mitsuhashi W, Dairi T, Kato N, Sassa T. Fusicoccins are biosynthesized by an unusual chimera diterpene synthase in fungi. Proc Natl Acad Sci USA. 2007;104:3084–3088. doi: 10.1073/pnas.0608426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedula LS, Jiang J, Zakharian T, Cane DE, Christianson DW. Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: Probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+B motif. Arch Biochem Biophys. 2008;469:184–194. doi: 10.1016/j.abb.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyerstahl P, Splittgerber U, Marshall H. Constitutents of the leaf essential oil of Hypericum perforatum L. from India. Flavour Fragrance J. 1995;10:365–370. [Google Scholar]
- Yoshikuni Y, Martin VJ, Ferrin TE, Keasling JD. Engineering cotton (+)-delta-cadinene synthase to an altered function: germacrene D-4-ol synthase. Chem Biol. 2006;13:91–98. doi: 10.1016/j.chembiol.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Zhao B, Lin X, Lei L, Lamb DC, Kelly SL, Waterman MR, Cane DE. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor A3(2) J Biol Chem. 2008;283:8183–8189. doi: 10.1074/jbc.M710421200. [DOI] [PMC free article] [PubMed] [Google Scholar]