Abstract
Pairs of short peptide strands can be induced to adopt antiparallel β-sheet secondary structure in aqueous solution via a macrocyclic constraint, as illustrated by many natural and designed peptides. We show that an analogous strategy is successful for creation of small units of parallel β-sheet secondary structure in aqueous solution. Cyclization in this case requires non-peptide segments for N-to-N and C-to-C interstrand linkage. Surprisingly, we find that only one of these segments need be preorganized.
Cyclization is a powerful strategy for inducing antiparallel β-sheet secondary structure in relatively short peptide segments. Biological examples feature cyclization via the backbone,1 as seen in gramicidin S and θ-defensin, and cyclization via side chains (disulfide formation),2 as seen in tachyplesins and protegrins. These natural prototypes have inspired the use of cyclization in β-sheet design efforts aimed at both structural and functional goals. For example, both backbone and side chain cyclization have been used to generate peptides that serve as spectroscopic references for the β-sheet conformations adopted by flexible, linear peptides.3 Cyclic β-sheet scaffolds have provided a fruitful basis for development of peptides with a variety of biological activities, including antibiotics, vaccine epitopes, RNA ligands and, perhaps most intriguingly, helix-mimetic inhibitors of protein-protein recognition.4
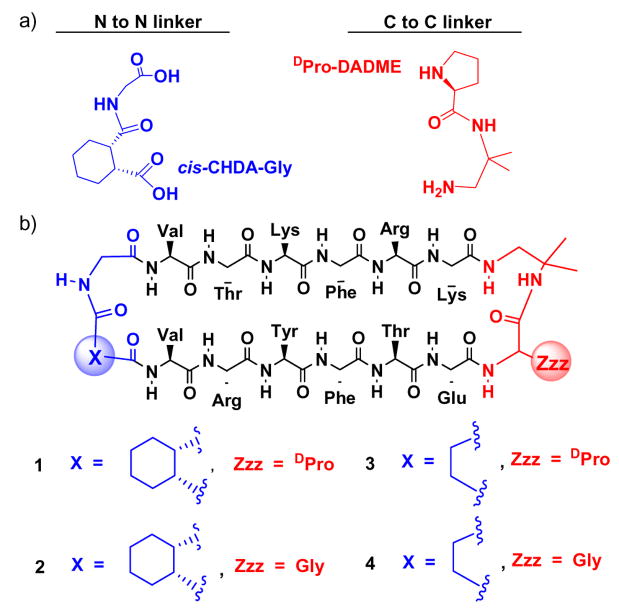
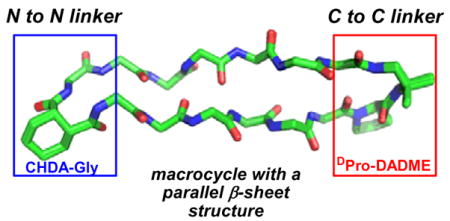
The demonstrated utility of cyclically enforced antiparallel β-sheet scaffolds raises the prospect that analogous parallel β-sheet scaffolds would be comparably useful. Biology does not offer a clear basis for achieving this structural goal in relatively small molecules. Interstrand disulfides seem to be incompatible with parallel β-sheet secondary structure, given the rarity of such crosslinks in proteins.5 β-Strand-forming segments must be linked N-terminus-to-C-terminus within peptides and proteins; therefore, covalent connection of β-strands in parallel orientation requires a peptidic linker that is at least as long as the β-strands themselves. This topological limitation has inspired many efforts to devise small non-peptide units that can be used to connect peptide segments in C-to-C or N-to-N fashion.6–8 Such units should ideally have a strong turn-forming propensity that can encourage β-sheet interactions between attached peptide segments. We have developed both preorganized C-to-C linkers and preorganized N-to-N linkers that promote (but do not enforce) parallel β-sheet formation between peptide segments in aqueous solution (Figure 1a).9 Here we evaluate these linkers in the context of backbone cyclization. Unexpectedly, we find that only one of the two turn units need to be preorganized to enforce a high level of parallel β-sheet folding.
Figure 1.
(a) Linkers used to promote parallel β-sheet secondary structure between attached peptide strands. (b) Macrocycles 1–4.
We previously showed that a linker containing D-proline and 1,2-diamino-1,1-dimethylethane (D-Pro-DADME) promotes parallel β-sheet formation between peptide segments attached via their C-termini,9a–b and that linkers containing cis-1,2-cyclohexanedicarboxylic acid and glycine (cis-CHDA-Gly) promote parallel β-sheet formation between peptide segments attached via their N-termini.9c Substituting L-Pro for D-Pro in the C-to-C linker abolished folding, for strands containing exclusively L-residues, but the two configurations of the cis-CHDA unit in the N-to-N linker (that is, the cis-CHDA-Gly unit shown in Fig. 1a and its enantiomer) displayed comparable promotion of parallel β-sheet. Macrocycles 1–4 contain varying combinations of these two types of preorganized linkers and flexible analogues, along with an invariant pair of hexapeptide strands. In 1, both linkers are preorganized to promote folding. In 2, however, only the N-to-N linker is preorganized, because the C-to-C linker contains Gly in place of D-Pro. A linear analogue containing the two strand segments connected via only the flexible Gly-DADME unit showed no evidence for folding,10 which confirms the importance of linker preorganization in the absence of a macrocyclic constraint. Both 1 and 2 contain one particular configuration of the cis-CHDA unit; the diastereomers with the other cis-CHDA configuration displayed very similar behavior.10 In 3, only the C-to-C linker is preorganized, because the N-to-N linker contains a succinyl unit in place of cis-CHDA. In 4, neither linker is preorganized.
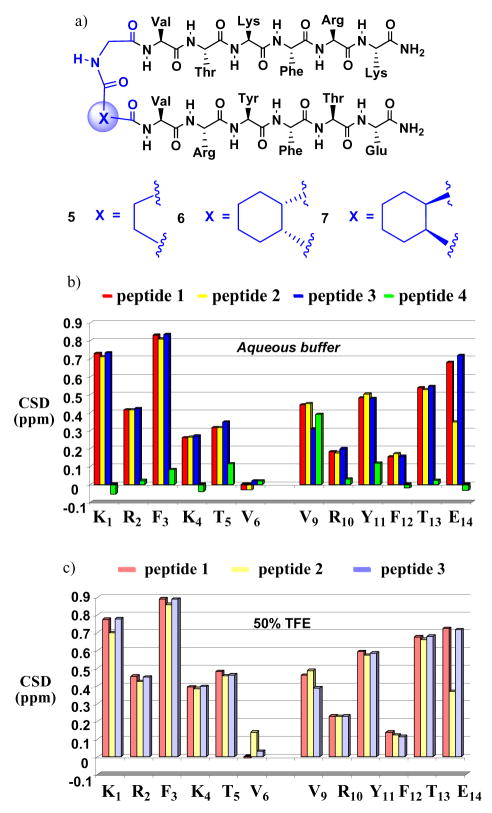
Chemical shifts observed for protons attached to amino acid residue α-carbons have proven to be useful site-specific indicators of secondary structure formation.11 β-Sheet secondary structure is suggested by sets of three or more sequential δCαH values that are downfield by ≥ 0.1 ppm relative to δCαH expected for the random coil state. Measurement of ΔδCαH = δCαH(observed) − δCαH(random coil), which is referred to as the “chemical shift deviation” (CSD), requires a source of “random coil” data. We used non-cyclic molecule 5 to provide these data (Figure 2a), because the flexible succinyl-Gly linker does not induce parallel β-sheet interactions between the attached strands. Indeed, δCαH values for 5 are very simil0ar to δCαH values that have been used to represent sequence-independent random coil references.12 Figure 2b shows ΔδCαH data for the 12 strand residues common to 1–4. These NMR data were acquired in 9:1 H2O:D2O containing 100 mM sodium acetate, pH = 3.8, with 2.5 mM peptide samples.13 DOSY measurements14 showed that peptide diffusion coefficients are invariant in 0.3 and 5 mM solutions, which suggests that there is little or no peptide aggregation under these conditions.
Figure 2.
(a) N-linked peptides 5–7. (b) ΔδCαH = δCαH(observed) − δCαH(random coil), or CSD, for α-amino acid residues of macrocycles 1–4 dissolved in aqueous buffer. (c) CSD for α-amino acid residues of macrocycles 1–4 dissolved in a 1:1 vol:vol solution of aqueous buffer and 2,2,2-trifluoroethanol (TFE).
The ΔδCαH data indicate extensive parallel β-sheet formation for 1–3, but not for 4, in aqueous buffer (Figure 2b). Among 1–3, 11 of 12 strand residues show ΔδCαH > 0.1 ppm, and the absolute values for each residue are very similar in these three cyclic peptides. The ΔδCαH values change only slightly in the presence of 50 vol % TFE (Figure 2c), which suggests that the antiparallel β-sheet populations for 1–3 are very high in pure aqueous buffer. In contrast, the ΔδCαH data for 4 suggest that this molecule forms little or no β-sheet secondary structure in aqueous solution. The behavior of 4 shows that merely placing the two strand segments in a macrocyclic context by using flexible linkers is not sufficient to induce parallel β-sheet folding; conformational preorganization of linking segments plays a vital role. However, the lack of significant distinction among 1–3 reveals a conclusion that we did not anticipate: only one of the two linkers must be preorganized in order to achieve maximum β-sheet promotion. The data suggest that the D-Pro-DADME (C-to-C) and cis-CHDA-Gly (N-to-N) linkers have comparable sheet-promoting propensities.
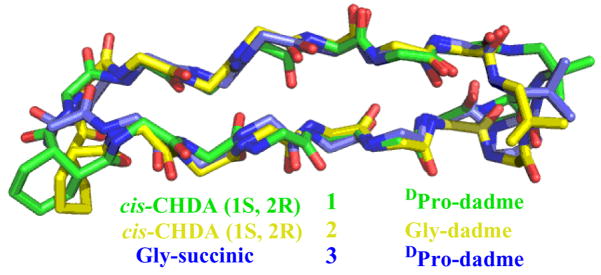
In order to examine the conformations adopted by 1–3 in greater detail, we used NOE-restrained dynamics to determine the structures in aqueous buffer.15 Superimposition of the 10 most favorable conformations identified by this approach for each macrocycle led to very good structural overlap (RMSD among backbone atoms = 0.035 ± 0.017 Å for 1, 0.284 ± 0.179 Å for 2 and 0.300 ± 0.186 Å for 3). Figure 3 shows an overlay of the most favorable conformation for each of 1–3 according to the NOE-restrained dynamics analysis. Each molecule forms a two-stranded parallel β-sheet, as intended. The β-strand segments overlap quite well, and the major deviations are seen in the linkers. In contrast to the many NOEs between protons from sequentially non-adjacent residues observed for 1–3, no medium- or long-range NOEs were detected for 4, in which both linkers are flexible.
Figure 3.
Overlay of NMR-derived conformations for macrocycles 1–3 in aqueous buffer. RMSD for backbone atoms in the strand segments is 0.659 Å for 1 vs. 3, and 0.691 for 2 vs. 3.
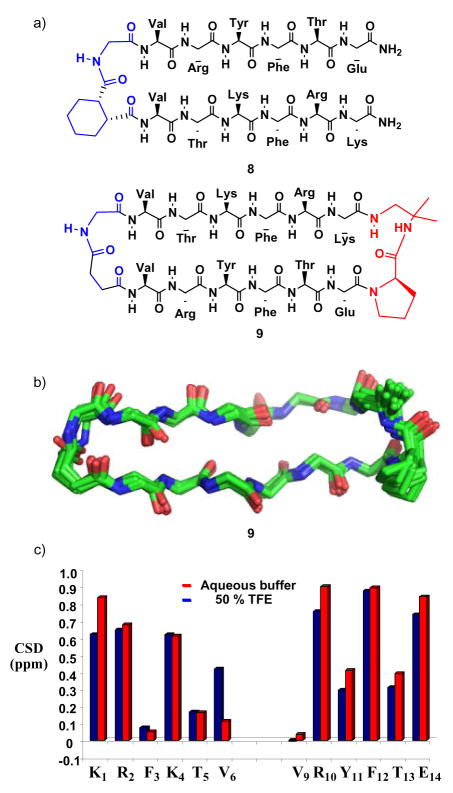
The two six-residue strand segments common to 1–4 were designed to be prone to parallel β-sheet formation, based on inter-strand neighbor preferences deduced by Fooks et al. from the protein structure database.16 Previously we showed that N-to-N linkage of these two strands via a cis-CHDA-Gly unit, as in 6 or 7, leads to significant population of parallel β-sheet secondary structure.9c The lack of detectable folding in analogue 5, in which cis-CHDA has been replaced by succinyl, shows the importance of linker preorganization for parallel β-sheet formation. However, our earlier study revealed that if the strand positions are swapped on a cis-CHDA-Gly linker, to generate 8, then no parallel β-sheet forms.9c The dramatic difference between strand-swapped isomers 6 and 8 shows that a cis-CHDA-Gly cannot enforce parallel β-sheet interactions between strands that have a low intrinsic propensity to pair in this way.
For design purposes, it would be very valuable to identify a strategy that induces parallel β-sheet secondary structure even when the strand segments have a low intrinsic propensity for parallel sheet interactions. We therefore examined macrocycle 9, which has the same strand juxtaposition as in 8 and features a preorganized D-Pro-DADME linker for C-to-C linkage and a flexible N-to-N linker. Macrocycle 9 displays numerous NOEs between protons on residues that are not adjacent in sequence; these data are consistent with parallel β-sheet secondary structure in the strand segments. NOE-restrained dynamics15 (Figure 4b) suggest a backbone conformation very similar to that of 3, which features the same pair of linkers. ΔδCαH data for 9 are consistent with high population of parallel β-sheet secondary structure in aqueous solution (Figure 4c). Thus, using a macrocyclic backbone to link two peptide strands in parallel orientation, with at least one linker appropriately preorganized, appears to be a robust strategy for inducing parallel β-sheet secondary structure.
Figure 4.
(a) Peptides 8 and 9. (b) Overlay of the 10 best structures for 9 in aqueous buffer obtained via NOE-restrained dynamics (see text for details). RMSD for the backbone atoms in the strand segments is 0.320 + 0.117 Å. (c) ΔδCαH = δCαH(observed) − δCαH(random coil), or CSD, for α-amino acid residues of macrocycle 9 dissolved in aqueous buffer or in a 1:1 vol:vol solution of aqueous buffer and 2,2,2-trifluoroethanol (TFE).
The results reported here provide new and apparently general guidelines for creating peptidic scaffolds that display substantial conformational stability in aqueous solution. There are well-established strategies for creating relatively short peptides that display high population of α-helix17 or antiparallel β-sheet18 secondary structure in aqueous solution; the approach documented above complements these strategies by providing parallel β-sheet secondary structure. Two non-peptide linkers are necessary to generate macrocycles that promote parallel strand interactions, and the finding that only one of these linkers need to be preorganized in cyclic systems is useful because the chiral element in the D-Pro-DADME linker is commercially available, while the chiral element in a cis-CHDA-Gly linker must be generated via asymmetric synthesis.
Supplementary Material
Acknowledgments
This research was supported by the NIH (GM61238). F. F. was supported in part by a MEC-Fulbright Post-Doctoral Fellowship. NMR spectrometers were purchased in part by grants from NIH and NSF.
Footnotes
Supporting Information Available: Experimental details, compound characterizations, and NMR data.
References
- 1.(a) Tishchenko GN, Adrianov VI, Vainstein BK, Woolfson MM, Dodson E. Acta Cryst. 1997;D53:151–159. doi: 10.1107/S0907444995000916. [DOI] [PubMed] [Google Scholar]; (b) Gallo SA, Wang W, Rawat SS, Jung G, Waring AJ, Cole AM, Lu H, Yan X, Daly NL, Craik DJ, Jiang S, Lehrer RI, Blumenthal R. J Biol Chem. 2006;281:18787. doi: 10.1074/jbc.M602422200. [DOI] [PubMed] [Google Scholar]
- 2.(a) Nakamura T, Furunaka H, Miyata T, Tokunagas F, Mutas T, Iwanagall S. J Biol Chem. 1988;263:16709. [PubMed] [Google Scholar]; (b) Kokryakov VN, Harwig SSL, Panyutich EA, Shevchenko AA, Aleshina GM, Shamova OV, Korneva HA, Lehrer RI. FEBS Letters. 1993;327:231. doi: 10.1016/0014-5793(93)80175-t. [DOI] [PubMed] [Google Scholar]
- 3.Syud FA, Espinosa JF, Gellman SH. J Am Chem Soc. 1999;121:11577. [Google Scholar]; (b) Tatko CD, Waters ML. J Am Chem Soc. 2002;124:9372. doi: 10.1021/ja0262481. [DOI] [PubMed] [Google Scholar]
- 4.(a) Robinson JA. Acc Chem Res. 2008;41:1278. doi: 10.1021/ar700259k. [DOI] [PubMed] [Google Scholar]; (b) Seneque O, Bourles E, Lebrun V, Bonnet E, Dunny P, Latour JM. Angew Chem Int. 2008;47:6888. doi: 10.1002/anie.200800677. [DOI] [PubMed] [Google Scholar]
- 5.Cootes AP, Curmi PM, Cunningham R, Donnelly C, Torda AE. Proteins. 1998;32:175. [PubMed] [Google Scholar]
- 6.(a) Khakshoor O, Nowick JS. Curr Opin Chem Biol. 2008;12:722. doi: 10.1016/j.cbpa.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Levin S, Nowick JS. J Am Chem Soc. 2007;129:13043. doi: 10.1021/ja073391r. [DOI] [PMC free article] [PubMed] [Google Scholar]; Nowick JS, Smith EM, Noronha G. J Org Chem. 1995;60:7386. [Google Scholar]; (c) Nowick JS, Insaf S. J Am Chem Soc. 1997;119:10903. [Google Scholar]; (d) Nowick JS. Acc Chem Res. 1999;32:287. [Google Scholar]
- 7.(a) Wagner G, Feigel M. Tetrahedron. 1993;49:10831. [Google Scholar]; (b) Ranganathan D, Haridas V, Kurur S, Thomas A, Madhusudanan KP, Nagaraj R, Kunwar AC, Sarma AVS, Karle IL. J Am Chem Soc. 1998;120:8448. [Google Scholar]; (c) Fisk JD, Powell DR, Gellman SH. J Am Chem Soc. 2000;122:5443. [Google Scholar]
- 8.For analysis of parallel β-sheet model systems in aqueous or mixed aqueous-organic solvents, see: (a) Kemp, D. S.; Blanchard, D. E.; Muendel, C. C. In Peptides-Chemistry and Biology; Smith, J., Rivier, J., Eds.; ESCOM: Leiden, 1992; p 319. (b) Junquera E, Nowick JS. J Org Chem. 1999;64:2527.Chitnumsub P, Fiori WR, Lashuel HA, Diaz H, Kelly JW. Bioorg Med Chem. 1999;7:39. doi: 10.1016/s0968-0896(98)00222-3.
- 9.(a) Fisk JD, Gellman SH. J Am Chem Soc. 2001;123:343. doi: 10.1021/ja002493d. [DOI] [PubMed] [Google Scholar]; (b) Fisk JD, Schmitt MA, Gellman SH. J Am Chem Soc. 2006;128:7148. doi: 10.1021/ja060942p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Freire F, Fisk JD, Peoples AJ, Ivancic M, Guzei IA, Gellman SH. J Am Chem Soc. 2008;130:7839. doi: 10.1021/ja802042c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Please see Supporting Information.
- 11.Wishart DS, Sykes BD, Richards FM. J Mol Biol. 1991;222:311. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]; (b) Wishart DS, Sykes BD, Richards FM. Biochemistry. 1992;31:1647. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 12.See http://andersenlab.chem.washington.edu/CSDb for a good source of δCαH (random coil) values..
- 13.COSY: Aue WP, Bartholdi E, Ernst RR. J Chem Phys. 1976;64:2229.TOCSY: Bax A, Davis DG. J Magn Reson. 1985;65:355.ROESY: Bothner-by AA, Stephens RL, Lee JM, Warren CD, Jeanloz RW. J Am Chem Soc. 1984;106:811.
- 14.(a) Cohen Y, Avram L, Frish L. Angew, Chem Int Ed. 2005;44:520. doi: 10.1002/anie.200300637. [DOI] [PubMed] [Google Scholar]; (b) Dehner A, Kessler H. Chem Bio Chem. 2005;6:1550. doi: 10.1002/cbic.200500093. [DOI] [PubMed] [Google Scholar]; (c) Altieri AS, Hinton DP, Byrd RA. J Am Chem Soc. 1995;117:7561. [Google Scholar]
- 15.Brüngerm AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Cristallogr D Biol Crystallogr. 1998;D54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Fooks HM, Martin ACR, Woolfson DN, Sessions RB, Hutchinson EG. J Mol Biol. 2006;356:32. doi: 10.1016/j.jmb.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabartty A, Baldwin RL. Adv Protein Chem. 1995;46:141. [PubMed] [Google Scholar]
- 18.(a) Gellman SH. Curr Opin Chem Biol. 1998;2:717. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]; (b) De Alba E, Rico M, Jiménez MA. Protein Sci. 1999;8:2234. doi: 10.1110/ps.8.11.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lacroix E, Kortemme T, de la Paz ML, Serrano L. Curr Opin Struct Biol. 1999;9:487. doi: 10.1016/s0959-440x(99)80069-4. [DOI] [PubMed] [Google Scholar]; (d) Searle MS, Ciani B. Curr Opin Struct Biol. 2004;14:458. doi: 10.1016/j.sbi.2004.06.001. [DOI] [PubMed] [Google Scholar]; (e) Hughes RM, Waters ML. Curr Opin Struc Biol. 2006;16:514–524. doi: 10.1016/j.sbi.2006.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.