Abstract
Presently there are no good assays for comparing somatic mutation frequencies and spectra between different vertebrate and invertebrate organisms. Here we describe a new lacZ mutation reporter system in D. melanogaster, which complements existing systems in the mouse. The results obtained with the new model indicate two-to threefold higher frequencies of spontaneous mutations than in the mouse, with most of the mutations characterized as large genome rearrangements.
Mutations are the prime substrate for natural selection in creating a wide variety of species well adapted to their environment. Random mutations, however, have generally adverse effects and are the cause of heritable disease, cancer and, possibly, aging1. Somatic mutagenesis is difficult to study in higher organisms, with most assays being indirect and based on alterations in phenotypic characteristics, such as the mouse or D. melanogaster spot tests2,3. Direct methods are available, but they are restricted to point mutations in restriction enzyme recognition sites4. In the past, we have generated transgenic mouse models harboring chromosomally integrated lacZ-plasmid constructs that can be recovered into Escherichia coli for the subsequent quantification and sequence characterization of a broad range of spontaneous mutations. Such systems do not exist for invertebrates, and information as to how the spontaneous mutation burden in somatic tissues of such organisms differs from those in mammals is absent.
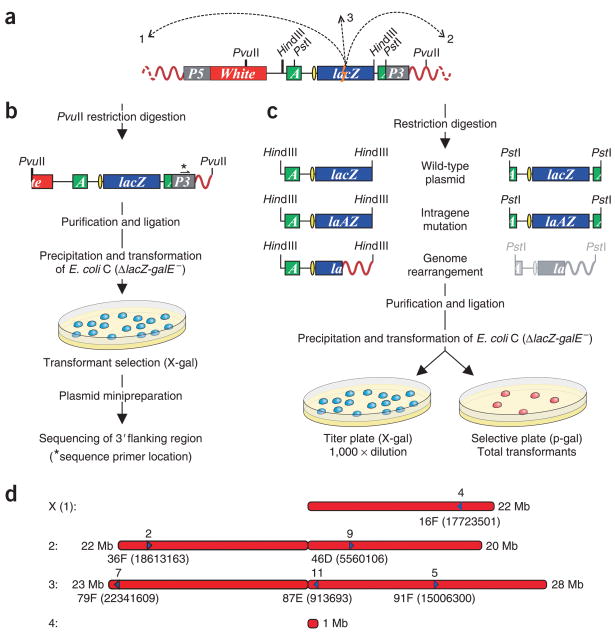
Here we generated a transgenic D. melanogaster animal model for studying a broad range of somatic mutations by inserting a lacZ plasmid reporter construct (pUR288-S; Supplementary Methods online and Supplementary Fig. 1 online) in a D. melanogaster chromosome as part of a pCasper transformation vector (Fig. 1). We made a total of six lines (2, 4, 5, 7, 9 and 11), each harboring one copy of the plasmid construct (Fig. 1a) integrated at a particular chromosomal location. We mapped each integration site more precisely by using unique restriction sites in the construct that allowed recovery of plasmids harboring a D. melanogaster flanking sequence (for example, PvuI; Fig. 1b).
Figure 1.
Schematic depiction of the pUR288-S mutation reporter model in D. melanogaster. (a) The integrated P element, containing the pUR288-S plasmid flanked by HindIII sites, is schematically depicted in the D. melanogaster genome (red curved lines). P5, 5′ end of pCasper; P3, 3′ end of pCasper; White, white selection marker; A, ampicillin-resistance gene; yellow ellipse, origin of replication. Dashed arrows represent the occurrence of a hypothetical genome rearrangement with one breakpoint in a lacZ reporter gene (lightning bolt) and one in the fly genome, 5′ (1) or 3′ (2) of the integration sites or on another chromosome (3). (b) Cloning of the fly 3′ flanking sequence of the integration site using PvuII digestion. (c) Possible outcomes of plasmid rescue. The pUR288-S plasmid can be rescued from transgenic fly lines by excision of genomic DNA with HindIII or PstI and recovered in the form of ampicillin-resistant colonies. (d) Physical map of the cloned integration sites of pUR288-S for the different lines of transgenic D. melanogaster. Cytological locations of the insertions are indicated. Arrows indicate the orientation of the pUR288-S reporter for each line, with the map position of each line indicated below the arrows.
Sequencing of this fly-specific fragment indicated the exact integration site and orientation in the D. melanogaster genome sequence database. The results indicate that the transgenic lines had the reporter plasmid integrated at different sites on chromosomes 1 (X), 2 and 3 (Fig. 1d). In all cases integration was in euchromatin, as is common for P element insertion5, with only noncoding sequences affected. The flanking sequences showed no particular characteristics that would suggest target-site preference (data not shown).
The specific configuration of the integrated construct permits plasmid rescue using either HindIII or PstI. Only rescue with HindIII allows the detection of genome rearrangements as ligation at a PstI site anywhere other than in the ampicillin-resistance gene will not yield ampicillin resistance. When the origin of replication and the ampicillin-resistance gene are not deleted, the upstream truncated plasmid sequence will result in a mutant colony after HindIII digestion (Fig. 1c). We first assessed spontaneous lacZ mutant frequencies and spectra from pools of 50 flies using HindIII for plasmid excision. The results for line 11 (randomly picked) indicated a mutant frequency in male flies of about 11 × 10−5 with a significantly higher mutant frequency of about 15 × 10−5 for females (P < 0.0001). We subsequently subdivided the mutations in ‘no-change’ and ‘size-change’ mutations. No-change mutations are point mutations that do not alter restriction enzyme patterns, whereas size-change mutations are mostly genome rearrangements that do alter the restriction enzyme pattern. In the fly, most mutations appeared to be size-change mutants.
Among 13 of the size-change mutants sequenced, only one appeared to be an internal deletion inside the pUR288-S reporter plasmid. All others were genome rearrangements, that is, lacZ-inactivating mutations with one breakpoint in the lacZ gene and the other elsewhere in the fly genome (Supplementary Table 1 online). Characterization of these 12 mutants showed that in eight of them the breakpoint was on the same chromosome harboring the pUR288-S construct in this line, that is, chromosome 3R. From the direction of the sequenced fragments we inferred that of these genome rearrangements, four were deletions, three were inversions, and one was complex and could represent a transposition event. The breakpoints of the remaining rearrangements were located on chromosome 2L (3 translocations) and X (1 translocation; Supplementary Table 1). A high rate of chromosomal rearrangements in the D. melanogaster germ line is well documented6. Of note, none of the somatic genome rearrangements observed in this present study were caused by obvious transpositions. Instead, it is more likely that they are a consequence of erroneous repair of spontaneous DNA double-strand breaks.
To confirm that all or most of the mutations had occurred in the fly and were not due to artifacts of E. coli, we did mock rescues using either PstI- or HindIII-linearized pUR288-S plasmid generated in E. coli, mixed with genomic DNA from nontransgenic flies. The results indicated a four- to sixfold higher mutant frequency of the construct rescued from the flies than the ones grown in E. coli (Supplementary Fig. 2 online), indicating that the former represents the natural mutation burden at this locus in this invertebrate organism.
To demonstrate that the plasmid system is capable of detecting not only spontaneous mutations but also induced mutations, we treated flies of line 11 with the powerful mutagen ethylnitrosourea (ENU). In this experiment, we grew males and females that were fed different doses of ENU (0.01, 0.05, 0.1 and 0.5 mM), and measured the mutant frequency at the lacZ locus in 1–2-day-old flies. The results indicated a dose-dependent increase in mutant frequency from 0.01 to 0.1 mM (Fig. 2a). No adult flies hatched from the larvae fed on 0.5 mM ENU. Sequence characterization indicated that the predominant mutation type found to be induced by the mutagen was a GC to AT transition, which is a signature mutation of ENU (data not shown).
Figure 2.
Mutant frequencies and spectra in D. melanogaster. (a) Frequencies of no-change and size-change mutant pUR288-S lacZ reporter genes recovered from D. melanogaster lacZ transgenic males or females after treatment with ENU. Error bars indicate either s.d. over three determinations (male) or the difference between two determinations (female) of total mutation frequencies. (b) Frequencies of no-change and size change mutations in different lines of pUR288-S flies. Each data point is the average of 5–16 independent determinations on the same line, with the error bars indicating s.d. over total mutation frequencies. *P < 0.0001.
To determine whether the sex of the flies or the position of the pUR288-S insertion in the pCasper vector had an effect on the lacZ somatic mutation frequency in the fly, we compared 1–7-day-old male and female flies of the six different transgenic lines. The results indicate that in male flies the mutant frequency was significantly lower than in females. We also observed some minor, albeit statistically significant variation as a function of the integration site (Fig. 2b). ANOVA analysis demonstrated significant effects on mutation frequency primarily owing to sex (F = 3817.7, P < 2.2 × 10−16) and secondarily owing to integration site (F = 8.2, P = 1.17 × 10−6) with nonsignificant interaction (F = 1.9, P = 0.09), indicating that there are differences from line to line, but sex-related differences are independent of these differences. The mutant spectra were not significantly different among the different lines. Characterization of at least 48 lacZ mutants per line and gender indicated that in all cases size change mutations were much more prominent than no-change mutations in all lines (Fig. 2b). In females the fraction of no-change mutations relative to size-change mutations appeared to be somewhat higher than in males.
The results thus far obtained indicate that the new D. melanogaster lacZ mutation reporter model is robust and can be readily used for comparative studies of somatic mutagenesis, for example, fly versus mouse, and in relation to various genetic and environmental factors. While validating the new model, the results obtained in this study are of fundamental interest for understanding spontaneous genomic instability in different higher organisms. First, although genomic integration site was found to significantly affect spontaneous mutation frequency, this effect was mainly due to one line (line 7) and overall mutation frequencies do not differ much from line to line. We cannot completely exclude that preference of transgenic integration for particular genomic environments underlies this uniform level of instability across the genome. However, we noticed no special characteristics of the sequence environments of the integration sites.
A second observation of general interest indicating a difference with mammals is the higher spontaneous mutation frequency in female than in male flies. Such a difference is not present in the mouse (data not shown) and also not known for human mutation frequencies at the HPRT locus in lymphocytes7. The difference in mutation frequency between the sexes is mainly due to a higher frequency of no-change mutations in the females (Fig. 2b). Most of such mutations are point mutations, which are mainly due to replication errors8. It is possible that female flies (which are bigger than males) undergo more cell divisions since hatching and therefore have a higher chance of replication errors. Preliminary results indicate a shorter life span for females than males in the D. melanogaster genetic background used in the experiments.
Finally, we observed a significantly higher somatic mutation burden in young flies as compared to young mice. Indeed, our results indicate a two- to threefold higher mutant frequency in D. melanogaster than in Mus musculus (ref. 9 and data not shown). Because of the about 14-fold larger size of the mouse genome, the mutation density in the fly genome (mutations per base pair) is far higher, that is, about 30–40-fold. This may be of physiological significance, especially because so many spontaneous mutations in D. melanogaster appeared to be large genome rearrangements, which are much more likely to have adverse effects than point mutations. It is possible that the high tolerance for somatic mutations of the fly genome relative to that of the mouse reflects differences in genome organization between these two species. For example, gene function in mammals may depend to a much greater extent on interactions with surrounding genes and regulatory sequences, often shared with other genes, than in D. melanogaster, in which genes are more autonomous with independent regulatory elements. In the mouse, a mutation burden as high as in the fly, with sizable fractions of genome rearrangements, could disrupt the many long-distance gene regulatory interactions and be unsustainable. The evolution of more complex species with longer life spans and more numerous cell divisions most likely also required the evolution of more sophisticated mechanisms for replication and repair to prevent the deleterious effects of genome rearrangements1.
Supplementary Material
Acknowledgments
We thank P. Kapahi for critical comments and valuable suggestions. This work was supported by a grant from the US National Institutes of Health (AG20438).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
AUTHOR CONTRIBUTIONS
J.V. conceived, designed and supervised the study (initially with M.E.T.D.), obtained the funding and took the primary role in writing the manuscript. A.M.G. designed and performed most of the experiments, with A.D., assisted by R.B. and E.P. M.L. generated the transgenic flies (assisted by L.C.) and supervised the final experiments. R.B.C. provided statistical support.
References
- 1.Vijg J. Aging of the Genome. Oxford University Press; Oxford: 2007. [Google Scholar]
- 2.Russell LB, Selby PB, von Halle E, Sheridan W, Valcovic L. Mutat Res. 1981;86:355–379. doi: 10.1016/0165-1110(81)90011-7. [DOI] [PubMed] [Google Scholar]
- 3.Kaya B, Marcos R, Yanikoglu A, Creus A. Mutat Res. 2004;557:53–62. doi: 10.1016/j.mrgentox.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Bielas JH, Loeb LA. Nat Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 5.Castro JP, Carareto CM. Genetica. 2004;121:107–118. doi: 10.1023/b:gene.0000040382.48039.a2. [DOI] [PubMed] [Google Scholar]
- 6.Ranz JM, Casals F, Ruiz A. Genome Res. 2001;11:230–239. doi: 10.1101/gr.162901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curry J, Karnaoukhova L, Guenette GC, Glickman BW. Genetics. 1999;152:1065–1077. doi: 10.1093/genetics/152.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busuttil RA, Rubio M, Dollé ME, Campisi J, Vijg J. DNA Repair (Amst) 2006;5:52–60. doi: 10.1016/j.dnarep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Dollé ME, et al. Mutat Res. 2006;596:22–35. doi: 10.1016/j.mrfmmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.